Abstract
The function of dendritic spines, postsynaptic sites of excitatory input in the mammalian central nervous system (CNS), is still not well understood. Although changes in spine morphology may mediate synaptic plasticity, the extent of basal spine motility and its regulation and function remains controversial. We investigated spine motility in three principal neurons of the mouse CNS: cerebellar Purkinje cells, and cortical and hippocampal pyramidal neurons. Motility was assayed with time-lapse imaging by using two-photon microscopy of green fluorescent protein-labeled neurons in acute and cultured slices. In all three cell types, dendritic protrusions (filopodia and spines) were highly dynamic, exhibiting a diversity of morphological rearrangements over short (<1-min) time courses. The incidence of spine motility declined during postnatal maturation, but dynamic changes were still apparent in many spines in late-postnatal neurons. Although blockade or induction of neuronal activity did not affect spine motility, disruption of actin polymerization did. We hypothesize that this basal motility of dendritic protrusions is intrinsic to the neuron and underlies the heightened plasticity found in developing CNS.
Dendritic spines are the major sites of excitatory input in mammalian central nervous system (CNS) neurons (1, 2), but their function is still not well understood. In recent years, dendritic spines have been shown to act as biochemical compartments (3–5) that could mediate synapse-specific plasticity (6, 7), perhaps through rapid changes in spine morphology (8). Indeed, recent studies have demonstrated that activity can elicit new spine-like protrusions (9, 10). Nevertheless, the question of whether spines are inherently motile remains controversial. In developing hippocampal neurons from both dissociated and slice cultures, dendritic filopodia, proposed to be precursors of mature spines, are highly dynamic (11, 12). However, after synapse formation in culture and in acute slices of mature hippocampus, spines appear to be relatively immotile (12, 13). In contrast, spines on hippocampal neurons in long-term cultures are extremely dynamic, even though these neurons bear synaptic contacts (14).
To determine the extent of motility of dendritic spines, its developmental regulation and mechanisms, we imaged three major classes of spiny neurons of the CNS: cerebellar Purkinje neurons, and pyramidal neurons from cortex and hippocampus. Acute or cultured slices were transfected with enhanced green fluorescent protein DNA, expressed under a cytomegalovirus promoter (CMV-EGFP), by using biolistic gene transfer (15). Labeled cells were then imaged by using a custom-built two-photon microscope (16). We find that in the cells examined at mid- to late-postnatal stages, the majority of dendritic spines are motile structures over time scales as short as a minute, and that motility subsides, although does not disappear, with increasing age of the cell. Spines are motile in both cultured and acute cortical slices of the same chronological age, and this motility is actin based, suggesting that motility may occur in vivo. Finally, spine motility is surprisingly unaffected by global changes in activity. Our results imply that spine motility is an intrinsic feature of CNS neurons.
Methods
Slices.
Brain slices were prepared from C57/B6J mice obtained from Taconic Farms or from our breeding colony. Sagittal (250-μm) or frontal (350-μm) cerebellar slices were prepared from postnatal day (P)10 mouse cerebella by using a tissue chopper. Slices of P19-P22 cerebella were prepared on a vibratome. Transversal cortical and hippocampal slices (300 μm) were prepared by using a tissue chopper. In contrast to Purkinje cells, cortical and hippocampal neurons did not survive well in culture when slices were prepared from late-postnatal mice. Therefore, cortical and hippocampal slices were prepared from P0-P1 mouse brains and cultured for up to 3 wk. Cultured slices were incubated on Millicell culture inserts (17) in serum-free medium [SFM; (18)] or in 10–25% horse serum medium (HSM; HyClone). Although greater numbers of cells were labeled in slices maintained in HSM, no difference in motility of spines was observed in slices kept in SFM or HSM; therefore, data from these experiments were pooled. Slices were transfected by using biolistic particle-mediated delivery (Bio-Rad hand-held gene gun) with 1-μm gold particles coated with CMV-EGFP vector (N. Heintz, Rockefeller University or CLONTECH) and incubated in 5% CO2 at 37°C. After 36–48 hr, slices were transferred to a heated chamber (37°C; Bioptechs, Butler, PA) and were perfused with artificial cerebrospinal fluid [ACSF; (16)].
Acute cortical slices (300 μm) were made from visual cortex from P11-P19 mice on a vibratome. Slices were transfected with gold beads coated with higher concentration of the same construct to produce rapid labeling of cells. Cells were imaged as early as 8 hr after preparation of slices.
Pharmacology.
For some experiments, Cytochalasin D (Sigma), 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) (Tocris Neuramin, Bristol, U.K.), (+)-2-methyl-4-carboxyphenylglycine (MCPG) (Tocris Neuramin), NiCl2 (Sigma), N-methyl-d-aspartate (NMDA) (Sigma), potentiation medium (19), glutamate, or d, l-2-amino-5-phosphonovalerate (APV) were included in the perfused ACSF medium after a control imaging period. Imaging was done either during the addition of drugs or after a 15- to 30-min incubation period.
Imaging, Image Processing, and Analysis.
Images were collected with a 40 × 0.8 NA water-immersion objective (Olympus, New Hyde Park, New York) by using a custom-built two-photon laser-scanning microscope consisting of a modified fluoview (Olympus) confocal microscope with a Ti/sapphire laser providing 790- to 850-nm 130-fs pulses at 75 MHz (Mira, Coherent Radiation, Palo Alto, CA) pumped by a solid-state source (Verdi, Coherent). Fluorescence was detected by using photomultiplier tubes (HC125–02, Hamamatsu, Ichinoko, Japan) in external whole-area detection mode, and images were acquired by using fluoview (Olympus) software. Images of spines were acquired at the highest digital zoom (x10), resulting in a nominal spatial resolution of 20 pixels per micron. Before time-lapse imaging, a 5- to 7-μm-deep Z-stack was collected above and below the plane of interest to visualize all dendritic structures. For time-lapse sequences, images were collected every 20–45 sec. At each time point, 2–5 focal planes 0.5–1 μm apart were scanned; these were later projected into a single image. Image processing and analysis was done with custom-written macros by using nih image. Images were aligned to correct for drift in the XY planes. Frames that drifted out of focus were discarded. To determine the percentage of motile spines, every clearly visible spine was scored as motile or not by an observer naive to the experimental condition. For calculations of motility index, images were thresholded to a single level throughout the entire sequence. Images were then binarized and outlined by using nih image macros. Seven frames representing maximal spine displacement were chosen and superimposed. The motility index is defined as the ratio of the difference between accumulated and smallest areas occupied by the spine divided by the average area of the spine, when the outlines of the spines in a time-lapse recording were superimposed digitally.
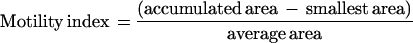 |
Electrophysiology.
Whole-cell recordings were made with an Axoclamp 2B (Axon Instruments, Foster City, CA) amplifier operating under current and voltage clamp. Resting potential (Vm) was held at −70 mV and was not corrected for junction potentials. Electrodes were filled with a solution containing (in mM): 5 NaCl, 10 KCl, 10 Hepes, 135 potassium methylsulfate, 2.5–4 MgATP, 0.3 NaGTP, and, in some cases, 100–200 μM calcium green-1 (Molecular Probes); resistances were 6–7 MΩ. Electrophysiological signals were digitized by using an analog-to-digital board and Superscope (InstruNet, GW Instruments, Somerville, MA). Action potentials were elicited by brief injections of depolarizing current through the somatic electrode.
Results
GFP-Labeled Cells in Acute and Cultured Slices Have Normal Morphology and Physiology.
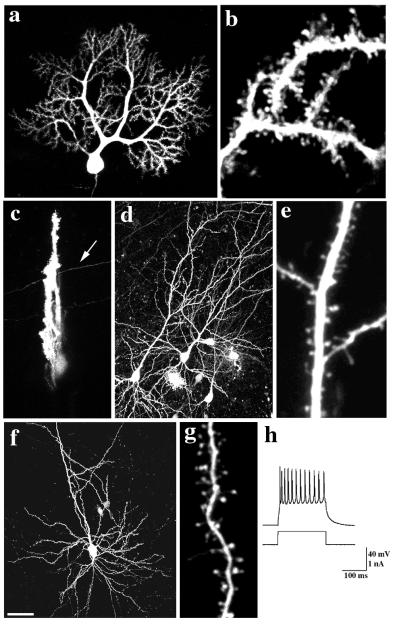
We first examined the effect of transfection on the neurons. Imaging of GFP-labeled cells revealed that transfected cells in long- and short-term cultures and acute slices had the stereotypic morphologies appropriate for the neuron type examined. Purkinje cells had elaborate dendritic trees studded with dendritic spines and filopodia (Fig. 1 a and b). Calbindin-D28k immunostaining of transfected Purkinje cells was indistinguishable from that of neighboring nontransfected cells (not shown). Hippocampal and cortical pyramidal cells (Fig. 1 d, e and f, g, respectively) had typical morphologies with a long apical dendrite and a tuft of basal dendrites bearing dendritic spines and filopodia. Whole-cell recordings of GFP-labeled cortical pyramidal cells in acute slices (12 hr after preparation) demonstrated action potentials of normal size and duration (>100 mV, <4 msec), in response to current injection at the soma (Fig. 1h, n = 5). We concluded that the transfection did not alter the morphology or physiology of the neurons.
Figure 1.
GFP-transfected cells in slices have normal morphology and physiology. GFP-labeled Purkinje cells from P 10 + 2 div (a and b) sagittal or (c) frontal slices. Note labeled parallel fiber (arrow) in the frontal slice (c). (d and e) GFP-labeled hippocampal (P0 + 11 div) and (f and g) cortical (P1 + 22 div) pyramidal neurons. Individual dendritic spines on (b) Purkinje, (e) hippocampal, and (g) pyramidal neurons are clearly resolved at high magnification. (h) Whole-cell recording of action potentials elicited from a GFP-labeled cortical pyramidal neuron (10-hr acute slice) by injection of a depolarizing current. Bar = 50 μm in a, c, d, and f; 5 μm in b, e, and g.
Spines Exhibit Different Types of Motility.
GFP-labeled cells were imaged for periods of 30–90 min with minimal photobleaching and no apparent photodamage. Time-lapse sequences of labeled cells revealed that the majority of dendritic protrusions were highly motile, with displacements as large as one spine head diameter, on time scales as short as 30 sec [Fig. 2; Table 1; supplemental video sequences 1–3; see the PNAS web site (www.pnas.org)]. The protrusions took the form of clearly shaped spines, filopodia, or intermediate morphologies. Because of the morphological plasticity observed and disagreement as to what constitutes a spine vs. a filopodium (9, 12, 20, 21), we opted not to use strict categories to describe these structures and instead, for this study, defined dendritic spines as any small protrusion that emerges from dendrites. We therefore include filopodia, as recognized traditionally by their slender shape and lack of a distinct head (20), within the same category as spines.
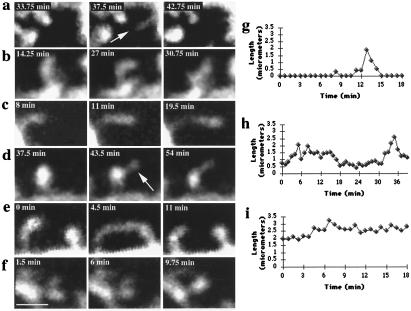
Figure 2.
Dendritic spines exhibit different types of morphological rearrangements. (a) A dendritic filopodium (arrow) appears and disappears (P). (b) Amorphous changes in the spine head (“morphing;” P). (c) Elongation of a spine (C). (d) Emergence of a filopodium (arrow) from a spine head (P). (e) Transient “touching” of neighboring spines (C). (f) Merging of split spine heads (P). (g) Length measurements of a transient dendritic spine, (h) a spine that elongates and retracts, and (i) a spine that elongates and remains elongated. Cellular origin of spines is denoted by: P, Purkinje; C, cortical pyramidal cells. Bar = 2 μm.
Table 1.
Quantification of different types of spine motility
| Cell type | Age | Spines* (No. cells) | % Motile spines† | % Spines emerge‡ | % Spines disappear | % Spines “morph” + other | % Spines elongate | % Filopodia from spine |
|---|---|---|---|---|---|---|---|---|
| Purkinje | P10 + 2div | 464 (14) | 73.1 | 3.6 | 1.7 | 45 | 15.2 | 12.9 |
| P(19–22) + 2div | 427 (7) | 45 | 0.2 | 0 | 32 | 9.5 | 3.5 | |
| Cortical pyramidal | P0 + (7–11)div | 217 (11) | 73.6 | 5.2 | 4.1 | 59 | 10.2 | 4.4 |
| P0 + (12–17)div | 205 (9) | 55.8 | 0.4 | 0.4 | 45.6 | 5.8 | 4.4 | |
| P0 + (18–23)div | 247 (9) | 51.1 | 2.9 | 1.7 | 40.7 | 5.6 | 4.8 | |
| P17–19 Acute | 141 (7) | 39.7 | 1.1 | 1.0 | 36.1 | 2.0 | 1.6 | |
| Hippocampal pyramidal | P0 + (9–12)div | 188 (9) | 63.7 | 5.8 | 1.1 | 49.4 | 10.1 | 4.2 |
*Scored spines include filopodia (see Discussion).
† All motile spines summed.
‡ Percent spines emerging and disappearing were calculated separately.
In Purkinje cells, most spines were persistent, with less than 5% of them appearing de novo or disappearing during the imaging period (Fig. 2a). Spines predominantly exhibited amorphous changes in shape (“morphing”); if headed, the shape of the head is particularly motile (Fig. 2b). Many spines elongated (Fig. 2 c, h, and i) or formed a thin process from the spine head (Fig. 2d). Similar forms of motility were observed in hippocampal and cortical neurons, with overall similar proportions of the major forms of motility (Table 1). Additional types of motility found in all three neuron types included “touching” of adjacent spines as well as merging or splitting of spine heads (Fig. 2 e and f). The former could reflect direct spine interaction, although small structures or space below the limit of our optical resolution could separate the spines.
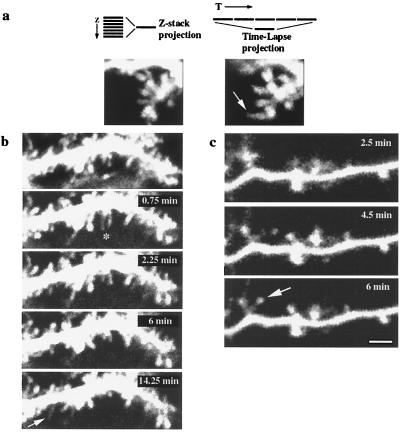
In spite of the fact that most of our time-lapse recordings involved Z-stacks, focusing artifacts could produce artifactual appearance of motility. To control for this, the projected image of an extended Z-stack taken at the beginning of a time-lapse sequence was compared with the projected image of the time-lapse sequence (Fig. 3a; see Methods). These controls showed that spines that appeared during the time-lapse sequence were not visible in the initial Z-stack projection, and spines that changed shape during the recording had a different morphology in the initial three-dimensional database. These controls therefore ruled out that focusing artifacts were producing the observed motility. As additional controls, neurons were imaged, fixed, and reimaged under identical conditions. Although small focal shifts were detected in fixed cells, spine motility was absent (not shown). Similarly, spine motility was abolished in neurons perfused with 45°C ACSF and reduced at 22°C (not shown), demonstrating that the movement is not because of Brownian motion but requires an optimal temperature of ≈37°C. In addition, motility was uncorrelated in neighboring spines under conditions of stable focus [supplemental video sequence number 5 (www.pnas.org)] and ceased when neurons were photodamaged. Also, spine motility was absent in some cells that were dialyzed with whole-cell recordings (A.M., unpublished observations). Finally, the developmental regulation of spine motility and its blockade by Cytochalasin D (see below) further demonstrated that motility was not an artifact of the imaging protocol.
Figure 3.
Spine motility does not result from focal-plane shifts, deafferentation, or slice-culture artifacts. (a Left) Projection of an “extended” 7-μm Z-stack (Purkinje cell, P10 + 2 div) spanning a volume above and below the plane of interest, collected before time-lapse imaging. (Right) Projection of several images from a time-lapse sequence into a single image. Note how the elongated spine that appears in the time-lapse projection (arrow) is not visible in the original “extended” Z-stack projection. (b) Spine motility in the frontal slices (Purkinje cell, P10 + 2 div), demonstrating retraction (∗) and appearance (arrow) of filopodia. (Top) A 7-μm Z-stack projection. (c) Time-lapse images from a cortical pyramidal neuron from an acute (10-hr) slice showing the appearance of a new spine (arrow). Bar = 2 μm.
In the cerebellum, we tested whether spine motility in sagittal slices could be a consequence of deafferentation, because parallel fiber afferents run orthogonal to the Purkinje cell dendritic tree. Purkinje cells were imaged in frontal slices, in which longer lengths of parallel fibers would be intact (Fig. 1c). In these slices, although the Purkinje cell was viewed “sideways” (Figs. 1c and 3b), spines were also motile, and motility appeared indistinguishable from the motility in sagittal slices (78 ± 16% motile spines; mean ± SD, 178 spines, 5 cells, Mann–Whitney U test, P > 0.1). Thus, the spine motility observed in Purkinje cells was not caused by loss of parallel fiber input.
We also inquired whether the motility was caused by culturing of the slices. To test this, we imaged neurons in transfected acute cortical slices as soon as 8 hr after the slices were made. We found that the motility in acute cortical slices was not significantly different from that of cultured slices (Fig. 3c; Table 1; 141 spines; 7 cells; Mann–Whitney U test; P = 0.2) and concluded that the motility is not produced by the culturing conditions.
Spine Motility Is Developmentally Regulated.
Because it is unclear whether spines on mature neurons are motile (12–14), we examined whether the motility we observed was developmentally regulated. In the cerebellum, spine motility during synaptogenesis (P10) was compared with motility of neurons from young adult animals (P19–P22). By P20, the external granular layer has disappeared, and the Purkinje cell dendritic tree is more highly branched and reaches the edge of the molecular layer. In addition, spines are more densely arrayed and have shorter stems and more rounded heads compared with those at the earlier age examined. We found a significant reduction in the number of spines exhibiting motility in P(19–22) + 2 days in vitro (div) Purkinje cells compared with P10 + 2 div [Fig. 4a, video sequence number 4 (see supplemental material, see www.pnas.org)]. Similarly, spine motility decreased in hippocampal and cortical cells with increasing time in culture, although the trend in the hippocampus was not as pronounced (Fig. 4 b and c). Interestingly, analysis of spines on cells in acute cortical slices revealed motility levels that matched the chronological age of the neuron and not the days spent in vitro (Fig. 4b; Table 1). In addition to an overall reduction in spine motility, emergence and retraction of dendritic protrusions was markedly reduced in mature neurons (Table 1). We conclude that spine motility is down-regulated in development, although a significant number of spines (40–50%) in the oldest ages observed (>P20) still moved.
Figure 4.
Developmental regulation of spine motility. (a) A histogram of the percent motility at two developmental stages in Purkinje cells. Note how motility is reduced from 73.1 ± 11.8% at P10 + 2 div (n = 14) to 45 ± 14.9% at P22 + 2 div (n = 7); Mann–Whitney U test; P < 0.001). Developmental changes in motility in (b) acute and cultured cortical or (c) cultured hippocampal neurons. Age is computed as the sum of postnatal age and days in vitro (cultured slices were made at P0–1). The line represents the best fit to data from cultured cortical neurons (regression ANOVA F < 0.0006).
Mechanisms of Spine Motility.
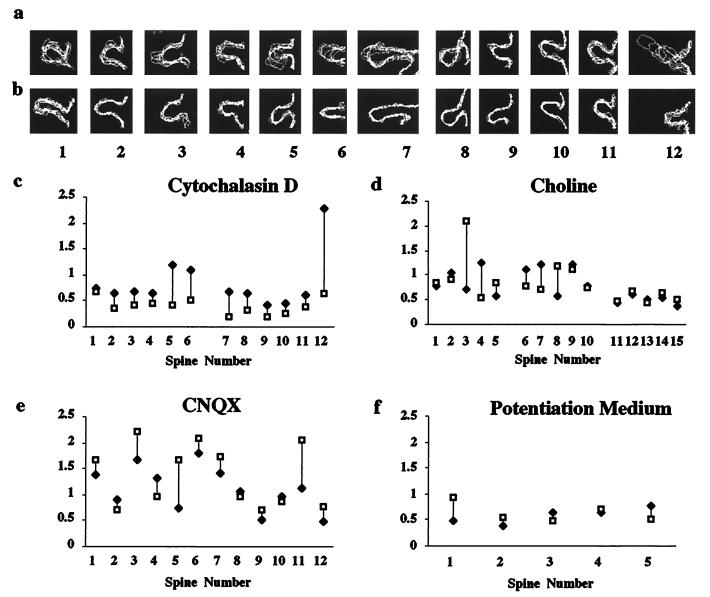
To explore mechanisms regulating spine motility, we investigated the roles of cell-intrinsic and cell-extrinsic factors, such as cytoskeletal components and neural activity, respectively. Actin is enriched in spines (22, 23) and underlies spine motility in cultured neurons (14). Bath application of Cytochalasin D (0.5–1 μg/ml), which blocks actin polymerization, inhibited spine motility, as assayed by either visual inspection or quantification of the motility index [14 Purkinje cells, 3 cortical neurons; video sequence number 4 [see supplemental material (www.pnas.org); Fig. 5 a–c]. We conclude, therefore, that spine motility in slices is actin based.
Figure 5.
Spine motility is regulated by actin polymerization but is not affected by blocking or inducing activity. Superimposed outlines of spines (labeled 1–6 for Purkinje and 7–16 for cortical pyramidal neurons) from time-lapse sequences before a and after b application of Cytochalasin D. Motility indexes of the spines demonstrate that Cytochalasin D (c) reduces spine motility (Mann–Whitney U test, P = 0.003). (d)Lack of effect of choline on the motility indexes of Purkinje cells (spines 1–10) and cortical pyramidal neuron (spines 11–15). (e) Lack of effect of blocking AMPA receptors on motility (Purkinje cell). (f) Lack of effect of stimulating neuronal activity with potentiation medium (hippocampal pyramidal neuron). P > 0.5 for d–f.
We next examined whether neural activity regulates spine motility by bath application of reagents that block or increase neuronal activity throughout the slice. Elimination of sodium currents by substitution of sodium with choline had no noticeable effect on spine motility (n = 8 cerebellum; n = 2 cortex; Fig. 5d). In Purkinje neurons, blocking AMPA or metabotropic glutamate receptors with CNQX (50 μM; n = 4; Fig. 5e) or (+)-2-methyl-4-carboxyphenylglycine, respectively (100 μM; n = 3) did not reduce or enhance the incidence of spine motility. Spine motility in Purkinje cells was also not affected by blocking calcium influx with NiCl2 (1 mM; n = 5) (16). In pyramidal neurons, blocking synaptic activity by nominally zero Ca2+ ACSF (n = 3; cortex) or l-2-amino-5-phosphonovalerate/CNQX (n = 2; hippocampus) had no effect. We also tested whether reagents that stimulate activity would alter spine motility. Application of N-methyl-d-aspartate (1–50 μM; n = 1 hippocampus, n = 1 cortex) or “potentiation” medium containing tetraethylammonium (19) did not affect spine motility (Fig. 5f; n = 3 hippocampus, n = 2 cortex). Finally, in all three cell types, spine motility was not altered by high concentrations of KCl (6–60 mM; n = 11 cerebellum, n = 3 cortex, n = 2 hippocampus) or application of glutamate (1 μM–1 mM; n = 6 cerebellum, n = 1 hippocampus) without causing cell death. These results indicate that blocking or eliciting activity does not dampen or accelerate the basal rate of motility.
Discussion
Using GFP transfection and two-photon microscopy, we showed that dendritic spine motility is a common feature of three major CNS neuron types in brain slices. We have encountered a large variety of morphological rearrangements that are regulated by actin. We therefore confirm in brain slices earlier predictions (8) and recent demonstrations (14) of actin-based spine motility. In addition to emergence and retraction of spines, spine-like structures undergo various changes in shape including elongation, “morphing,” splitting and merging of spine heads, and extension of filopodia from existing spines. The dynamic nature of spine form suggests that classifications of spines based on static images (20) represent a repertoire of morphologies that a single spine can exhibit over time.
The extent to which spine motility described in this study reflects spine behavior in vivo is still not known. Alterations in spine and synapse numbers have been reported to occur after slice preparation (24), although these changes occur in the first few hours after slice preparation, plateauing thereafter. Although the motility described here could be altered because of our experimental models, the fact that spine motility is developmentally regulated and is controlled by a specific biochemical mechanism strongly suggests that it is present in vivo.
Although recent studies have highlighted the role of synaptic activity in induction of new dendritic protrusions (9, 10), the role of activity in the regulation of basal motility of spines has not yet been addressed. We show that bath application of reagents that block or induce activity does not affect the incidence of spine motility. Given the rich variety of forms of motility that we detect, together with the many possible types of neuronal activity, we cannot exclude that particular activation of synapses influences particular forms of motility. Also, prolonged (25) rather than short-term, as well as local (9, 10) rather than global, differences in activity might affect spine motility. Nevertheless, on the basis of the lack of effect of neural activity manipulations on spine motility, we conclude that there is a basal level of spine motility that is independent of neuronal activity. The apparent lack of regulation of basal spine motility by activity suggests that spine motility is an intrinsic property of neuronal cells, although it could still be modulated by interactions with factors extrinsic to the cells (26–28). This view of spine motility parallels recent evidence for intrinsic, activity-independent spinogenesis in Purkinje cells (29).
Our data show that dendritic spine motility is developmentally regulated in all cell types examined, in that the number of motile spines is reduced with age, supporting previous studies from hippocampal cells (11, 12). In addition, the number of new protrusions is also decreased in mature cells. Our results, however, differ from previous studies, in that we detect a large number (>40% in all three cell types) of spine-like structures that continue to be motile in slices from juvenile animals. Why are spines less motile in adult neurons? One possibility is that the increased number of synaptic contacts on dendritic protrusions in mature cells stabilizes the spines (12). However, in our study, motility of dendritic spines was comparable in both more and less deafferented situations (sagittal cerebellar vs. frontal cerebellar slices or hippocampal slices), suggesting that basal spine motility may not depend on synaptic contact. Moreover, dendritic filopodia can receive synapses (21, 30), implying that synaptic contacts per se do not dampen spine motility. Localization of imaged spines by electron microscopy will test definitively how synaptic contact relates to spine motility. It is also possible that mature cells undergo an intrinsic aging process that down-regulates the motility of dendritic spines, or that spines become physically constrained as development proceeds, perhaps because of myelination of the neuropil (31).
What is the function of this intrinsic basic motility of dendritic spines? Motility could enable spines to explore their environment, probing for axonal terminals (12). Alternatively, it could result in rearrangements of protein scaffolding in a spine, affecting receptor targeting (32–34). Spine motility could also alter synapse function, either by changing the dimensions of the synaptic cleft (35, 36) or by modulating the number of postsynaptic receptors. On the basis of the developmental regulation of the motility, we hypothesize that the heightened motility in developing CNS is related to the increased plasticity in the developing brain. Indeed, in our cortical preparations, the ages at which the motility is down-regulated correspond to the end of the critical period for the monocular deprivation in mouse area V1 (37). The reduced motility of spines with increasing age could be a key factor that makes circuit rearrangements in the mature CNS less feasible.
Supplementary Material
Acknowledgments
We are grateful to Dr. N. Heintz for introducing us to the biolistics and for the initial CMV-EGFP construct. We thank Dr. D. Toran-Allerand for the use of the gene-gun, F. Hamzei-Sichani and Gül Dölen for help, and Drs. M. Morrison and S. Siegelbaum for discussions. This work was supported by National Institutes of Health grants National Research Service Award NS10370 (A.D.), R01 NS16951 (C.A.M.), and R01 EY111787 (R.Y.), and the Human Frontiers Science Program, EJLB, and the Arnold and Mabel Beckman Foundations (R.Y.).
Abbreviations
- CNS
central nervous system
- ACSF
artificial cerebrospinal fluid
- CNQX
6-cyano-7-nitroquinoxaline-2,3-dione
- Pn
postnatal day n
- CMV-EGFP
cytomegalovirus promoter enhanced green fluorescent protein
- div
days in vitro
Note Added in Proof
A recent paper by Kaech et al. (38) shows that spines on hippocampal cells in slices are motile on a spatiotemporal scale similar to the one reported here.
References
- 1.Cajal R Y. Rev Trimest Histol Norm Pat. 1888;1:1–10. [Google Scholar]
- 2.Gray E G. J Anat. 1959;83:420–433. [PMC free article] [PubMed] [Google Scholar]
- 3.Guthrie P B, Segal M, Kater S B. Nature (London) 1991;354:76–80. doi: 10.1038/354076a0. [DOI] [PubMed] [Google Scholar]
- 4.Muller W, Connor J A. Nature (London) 1991;354:73–76. doi: 10.1038/354073a0. [DOI] [PubMed] [Google Scholar]
- 5.Yuste R, Denk W. Nature (London) 1995;375:682–684. doi: 10.1038/375682a0. [DOI] [PubMed] [Google Scholar]
- 6.Wickens J. Prog Neurobiol. 1988;31:507–528. doi: 10.1016/0301-0082(88)90013-5. [DOI] [PubMed] [Google Scholar]
- 7.Koch C, Zador A. J Neurosci. 1993;13:413–422. doi: 10.1523/JNEUROSCI.13-02-00413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crick F. Trends Neurosci. 1982;5:44–46. [Google Scholar]
- 9.Maletic-Savatic M, Malinow R, Svoboda K. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- 10.Engert F, Bonhoeffer T. Nature (London) 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- 11.Dailey M E, Smith S J. J Neurosci. 1996;16:2983–2994. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziv N E, Smith S J. Neuron. 1996;17:91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
- 13.Hosokawa T, Bliss T V P, Fine A. NeuroReport. 1992;3:477–480. doi: 10.1097/00001756-199206000-00005. [DOI] [PubMed] [Google Scholar]
- 14.Fischer M, Kaech S, Knutti D, Matus A. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 15.Arnold D, Feng L, Kim J, Heintz N. Proc Natl Acad Sci USA. 1994;91:9970–9974. doi: 10.1073/pnas.91.21.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuste R, Majewska A, Cash S, Denk W. J Neurosci. 1999;19:1–12. doi: 10.1523/JNEUROSCI.19-06-01976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoppini L, Buchs P-A, Muller D. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 18.Baptista C A, Hatten M E, Blazeski R, Mason C A. Neuron. 1994;12:243–260. doi: 10.1016/0896-6273(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 19.Hosokawa T, Rusakov D A, Bliss T V P, Fine A. J Neurosci. 1995;15:5560–5573. doi: 10.1523/JNEUROSCI.15-08-05560.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris K M, Kater S B. Annu Rev Neurosci. 1994;17:341–371. doi: 10.1146/annurev.ne.17.030194.002013. [DOI] [PubMed] [Google Scholar]
- 21.Fiala J C, Feinberg M, Popov V, Harris M. J Neurosci. 1998;18:8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matus A, Ackermann M, Pehling G, Byers H R, Fujiwara K. Proc Natl Acad Sci USA. 1982;79:7590–7594. doi: 10.1073/pnas.79.23.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fifkova E, Delay R J. J Cell Biol. 1982;95:345–350. doi: 10.1083/jcb.95.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirov S A, Sorra K E, Harris K M. J Neurosci. 1999;19:2876–2886. doi: 10.1523/JNEUROSCI.19-08-02876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinney R A, Capogna M, Durr R, Gahwiler B H, Thompson S M. Nat Neurosci. 1999;2:44–49. doi: 10.1038/4548. [DOI] [PubMed] [Google Scholar]
- 26.Shimada A, Mason C A, Morrison M E. J Neurosci. 1998;18:8559–8570. doi: 10.1523/JNEUROSCI.18-21-08559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson Horch H, Kruttgen A, Portbury S D, Katz L C. Neuron. 1999;23:353–364. doi: 10.1016/s0896-6273(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 28.Woolley C S, McEwen B S. J Neurosci. 1994;14:7680–7687. doi: 10.1523/JNEUROSCI.14-12-07680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bravin M, Morando L, Vercelli A, Rossi F, Strata P. Proc Natl Acad Sci USA. 1999;96:1704–1709. doi: 10.1073/pnas.96.4.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papa M, Bundman M C, Greenberger V, Segal M. J Neurosci. 1995;15:1–11. doi: 10.1523/JNEUROSCI.15-01-00001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobson M. Developmental Neurobiology. New York: Plenum; 1978. p. 166. [Google Scholar]
- 32.Rosenmund C, Westbrook G L. Neuron. 1993;10:805–814. doi: 10.1016/0896-6273(93)90197-y. [DOI] [PubMed] [Google Scholar]
- 33.Allison D A, Gelfand V I, Spector I, Craig A M. J Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim C, Lisman J E. J Neurosci. 1999;19(11):4314–4324. doi: 10.1523/JNEUROSCI.19-11-04314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang L, Hung C P, Schuman E M. Neuron. 1998;20:1137–1175. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- 36.Liu G, Choi S, Tsien R W. Neuron. 1999;22:395–409. doi: 10.1016/s0896-6273(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 37.Antonini A, Fagiolini M, Stryker M P. J Neurosci. 1999;19:4388–4406. doi: 10.1523/JNEUROSCI.19-11-04388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaech S, Brinkhaus H, Matus A. Proc Natl Acad Sci USA. 1999;96:10433–10437. doi: 10.1073/pnas.96.18.10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.