Abstract
Background
Polypodium hydriforme is a parasite with an unusual life cycle and peculiar morphology, both of which have made its systematic position uncertain. Polypodium has traditionally been considered a cnidarian because it possesses nematocysts, the stinging structures characteristic of this phylum. However, recent molecular phylogenetic studies using 18S rDNA sequence data have challenged this interpretation, and have shown that Polypodium is a close relative to myxozoans and together they share a closer affinity to bilaterians than cnidarians. Due to the variable rates of 18S rDNA sequences, these results have been suggested to be an artifact of long-branch attraction (LBA). A recent study, using multiple protein coding markers, shows that the myxozoan Buddenbrockia, is nested within cnidarians. Polypodium was not included in this study. To further investigate the phylogenetic placement of Polypodium, we have performed phylogenetic analyses of metazoans with 18S and partial 28S rDNA sequences in a large dataset that includes Polypodium and a comprehensive sampling of cnidarian taxa.
Results
Analyses of a combined dataset of 18S and partial 28S sequences, and partial 28S alone, support the placement of Polypodium within Cnidaria. Removal of the long-branched myxozoans from the 18S dataset also results in Polypodium being nested within Cnidaria. These results suggest that previous reports showing that Polypodium and Myxozoa form a sister group to Bilateria were an artifact of long-branch attraction.
Conclusion
By including 28S rDNA sequences and a comprehensive sampling of cnidarian taxa, we demonstrate that previously conflicting hypotheses concerning the phylogenetic placement of Polypodium can be reconciled. Specifically, the data presented provide evidence that Polypodium is indeed a cnidarian and is either the sister taxon to Hydrozoa, or part of the hydrozoan clade, Leptothecata. The former hypothesis is consistent with the traditional view that Polypodium should be placed in its own cnidarian class, Polypodiozoa.
Background
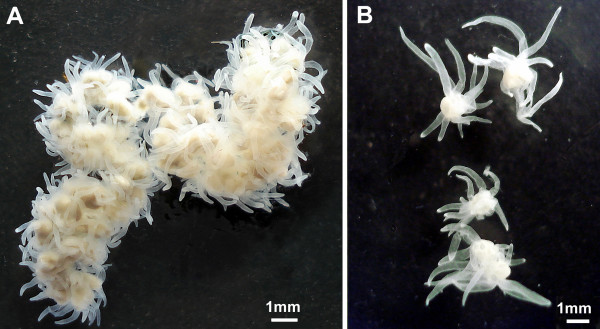
Polypodium hydriforme is an endocellular parasite whose unusual life cycle, peculiar morphology, and high rates of DNA evolution, have led to much controversy regarding its phylogenetic position within metazoans [1-5]. Polypodium spends most of its life inside the oocytes of acipenseriform fishes (sturgeons and paddlefish). During this time, Polypodium develops from a binucleate cell into an inside-out planuliform larva and then into an elongate inside-out stolon; the epidermal cell layer is located internal to the body and the gastrodermis is located externally [6-8]. The embryo, larva and stolon are surrounded by a protective polyploid cell, which also functions in digestion [7]. Just prior to host spawning, Polypodium everts to the normal position of cell layers, revealing tentacles scattered along the stolon. During eversion, the yolk of the host oocyte fills the gastral cavities of the parasite, supplying the future free-living stage with nutrients [6,7]. Finally, upon emerging from the host egg in fresh water, the free-living stolon (Figure 1A) fragments into individual medusoid-like forms (Figure 1B) that go on to multiply by means of longitudinal fission, form sexual organs, and ultimately infect host fish with their gametophores [6-9].
Figure 1.
Polypodium hydriforme. A) Stolon stage just after emerging from the host oocyte. B) Four specimens of free-living Polypodium with 12 tentacles. Photos by E. Raikova.
Two conflicting hypotheses have been proposed regarding the phylogenetic placement of Polypodium. The first, more traditional hypothesis is that Polypodium is a cnidarian. Some have suggested it is nested within a derived group of hydrozoans, the Narcomedusae [10-13] or the cnidarian class Scyphozoa [14]; while others have suggested it belongs to a separate cnidarian class, Polypodiozoa [1,15,16]. The assignment of Polypodium to Cnidaria is based primarily on morphological evidence, most notably the fact that Polypodium possesses nematocysts [17,18], the stinging structures characteristic of all cnidarians. In addition, the presence of tentacles and overall body-plan organization of Polypodium are reminiscent of cnidarians, although it is unclear if the adult free-living stage is homologous to a polyp or medusa stage. This hypothesis is supported by a cladistic analysis of small subunit nuclear ribosomal DNA (18S rDNA) sequences in conjunction with morphological characters (including nematocysts) [2]. In this study, Polypodium falls within the medusozoan clade of cnidarians, although the non-cnidarian placozoan, Trichoplax [19,20], also fell within this clade, rendering Cnidaria paraphyletic.
The second hypothesis is that Polypodium is the sister taxon to Myxozoa, a diverse group of parasites in aquatic animals, and that Polypodium + Myxozoa is the sister group to Bilateria [2-4]. This hypothesis is derived from cladistic analyses utilizing 18S rDNA sequences [2-4]. However, because Polypodium and myxozoans have unusually high divergence rates in their 18S rDNA sequences, these cladistic analyses have been criticized by a number of authors who suggest that the data might be unduly affected by long-branch attraction (LBA) [5,21,22]. Despite some attempts to overcome the effects of LBA through the use of a maximum likelihood (ML) approach [21-23] and pruning long branches [5,22], these results have been largely silent on the placement of Polypodium. For instance, Kim et al. [22] applied a maximum likelihood approach to 18S rDNA sequence data and found that myxozoans and Polypodium did not group together. Instead, Polypodium was part of an unresolved polytomy that included several cnidarian lineages and Trichoplax, as well as myxozoans + Bilateria. Most recently, Jimenez-Guri et al. [24] utilized multiple protein-coding gene sequences in a ML analysis and found the myxozoan, Buddenbrockia plumatellae nested within cnidarians. Unfortunately, this study had relatively limited sampling of cnidarians and did not include Polypodium.
In an attempt to resolve this controversy, we sequenced an additional marker in Polypodium, a partial gene sequence of the large nuclear ribosomal unit (28S rDNA), and greatly expanded the taxonomic sampling of cnidarian sequences. Using this approach, we provide evidence that Polypodium is nested within Cnidaria and does not group with myxozoans.
Results
Sampled taxa
All taxa used in this study are arranged taxonomically in Table 1. 155 sequences were obtained from GenBank. 45 new cnidarian sequences for 18S and 59 for 28S (including 2 new 18S and 2 new partial 28S from Polypodium taxa) were generated for this study and deposited in GenBank (see Table 1 for accession numbers). Polypodium hydriforme sequences were obtained from both North American and Eurasian hosts. Eurasian samples were collected from two individuals of Acipenser ruthenus. North American samples were collected from Polyodon spathula and Scaphirhynchus platorynchus. This is the first reported presence of Polypodium infection in Scaphirhynchinae. While Polypodium was recovered from the oocytes of S. platorynchus, the sample from which we extracted sequence data was found externally attached to its presumed host. More specific collection data for Polypodium specimens are associated with each sequence submitted to GenBank (see Table 1 for accession numbers).
Table 1.
Taxon and sequence list
| Accession numbers | ||||
|---|---|---|---|---|
| Higher classification | Taxon ID | 28S | 18S | Voucher |
| Bilateria | ||||
| Annelida | Proceraea cornuta | AF212165 | AF212179 | |
| Annelida | Urechis caupo | AF342804 | AF342805 | |
| Arthropoda | Limulus polyphemus | AF212167 | U91490 | |
| Arthropoda | Tenebrio sp./Tenebrio molitor | AY210843 | X07801 | |
| Brachiopoda | Phoronis vancouverensis | AF342797 | U12648 | |
| Chordata | Oncorhynchus sp./O. kisutch | U34341 | AF030250 | |
| Chordata | Petromyzon marinus | AF061798 | M97575.1 | |
| Chordata | Raja schmidti | AF278683 | AF278682 | |
| Chordata | Triakis semifasciata | AF212182 | AF212180 | |
| Echinodermata | Strongylocentrotus purpuratus | AF212171 | L28056. | |
| Hemichordata | Cephalodiscus gracilis | AF212172 | AF236798 | |
| Hemichordata | Harrimania sp. | AF212173 | AF236799 | |
| Hemichordata | Ptychodera flava | AF212176 | AF278681 | |
| Hemichordata | Ptychoderidae | AF278684 | D14359 | |
| Hemichordata | Saccoglossus kowalevskii | AF212175 | L28054 | |
| Kinorhyncha | Pycnophyes sp.Tjarno | AY859597 | AY859598 | |
| Mollusca | Parvicardium minimum | DQ279966 | DQ279942 | |
| Mollusca | Placopecten magellanicus | AF342798 | X53899 | |
| Nematoda | Caenorhabditis elegans | X03680 | X03680 | |
| Nematomorpha | Chordodes morgani | AF342787 | AF036639 | |
| Nemertea | Amphiporus sp. | AF342786 | AF119077 | |
| Nemertodermatida | Meara stichopi | AY157605 | AF119085 | |
| Onychophora | Peripatus sp. | AY210836 | AY210837 | |
| Platyhelminthes | Diclidophora denticulata | AY157169 | AJ228779 | |
| Platyhelminthes | Stenostomum leucops | AY157151 | D85095 | |
| Platyhelminthes | Stylochus zebra | AF342800 | AF342801 | |
| Priapulida | Priapulus caudatus | AY210840 | Z38009 | |
| Sipuncula | Phascolopsis gouldii | AF342795 | AF342796 | |
| Tardigrada | Milnesium.sp.\M. tardigradum | AY210826 | U49909 | |
| Urochordata | Styela plicata | AF158724 | L12444 | |
| Urochordata | Thalia democratica | AF158725 | D14366 | |
| Cnidaria | ||||
| Polypodiozoa | Polypodium (Host: Acipenser ruthenus) | EU272585 | EU272630 | |
| Polypodiozoa | Polypodium (Host: Polyodon spathula) | EU272629 | ||
| Polypodiozoa | Polypodium (Host:Scaphirhynchus platorynchus) | EU272586 | ||
| Anthozoa, Antipatharia | Antipathes galapagensis | AY026365 | AF100943 | |
| Anthozoa, Scleractinia | Montastraea franksi | AY026375 | AY026382 | |
| Cubozoa, Carybdeidae | Carybdea rastonii | AY920787 | AF358108 | |
| Cubozoa, Carybdeidae | Darwin carybdeid sp. | AY920788 | AF358105 | |
| Cubozoa, Carybdeidae | Tripedalia cystophora | EU272595 | EU272637 | |
| Cubozoa, Chirodropidae | Chironex fleckeri | AY920785 | AF358104 | |
| Cubozoa, Chirodropidae | Chiropsalmus sp. | AY920786 | AF358103 | |
| Hydrozoa, Capitata | Dipurena ophiogaster | EU272560 | EU272615 | KUNHM 2803 |
| Hydrozoa, Capitata | Ectopleura dumortieri | EU272561 | EU272616 | |
| Hydrozoa, Capitata | Euphysora bigelowi | EU272563 | EU272618 | KUNHM 2829 |
| Hydrozoa, Capitata | Moerisia sp. | AY920801 | AF358083 | |
| Hydrozoa, Capitata | Pennaria disticha | EU272581 | AY920762 | |
| Hydrozoa, Capitata | Polyorchis penicillatus | AF358090 | ||
| Hydrozoa, Capitata | Porpita sp. | AY920803 | AF358086 | |
| Hydrozoa, Capitata | Ralpharia gorgoniae | EU272590 | EU272633 | KUNHM 2778 |
| Hydrozoa, Capitata | Scrippsia pacifica | AY920804 | AF358091 | |
| Hydrozoa, Capitata | Solanderia ericopsis | EU272593 | EU272636 | MHNG INVE29593 |
| Hydrozoa, Capitata | Velella sp. | EU272597 | AF358087 | |
| Hydrozoa, Capitata | Zanclea prolifera | EU272598 | EU272639 | KUNHM 2793 |
| Hydrozoa, Capitata | Zyzzyzus warreni | EU272599 | EU272640 | KUNHM 2777 |
| Hydrozoa, Capitata | Candelabrum cocksii | AY920796 | AY920758 | MHNG INVE29531 |
| Hydrozoa, Capitata | Cladocoryne floccosa | EU272551 | EU272608 | |
| Hydrozoa, Filifera | Bimeria vestita | EU272548 | EU272605 | |
| Hydrozoa, Filifera | Bougainvillia carolinensis | EU272549 | EU272606 | |
| Hydrozoa, Filifera | Brinckmannia hexactinellidophila | EU272550 | EU272607 | MHNG INVE38148 |
| Hydrozoa, Filifera | Clava multicornis | EU272552 | EU272609 | |
| Hydrozoa, Filifera | Clavactinia gallensis | EU272553 | EU272610 | MHNG INVE33470 |
| Hydrozoa, Filifera | Cordylophora caspia | EU272556 | EU272612 | |
| Hydrozoa, Filifera | Corydendrium sp. | EU272557 | EU272613 | KUNHM 2764 |
| Hydrozoa, Filifera | Dicoryne conybearei | EU272559 | EU272614 | MHNG INVE32949 |
| Hydrozoa, Filifera | Eudendrium.racemosum | EU272562 | EU272617 | |
| Hydrozoa, Filifera | Fabienna sphaerica | AY920797 | AY920767 | |
| Hydrozoa, Filifera | Garveia annulata/Garveia sp. | EU272564 | AY920766 | KUNHM 2860 |
| Hydrozoa, Filifera | Hydra circumcincta | AY026371 | AF358080 | |
| Hydrozoa, Filifera | Hydractinia symbiolongicarpus | EU272568 | EU272621 | |
| Hydrozoa, Filifera | Hydrichthella epigorgia | EU272569 | EU272622 | KUNHM 2665 |
| Hydrozoa, Filifera | Hydrichthys boycei | EU272570 | MHNG INVE37417 | |
| Hydrozoa, Filifera | Koellikerina fasciculata | EU272571 | EU272623 | |
| Hydrozoa, Filifera | Leuckartiara octona | EU272573 | EU272624 | |
| Hydrozoa, Filifera | Lizzia blondina | EU272574 | EU272625 | |
| Hydrozoa, Filifera | Pachycordyle pusilla | EU272579 | EU272627 | MHNG INVE32953 |
| Hydrozoa, Filifera | Pandea sp. | EU272580 | AY920765 | |
| Hydrozoa, Filifera | Podocoryne carnea | AY920802 | AF358092 | |
| Hydrozoa, Filifera | Proboscidactyla ornata | EU272587 | EU272631 | KUNHM 2767 |
| Hydrozoa, Filifera | Pruvotella grisea | EU272588 | EU272632 | MHNG INVE34436 |
| Hydrozoa, Filifera | Rathkea octopunctata | EU272591 | EU272634 | KUMIP 314321 |
| Hydrozoa, Filifera | Rhizogeton nudus | EU272592 | EU272635 | MHNG INVE35757 |
| Hydrozoa, Filifera | Turritopsis dohrnii | EU272596 | EU272638 | MHNG INVE29753 |
| Hydrozoa, Leptothecata | Abietinaria filicula | EU272540 | EU272600 | MHNG INVE29947 |
| Hydrozoa, Leptothecata | Aglaophenia tubiformis | EU272543 | EU272601 | MHNG INVE29967 |
| Hydrozoa, Leptothecata | Amphisbetia minima | EU272544 | EU272602 | MHNG INVE25071 |
| Hydrozoa, Leptothecata | Anthohebella parasitica | EU272545 | EU272603 | MHNG INVE29762 |
| Hydrozoa, Leptothecata | Clytia noliformis | EU272554 | EU272611 | |
| Hydrozoa, Leptothecata | Halecium muricatum | EU272565 | EU272619 | MHNG INVE29028 |
| Hydrozoa, Leptothecata | Halopteris minuta | EU272567 | EU272620 | MHNG INVE25073 |
| Hydrozoa, Leptothecata | Melicertum octocostatum | EU272575 | AY920757 | USNM 1073342 |
| Hydrozoa, Leptothecata | Octophialucium indicum | EU272577 | EU272626 | MHNG INVE29970 |
| Hydrozoa, Leptothecata | Plumularia setacea | EU272583 | EU272628 | MHNG INVE36298 |
| Hydrozoa, Siphonophorae | Agalma elegans | EU272542 | AY937313 | YPM 35029 |
| Hydrozoa, Siphonophorae | Apolemia sp. | EU272546 | AY937331 | YPM 35090 |
| Hydrozoa, Siphonophorae | Cordagalma cordiforme | EU272555 | AY937317 | YPM 35032 |
| Hydrozoa, Siphonophorae | Halistemma rubrum | EU272566 | AY937358 | YPM 35359 |
| Hydrozoa, Siphonophorae | Nanomia bijuga | EU272576 | AY937338 | YPM 35043 |
| Hydrozoa, Siphonophorae | Nectopyramis sp. | AY026377 | AF358068 | |
| Hydrozoa, Siphonophorae | Physophora hydrostatica | EU272582 | AY937342 | YPM 35046 |
| Hydrozoa, Siphonophorae | Sulculeolaria quadrivalvis | EU272594 | AY937353 | YPM 35357 |
| Hydrozoa, Stylasteridae | Crypthelia cryptotrema | EU272558 | EU272641 | USNM1027758 |
| Hydrozoa, Stylasteridae | Lepidopora microstylus | EU272572 | EU272644 | USNM1027724 |
| Hydrozoa, Stylasteridae | Pseudocrypthelia pachypoma | EU272589 | EU272643 | USNM1027728 |
| Hydrozoa, Stylasteridae | Adelopora crassilabrum | EU272541 | EU272642 | USNM1027760 |
| Hydrozoa, Trachylina | Limnocnida tanganyicae | AY920795 | AY920755 | |
| Hydrozoa, Trachylina | Maeotias marginata | EU247810 | ||
| Hydrozoa, Trachylina | Olindias phosphorica | EU247808 | AY920753 | MHNG INVE29811 |
| Scyphozoa, Coronatae | Atolla vanhoeffeni | AY026368 | AF100942 | |
| Scyphozoa, Coronatae | Nausithoe rubra | AY920776 | AF358095 | |
| Scyphozoa, Rhizostomea | Catostylus sp. | AY920777 | AF358100 | |
| Scyphozoa, Semaeostomeae | Chrysaora melanaster | AY920780 | AF358099 | |
| Scyphozoa, Semaeostomeae | Aurelia sp. | EU272547 | EU272604 | |
| Scyphozoa, Semaeostomeae | Phacellophora camtschatica | AY920778 | AF358096 | |
| Staurozoa, Stauromedusae | Craterolophus convolvulus | AY920781 | AY845344 | |
| Staurozoa, Stauromedusae | Haliclystus octoradiatus | AH014894 | AY845346 | |
| Staurozoa, Stauromedusae | Haliclystus sanjuanensis | AY920782 | AF358102 | |
| Myxozoa | ||||
| Malacosporea | Buddenbrockia plumatellae | AJ937883 | ||
| Myxosporea | Henneguya salminicola | AY302726 | ||
| Myxosporea | Kudoa trifolia | AM490336 | AM183300 | |
| Myxosporea | Kudoa unicapsula | AM490335 | AM490334 | |
| Myxosporea | Myxobolus cerebralis | EF370481 | ||
| Myxosporea | Myxobolus dogieli | EU003978 | ||
| Myxosporea | Parvicapsula limandae | EF429096 | ||
| Outgroups | ||||
| Choanoflagellida | ||||
| Codonosigidae | Monosiga brevicollis | AY026374 | AF084618 | |
| Salpingoecidae | Salpingoeca infusionum | AY026380 | AF100941 | |
| Ctenophora, | ||||
| Cyclocoela | Beroe ovata | AY026369 | AF293694 | |
| Cyclocoela | Mnemiopsis leidyi | AY026373 | AF293700 | |
| Typhlocoela | Pleurobrachia bachei | AY026378 | AF293677 | |
| Fungi | ||||
| Ascomycota | Candida albicans | X70659 | X53497 | |
| Ascomycota | Saccharomyces cerevisiae | J01355 | M27607 | |
| Basidiomycota | Tricholoma matsutake | U62964 | U62538.1 | |
| Mucoromycotina | Mucor racemosus | AJ271061 | AJ271061 | |
| Porifera, | ||||
| Calcarea | Leucosolenia sp. | AY026372 | AF100945 | |
| Demospongia | Mycale fibrexilis | AY026376 | AF100946 | |
| Demospongia | Suberites ficus | AY026381 | AF100947 | |
A complete list of sequences used in the analyses with GenBank accession numbers and museum voucher numbers. Bold numbers indicate new sequences generated for this study. KUMIP = University of Kansas Museum of Invertebrate Paleontology, KUNHM = University of Kansas Natural History Museum, MHNG = Muséum d'histoire naturelle de Genève, YPM = Yale Peabody Museum, USNM = US National Museum of Natural History.
All Polypodium sequences were newly generated for this study. We did not include the previously published 18S Polypodium sequence (GenBank accession number U37526) because of concern over the quality of the sequence which included a number of ambiguities. Furthermore, while the two new Polypodium 18S sequences (from hosts Acipenser ruthensus and Polyodon spathula) differed from each other by a total of 8 sites they differed from #U37526 by 77 and 83 sites respectively. These differences included a large number of insertions and deletions. The two new 28S sequences (from hosts Acipenser ruthensus and Scaphirhynchus platorynchus) only differed from each other by 2 sites.
Position of Polypodium
The complete combined dataset of 18S rDNA and partial 28S rDNA contains 4842 characters, 2901 of which are variable and 2124 parsimony informative. Both the ML and parsimony topologies reconstructed from the combined dataset suggest that Polypodium is nested within a monophyletic Cnidaria, and myxozoans are the sister taxon to bilaterians (Figure 2). The ML bootstrap values supporting a monophyletic Cnidaria (including Polypodium), a monophyletic Medusozoa (including Polypodium) and the Polypodium + hydrozoan clade are 73, 67 and 73 respectively (Figure 2A, and Additional file 1). Parsimony analysis of the combined dataset differs from that of ML in that Polypodium is nested within a group of hydrozoans, the leptothecates (Figure 2B). The parsimony bootstrap values supporting a monophyletic Cnidaria and Hydrozoa, with Polypodium nested within these clades are 50 and 51 respectively (Figure 2B). The clade nested within hydrozoans, that includes Polypodium + leptothecates is weakly supported in the sub-sampling tests with a bootstrap value of less than 50.
Figure 2.
Phylogenetic hypotheses of relationships among 126 metazoan taxa, based on a combined analysis of nearly complete 18S and partial 28S rDNA sequences. Arrow indicates Polypodium taxa. A) Maximum likelihood topology. The assumed model (GTR+I + G) has six substitutions rates estimated from the data (A-C, 1.1786; A-G, 3.3654; A-T, 1.7283; C-G, 0.7403; C-T, 4.7803; G-T, 1.0000), an assumed proportion of invariant sites (0.1692) and a gamma shaped parameter or (0.5584). The length of the bar indicates 0.1 substitutions per site. Bootstrap values for this topology are indicated on the cladogram in Additional file 1. B) Strict consensus of 32 trees of length 25141 from a parsimony analyses. Bootstrap values of 50 or greater are indicated.
The analyses using partial 28S rDNA sequences alone (129 sampled taxa) contains 1756 characters, 1196 of which are parsimony informative. The ML topology using this dataset reveals Polypodium nested within Cnidaria, specifically within leptothecate hydrozoans, (Additional file 2). This analysis however fails to recover a monophyletic Cnidaria, as the anthozoans are placed outside the Cnidaria + Bilateria clade. Analysis of the 18S rDNA dataset alone (132 taxa, 3038 characters, 1469 parsimony informative) under both optimality criteria conflicts with the combined and partial 28S topologies. The 18S rDNA topology for both criteria place Polypodium at the base of Bilateria (Figure 3A, Additional files 3, 4 and 5). However, the ML topology also reflects a sister relationship between Polypodium and myxozoans (Figure 3A and Additional file 3A) while the parsimony topology does not (Additional files 4 and 5). Moreover, under parsimony criteria the position of myxozoans is dependent upon how gaps are coded: if gaps are coded as a fifth character state, myxozoans are placed as a highly derived clade of bilaterians (Additional file 4); if gaps are coded as missing, myxozoans are placed as sister to all metazoans (Additional file 5). The 18S analysis showing placement of Polypodium with Bilateria, and more specifically as sister to myxozoans, is consistent with previously reported studies using the same marker [2-4], but raises similar concerns of long-branch attraction [5].
Figure 3.
ML topologies of metazoan relationships of nearly complete 18S rDNA sequences. Arrow indicates Polypodium taxa. Bootstrap values for both topologies are indicated on the cladograms in Additional file 3. A) 132 taxa including 6 myxozoan taxa and two Polypodium taxa. The assumed model (GTR+I + G) has six substitutions rates estimated from the data (A-C, 1.4071; A-G, 3.3470; A-T, 1.6901; C-G, 0.84888; C-T, 4.7638; G-T, 1.0000), an assumed proportion of invariant sites (0.1757) and a gamma shaped parameter or (0.5837). B) Same dataset as (A) but with the 6 myxozoan taxa removed. The assumed model (GTR+I + G) has six substitutions rates estimated from the data (A-C, 1.4115; A-G, 3.3559; A-T, 1.7502; C-G, 0.8342; C-T, 4.8554; G-T, 1.0000), an assumed proportion of invariant sites (0.2464) and a gamma shaped parameter or (0.6326). The length of the bar indicates 0.1 substitutions per site.
Test of long-branch attraction
Myxozoans and Polypodium have unusually high rates of evolution in their 18S and 28S rDNA sequences relative to the other sampled taxa. To investigate the influence of myxozoans on the placement of Polypodium, we removed the myxozoans from our three datasets and re-ran each analysis. Under the ML analysis of 18S rDNA, the removal of myxozoans results in the placement of Polypodium nested within Cnidaria (Figure 3B and Additional file 3B). This result suggests that the placement of Polypodium at the base of bilaterians in the 18S analysis (Figure 3A) was indeed an artifact of LBA. The placement of Polypodium within Cnidaria was not effected by the removal of myxozoans in the 28S (Additional file 6) and combined datasets (Additional file 7).
To investigate the possible role of LBA on myxozoan placement, we removed Polypodium from the combined ML analyses and found that it did not affect the position of Myxozoa at the base of the Bilateria (not shown). Given that bilaterians also form long branches, we tried removing all bilaterian sequences in the combined ML analysis. This resulted in a Myxozoa + Polypodium clade nested within Cnidaria (not shown). However, when Polypodium and bilaterians were removed, myxozoans fell outside the cnidarians (not shown). Similar effects of myxozoan placement to long-branches were also found in parsimony analyses of the combined dataset (not shown).
Discussion
Polypodium is a cnidarian
Our metazoan dataset of 18S and partial 28S rDNA sequences, with a large taxonomic sample of cnidarians, places Polypodium within a monophyletic Cnidaria. This accords with the fact that Polypodium possesses nematocysts [17,18] and a cnidarian-like body plan [7-9,12]. The precise placement of Polypodium within Cnidaria is less certain. The ML combined analysis places Polypodium as sister to Hydrozoa (Figure 2A), a hypothesis consistent with the suggestion that Polypodium be considered a separate class of cnidarians, Polypodiozoa [1]. By contrast, the combined parsimony analysis (Figure 2B) and the ML analyses of 28S alone (Additional file 2 and 6) place Polypodium within the hydrozoan clade Leptothecata. Given that leptothecates have relatively high rates of evolution within hydrozoans, one possible explanation for the conflicting hypotheses is that the placement of Polypodium within leptothecates is an artifact of LBA and that the combined data, in conjunction with the ML approach (Figure 2A), overcame this localized LBA artifact.
Evolution of Polypodium life-history characters
Although the fresh water habitat of Polypodium is unusual for cnidarians, it is not unheard of, especially within hydrozoans. For instance, the model organism Hydra and the jellyfish Craspedacusta are both exclusively fresh-water hydrozoans. Hydra and Craspedacusta are distantly related [25] and our analyses do not indicate a close phylogenetic affinity of Polypodium to either of the clades containing these taxa. Thus, it appears that in the evolution of cnidarians, invasion to fresh-water habitats has happened at least three separate times.
Although Polypodium is the only known intracellular cnidarian parasite, other cnidarians have adopted parasitic life-styles [11,26-29]. For example, parasites belonging to the Narcomedusae (Hydrozoa) have been reported to live in the stomach cavities of other narcomedusae [11,27] and anthomedusae [27]. In addition, the sea anemone Edwardsiella lineata parasitizes the stomach cavity of the ctenophore Mnemiopsis leidyi [28] and the anemone Peachia quinquecapitata is reported to parasitize the stomachs of hydromedusa [29].
Effects of long-branch attraction
The well-documented effects of long-branch attraction artifacts (reviewed in Bergsten [30]) are particularly concerning when investigating relationships amongst early-diverging metazoans, where rates between lineages vary greatly [22]. Suggestions for avoiding LBA artifacts include choice of appropriate markers [31,32], increased taxonomic sampling to effectively break up long branches [33,34] and utilization of best-fit models that incorporate rate variation [21-23]. Previous conflicting reports that show Polypodium and myxozoans form a sister taxon to Bilateria [2-4] can be explained by limited taxon sampling and an inadequate number of informative characters in their analyses, both of which confound long-branch problems. In this study, the increased taxonomic sampling of cnidarians and the addition of 28S rDNA sequence data proved critical to placing the highly divergent Polypodium taxon within Cnidaria. The choice of optimality criteria (ML vs. parsimony) both supported Polypodium as a cnidarian but did affect the placement within Cnidaria.
Polypodium and Myxozoa
Our analyses are inconclusive in the placement of Myxozoa within metazoans. We found that myxozoans consistently grouped with long-branched taxa and that removal of long-branches resulted in myxozoans being placed to the next longest branch. For example myxozoans group with Polypodium in the absence of Bilateria and group with Bilateria in the absence of Polypodium (not shown).
Jimenez-Guri et al. [24] sampled the myxozoan, Buddenbrockia, and found it to fall within Cnidaria, as the sister group to two hydrozoan representatives and a single scyphozoan. Previous studies have suggested a sister group relationship between cnidarians and myxozoans [2-4], and some morphological evidence has been used to support this view [35]. Although our present study does not support this relationship, further investigation is merited. Myxozoans are a highly diverse group (reviewed in Kent et al. [36]) that comprise two clades, the Myxosporea and the Malacosporea [37]. We were only able to include 28S rDNA sequences from myxosporeans, although the malacosporean Buddenbrockia was included in our 18S analysis and found to group with other myxozoans and outside of Cnidaria. Future studies with a comprehensive sampling of myxozoans together with Polypodium, in a dataset that includes a large taxonomic sampling of cnidarians, should shed further light on the relationships between myxozoans and Polypodium.
Conclusion
Although previous molecular phylogenetic hypotheses conflicted with the traditional interpretation of cnidarian affinity for Polypodium, the molecular evidence we present, using an augmented dataset, ultimately confirms and reconciles this traditional hypothesis and suggests that Polypodium is indeed a cnidarian. This study also reaffirms the importance to large taxonomic sampling and inclusion of additional informative characters for avoiding long-branch attraction artifacts.
Methods
DNA isolation, amplification and sequencing
Genomic DNA was extracted using Qiagen DNeasy kits according to manufacturer's protocol (QIAGEN Inc., Mississauga, ON) or a standard phenol/chloroform protocol. The latter method involved tissue digestion with proteinase K (20 mg/ml) in a lysis buffer (20 mM Tris-CL pH 8.0, 5 mM EDTA pH 8.0, 400 mM NaCl, 2%SDS), extraction with phenol/chloroform (1:1), precipitation with 2.5 vol. 95% EtOH, and elution in TE or H2O.
An approximately 1.8 kb portion of the gene coding for 18S was amplified and sequenced with universal eukaryotic primers as described by Medlin et al. [38], with the annealing temperature modified to 57°C. With the exception of Polypodium samples, a nearly complete, roughly 3 kb portion of the gene coding for 28S was amplified and sequenced with an approach modified from that reported in Collins et al. [25]. 28S was directly amplified in two fragments with combinations of primers F63mod+R2077sq and F1379+R3264 from Medina et al. [39] or newly developed medusozoan specific primers F97+R2084 and F1383+R3238 (F97: CCYYAGTAACGGCGAGT, R2084: AGAGCCAATCCTTTTCC, F1383: GGACGGTGGCCATGGAAGT, and R3238: SWACAGATGGTAGCTTCG). Amplifications of 28S were conducted with the following thermal profile: 4 minutes at 94°C; 30 cycles of 30 seconds at 94°C, 1 minute at 45°C, and 3 minutes at 72°C; and 10 minutes at 72°C. For Polypodium, a portion of the 5' end of 28S (approx. 0.8–1.0 kbps) was amplified using two universal metazoan primers (fw1and rev2) as reported by Sonnenberg et al. [40]. Sequencing was carried out using amplification primers and F635sq and R635sq from Medina et al. [39].
All gene fragments were purified and sequenced by Cogenics, Inc. (Houston, TX) and assembled and edited using Sequencher v4.5 (Gene Code Co., 2005). Sequences for each marker were aligned using the program MUSCLE [41]. The 28S sequence alignment was then trimmed to reflect only that region which included sequence data for Polypodium. This trimmed 28S dataset was analyzed separately and used in conjunction with the complete 18S sequences to create the combined dataset.
Phylogenetic analyses
Phylogenetic analyses were performed using both maximum likelihood (ML) and parsimony criteria. ML searches were performed using GARLI v0.951.OsX-GUI [42] under an assumed GTR model with rates estimated from the data. The assumed model of nucleotide substitution was selected by using the Akaike Information Criterion (AIC) as implemented in ModelTest [43]. Each run was repeated 10 times from random starting trees using default termination conditions. Each run gave identical topologies and similar likelihood scores. 100 bootstrap replications were run in GARLI v0.951.0sX-GUI [42] under the same parameters.
To assess the effect that omitting length-variable regions has on topology, we removed these regions from the combined dataset, using the less stringent settings of Gblocks [44]. This dataset contained 126 metazoan taxa, 2415 characters, 1391 of which are parsimony informative. We found that removal of length-variable regions had no effect on the placement of Polypodium and minimal effect on overall topology in our combined ML analyses (Additional file 8). Therefore we performed all other analyses with the complete datasets, including the more variable regions.
Parsimony analyses were performed using TNTv.1.1 [45]. Separate tree searches were performed with gaps coded as missing and gaps coded as a fifth state. However, with one exception (see results for myxozoan placement with 18S data) there was no significant difference in topology. Numerous search methods available in TNT were utilized to search the tree space but the following approach was found to consistently recover trees with minimum lengths from our datasets. The implemented search was a driven new technology search with a random seed of 0 (where 0 = time). Default settings for sectorial searches (RSS and CSS) and tree fusing were used [46], with 5 replicates per repetition, and a requirement that the global optimum be found 20 times. TBR branch swapping was performed on the resulting trees and a strict consensus was calculated. TNT was used to calculate standard bootstrap values (1000 replicates). Alignments and trees for 18S, 28S and combined datasets have been submitted to TreeBASE http://www.treebase.org/treebase/index.html.
Authors' contributions
This study was inspired by the work of EVR and originally conceived by EVR, AL and AGC. NME performed most of the data collection and submission of the new sequences. NME and PC performed the analyses. PC took the lead in organizing the study and drafting the manuscript with contributions from NME. All other authors provided comments and suggestions and approved the final manuscript.
Supplementary Material
ML topology of relationships based on combined data. ML topology identical to Figure 1A but as a cladogram showing bootstrap values.
ML topology of relationships based on partial 28S rDNA sequences. Relationships of 128 metazoan taxa were analyzed with partial 28S rDNA sequences.
ML topologies of relationships based on 18S data with and without myxozoans. ML topologies identical to Figure 3 but as cladograms showing bootstrap values.
Parsimony topology of relationships based on 18S rDNA sequences. This parsimony analysis of 18S rDNA sequences included 132 taxa with gaps coded as a fifth state.
Parsimony topology of relationships based on 18S rDNA sequences. This parsimony analysis of 18S rDNA sequences included 132 taxa with gaps coded as missing.
ML topology of relationships excluding myxozoans, based on partial 28S rDNA data. This ML analysis of partial 28S rDNA sequences excluded myxozoan taxa.
ML topology of relationships excluding myxozoans, based on combined data. This ML analysis of partial 28S rDNA and 18S sequences excluded myxozoan taxa.
ML topology of relationships based on combined data. This analysis of 126 metazoan taxa was based on combined 18S and partial 28S rDNA with length variable sequences removed.
Contributor Information
Nathaniel M Evans, Email: evansnat@ku.edu.
Alberto Lindner, Email: alindner@usp.br.
Ekaterina V Raikova, Email: raikova@swipnet.se.
Allen G Collins, Email: COLLINSA@si.edu.
Paulyn Cartwright, Email: pcart@ku.edu.
Acknowledgements
We thank S. Ash Bullard, Jan Dean, Bobby Reed and Ron Bruch for contributions of Polypodium specimens. We also thank Peter Schuchert, Casey Dunn and Michael Dawson for contributions of other cnidarian specimens. NME acknowledges the instructors at the 2007 Ohio State Cladistics workshop, especially P. Goloboff, for help with TNT. We thank M. Holder for discussions, B. Bentlage, B. Lieberman, A. Nawrocki, and five anonymous reviewers for helpful comments on earlier versions of this manuscript. This work was supported by grants from NSF AToL EF-0531779 (to PC and AGC) and support for AL from NSF (PEET DEB-9978086) and FAPESP (06/02960-8/05821-9/60327-0).
References
- Raikova EV. In: Porifera and Cnidaria Contemporary state and perspectives of investigations. Koltum VM, Stepanjants SD, editor. Leningrad: Zoological Institute of Academy of Sciences of USSR; 1988. On the systematic position of Polypodium hydriforme Ussov, (Coelenterata) pp. 116–122. [Google Scholar]
- Siddall ME, Martin DS, Bridge D, Cone DM, Desser SS. The demise of a phylum of protests: Phylogeny of Myxozoa and other parasitic Cnidaria. J Parasitol. 1995;81:961–967. doi: 10.2307/3284049. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Whiting MF. Long-branch abstractions. Cladistics. 1999;15:9–24. doi: 10.1111/j.1096-0031.1999.tb00391.x. [DOI] [Google Scholar]
- Zrzavý J, Hypša V. Myxozoa, Polypodium, and the origin of the Bilateria: The phylogenetic position of "Endocnidozoa" in the light of the rediscovery of Buddenbrockia. Cladistics. 2003;19(3):164–169. doi: 10.1016/S0748-3007(03)00007-0. [DOI] [Google Scholar]
- Hanelt B, Van Schyndel D, Adema CM, Lewis L, Loker ES. The phylogenetic position of Rhopalura ophiocomae (Orthonectida) based on 18S ribosomal DNA sequence analysis. Mol Biol Evol. 1996;13:1187–1191. doi: 10.1093/oxfordjournals.molbev.a025683. [DOI] [PubMed] [Google Scholar]
- Raikova EV. Life cycle and systematic position of Polypodium hydriforme Ussov (Coelenterata), a cnidarian parasite of the eggs of Acipenseridae. Publ Seto Mar Biol Lab. 1973;20:165–173. [Google Scholar]
- Raikova EV. Morphology, ultrastructure and development of the parasitic larva and its surrounding trophamnion of Polypodium hydriforme Ussov (Coelenterata) Cell Tissue Res. 1980;206(3):487–500. doi: 10.1007/BF00237977. [DOI] [PubMed] [Google Scholar]
- Raikova EV. Life cycle, cytology, and morphology of Polypodium hydriforme, a coelenterate parasite of the eggs of acipenseriform fishes. J Parasitol. 1994;80(1):1–22. doi: 10.2307/3283338. [DOI] [PubMed] [Google Scholar]
- Raikova EV, Suppes VC, Hoffmann GL. The parasitic coelenterate, Polypodium hydriforme Ussov, from the eggs of the American acipenseriform Polyodon spathula. J Parasitol. 1979;65(5):804–810. doi: 10.2307/3280366. [DOI] [PubMed] [Google Scholar]
- Berrill NJ. Development and medusa-bud formation in the Hydromedusae. Quart Rev Biol. 1950;25:292–316. doi: 10.1086/397707. [DOI] [PubMed] [Google Scholar]
- Bouillon J. Considérations sur les dévéloppement des narcoméduses et sur leur position phylogenétique. Indo-Malayan Zool. 1987;4:189–278. [Google Scholar]
- Hyman L. The invertebrates. I. Protozoa through Ctenophora. New York and London: McGraw-Hill; 1940. [Google Scholar]
- Schimkevitch VM. Essay on contemporary state of the problem of the development of Hydrozoa. Vestnik Estestvoznanija. 1890;4:171–176. [Google Scholar]
- Lipin A. Geschlechtliche Form, Phylogenie und systematische Stellung von Polypodium hydriforme Ussov. Zool Jahrb Anat. 1925;47:541–635. [Google Scholar]
- Bouillon J, Medel MD, Pagès F, Gili JM, Boero F, Gravili C. Fauna of the Mediterranean Hydrozoa. Scientia Marina. 2004;68(Suppl 2):5–438. [Google Scholar]
- Bouillon J, Gravili c, Pagès F, Gili JM, Boero F. An introduction to Hydrozoa. Paris: Publications Scientifiques du Muséum, Paris; 2006. [Google Scholar]
- Raikova EV. Fine structure of the nematocytes of Polypodium hydriforme Ussov (Cnidaria) Zoologica Scripta. 1990;19(1):1–11. doi: 10.1111/j.1463-6409.1990.tb00236.x. [DOI] [Google Scholar]
- Ibragimov A, Raikova E. Nematocysts of Polypodium hydriforme, a cnidarian parasite of acipenseriform fishes. Hydrobiologia. 2004;531:165–171. doi: 10.1007/s10750-004-2651-y. [DOI] [Google Scholar]
- Dellaporta SL, Xu A, Sagasser S, Jakob W, Moreno MA, Buss LW, Schierwater B. Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum. Proc Natl Acad Sci USA. 2006;103(23):8751–8756. doi: 10.1073/pnas.0602076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AG. Phylogeny of Medusozoa and the evolution of cnidarian life cycles. J Evol Biol. 2002;15(3):418–432. doi: 10.1046/j.1420-9101.2002.00403.x. [DOI] [Google Scholar]
- Huelsenbeck JP. Is the Felsenstein zone a fly trap? Syst Biol. 1997;46:247–264. doi: 10.2307/2413636. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim W, Cunningham CW. A new perspective on lower metazoan relationships from 18S rDNA sequences. Mol Biol Evol. 1999;16:423–427. doi: 10.1093/oxfordjournals.molbev.a026124. [DOI] [PubMed] [Google Scholar]
- Cunningham CW, Zhu H, Hillis DM. Best-fit maximum likelihood models for phylogenetic inference: Empirical tests with known phylogenies. Evolution. 1998;52:978–987. doi: 10.2307/2411230. [DOI] [PubMed] [Google Scholar]
- Jimenez-Guri E, Philippe H, Okamura B, Holland PWH. Buddenbrockia is a cnidarian worm. Science. 2007;317(5834):116–118. doi: 10.1126/science.1142024. [DOI] [PubMed] [Google Scholar]
- Collins AG, Schuchert P, Marques AC, Jankowski T, Medina M, Schierwater B. Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst Biol. 2006;55(1):97–115. doi: 10.1080/10635150500433615. [DOI] [PubMed] [Google Scholar]
- Osborn DA. Cnidarian "parasites" on Solmissus incisa, a Narcomedusa. Sci Mar. 2000;64:157–163. [Google Scholar]
- Pagès F, Corbera J, Lindsay D. Piggybacking pycongonids and parasitic narcomedusae on Pandea rubra (Anthomedusae, Pandeidae) Plankton Benthos Res. 2007;2(2):83–90. doi: 10.3800/pbr.2.83. [DOI] [Google Scholar]
- Bumann D, Puls G. Infestation with larvae of the sea anemone Edwardsia lineata affects nutrition and growth of the ctenophore Mnemiopsis leidyi. Parasitology. 1996;113:123–128. [Google Scholar]
- Spaulding JG. The life cycle of Peachia quinquecapitata, an anemone parasitic on medusae during its larval development. Biol Bull. 1972;143(2):440–453. doi: 10.2307/1540065. [DOI] [Google Scholar]
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- Rokas A, King N, Finnerty J, Carroll SB. Conflicting phylogenetic signals at the base of the metazoan tree. Evol Dev. 2003;5(4):346–359. doi: 10.1046/j.1525-142X.2003.03042.x. [DOI] [PubMed] [Google Scholar]
- Xiang QY, Moody ML, Soltis DE, Fan C, Soltis PS. Relationships within Cornales and circumscription of Cornaceae – matK and rbcL sequence data and effects of outgroups and long branches. Mol Phyl Evol. 2002;24:35–57. doi: 10.1016/S1055-7903(02)00267-1. [DOI] [PubMed] [Google Scholar]
- Hillis DM. Taxonomic sampling, phylogenetic accuracy, and investigator bias. Syst Biol. 1998;47:3–8. doi: 10.1080/106351598260987. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 2002;51:588–598. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
- Raikova EV. Cytomorphological characters of Polypodium hydriforme and problems of myxozoan and cnidarian phylogeny. Tsitologiia. 2005;47(10):933–939. [PubMed] [Google Scholar]
- Kent ML, Andree KB, Bartholomew JL, El-Matbouli M, Desser SS, Devlin RH, Feist SW, Hedrick RP, Hoffmann RW, Khattra J, Hallett SL, Lester RJG, Longshaw M, Oswaldo P, Siddall ME, Xiao C. Recent advances in our knowledge of the Myxozoa. J Eukaryot Microbiol. 2001;48(4):395–413. doi: 10.1111/j.1550-7408.2001.tb00173.x. [DOI] [PubMed] [Google Scholar]
- Canning EU, Curry A, Feist SW, Longshaw M, Okamura B. A new class and order of myxozoans to accommodate parasites of bryozoans with ultrastructural observations on Tetracapsula bryosalmonae (PKX Organism) J Eukaryot Microbiol. 2000;47(5):456–468. doi: 10.1111/j.1550-7408.2000.tb00075.x. [DOI] [PubMed] [Google Scholar]
- Medlin LH, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Medina M, Collins AG, Silberman JD, Sogin ML. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Natl Acad Sci USA. 2001;98(18):9707–9712. doi: 10.1073/pnas.171316998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg R, Nolte A, Tautz D. An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Front Zool. 2007;4(1):6. doi: 10.1186/1742-9994-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin: The University of Texas; 2006. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 2000;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Goloboff P, Farris J, Nixon K. TNT: Tree Analysis Using New Technology. 1.1. Tucuman, Argentina: Published by the authors; 2003. [Google Scholar]
- Goloboff P. Analyzing large data sets in reasonable times: Solutions for composite optima. Cladistics. 1999;14:415–428. doi: 10.1111/j.1096-0031.1999.tb00278.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ML topology of relationships based on combined data. ML topology identical to Figure 1A but as a cladogram showing bootstrap values.
ML topology of relationships based on partial 28S rDNA sequences. Relationships of 128 metazoan taxa were analyzed with partial 28S rDNA sequences.
ML topologies of relationships based on 18S data with and without myxozoans. ML topologies identical to Figure 3 but as cladograms showing bootstrap values.
Parsimony topology of relationships based on 18S rDNA sequences. This parsimony analysis of 18S rDNA sequences included 132 taxa with gaps coded as a fifth state.
Parsimony topology of relationships based on 18S rDNA sequences. This parsimony analysis of 18S rDNA sequences included 132 taxa with gaps coded as missing.
ML topology of relationships excluding myxozoans, based on partial 28S rDNA data. This ML analysis of partial 28S rDNA sequences excluded myxozoan taxa.
ML topology of relationships excluding myxozoans, based on combined data. This ML analysis of partial 28S rDNA and 18S sequences excluded myxozoan taxa.
ML topology of relationships based on combined data. This analysis of 126 metazoan taxa was based on combined 18S and partial 28S rDNA with length variable sequences removed.