Abstract
We previously found increased erythropoietin receptor (EPO-R) protein levels in vigorously growing canine lungs after pneumonectomy (PNX), suggesting a role for paracrine EPO signaling in lung growth and remodeling. Now we find that sense and antisense EPO-R transcripts (sEPO-R and asEPO-R, respectively) are concordantly up-regulated in the post-PNX remaining lung, leading to the hypothesis that sEPO-R and asEPO-R interactions enhance EPO signaling during lung growth. We cloned a canine asEPO-R cDNA, which is fully complementary to the sense strand of the EPO-R gene from 2.5kb 3′ to the sense stop codon, and extends into the 5′ UTR of the sEPO-R transcript. Both asEPO-R and sEPO-R transcripts colocalize with EPO-R protein in the same lung cells. In cultured human embryonic kidney (HEK293) cells, transfection with sEPO-R (+FLAG tag) cDNA alone increased EPO-R protein expression (anti-EPO-R and anti-FLAG). At constant sEPO-R cDNA levels, cotransfection with escalating asEPO-R cDNA further increased recombinant EPO-R protein expression. The asEPO-R transcript harbors two putative opening reading frames (ORFs). Separate transfection of each asEPO-R ORF cDNA resulted in differential stimulatory effects on EPO-R protein expression. We conclude that both sEPO-R and asEPO-R transcripts contribute to in vivo up-regulation of EPO-R protein expression in the post-PNX remaining lung. This demonstrates synergism between sense–antisense EPO-R transcripts in response to physiological stimulation in a robust model of induced lung growth.
Keywords: compensatory lung growth, pneumonectomy
Erythropoietin (EPO), produced mainly in the kidney and fetal liver and stimulated by hypoxic or anemic stress, binds to its receptor (EPO-R) on bone marrow erythroid progenitor cells to inhibit apoptosis and promote erythrocyte differentiation (1). Both EPO and EPO-R have widespread paracrine/autocrine function in nonhematopoietic tissue (2), including prevention of apoptosis, facilitation of differentiation and angiogenesis, and protection against ischemic or hypoxic injury (3–5). EPO signaling is essential for normal organ development (6), possibly by coordinating recruitment and homing of endothelial progenitor cells (7). The modulators of EPO-R expression remain poorly defined. Several DNA motifs including GATA-1, SP-1 binding sites have been identified as essential to EPO-R promoter activity (8), but their physiological significance is unknown. We previously found increased EPO-R protein expression in actively growing postnatal canine lungs (9). After one lung is surgically removed by pneumonectomy (PNX), the remaining lung undergoes accelerated enlargement, growth, and remodeling, which eventually restores alveolar tissue volume, surface area, and gas exchange to normal values (10). During post-PNX compensation, expression of EPO-R mRNA and protein, its upstream transcriptional regulators, and downstream targets are up-regulated in the remaining lung (9, 11). These findings led us to examine how EPO-R expression is regulated after PNX. We detected the presence and concordant elevation of both sense and antisense EPO-R transcripts (sEPO-R and asEPO-R, respectively) in the remaining lung. This begs the question of whether interactions between asEPO-R and sEPO-R transcripts enhance EPO-R expression during compensatory lung growth. To address this, we cloned the canine asEPO-R cDNA, localized sEPO-R and asEPO-R transcripts as well as EPO-R protein in the lung, and characterized the regulation of EPO-R protein expression by asEPO-R in cultured cells.
Results
Endogenous asEPO-R Transcripts.
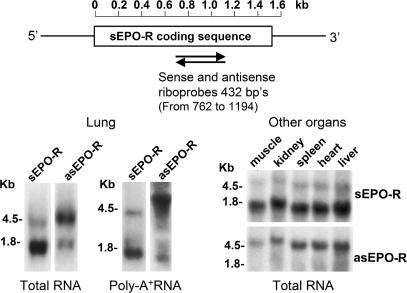
We detected asEPO-R transcript by RNA blot in normal canine lung using a sense riboprobe (Fig. 1). The major asEPO-R transcript is ≈4.5 kb, with a minor asEPO-R transcript of ≈1.6–1.8 kb. The major sEPO-R transcript is ≈1.7 kb, with a minor transcript of ≈4.0 kb (Fig. 1 Lower left). Both asEPO-R and sEPO-R transcripts are present in other canine organs (muscle, kidney, spleen, heart, and liver) (Fig. 1 Lower right) and in 12 major human tissues (HB-2010; Origene) (data not shown).
Fig. 1.
RNA blot of sense and antisense erythropoietin receptor (sEPO-R and asEPO-R, respectively). (Upper) Location of riboprobes. (Lower) Typical RNA blots labeled with sense or antisense probes under high stringency using either total (20 μg per lane) or polyA+ (1 μg per lane) RNA from lung and total RNA (20 μg per lane) from five other canine organs.
Regulated mRNA and Protein Expression in post-PNX Lung Growth.
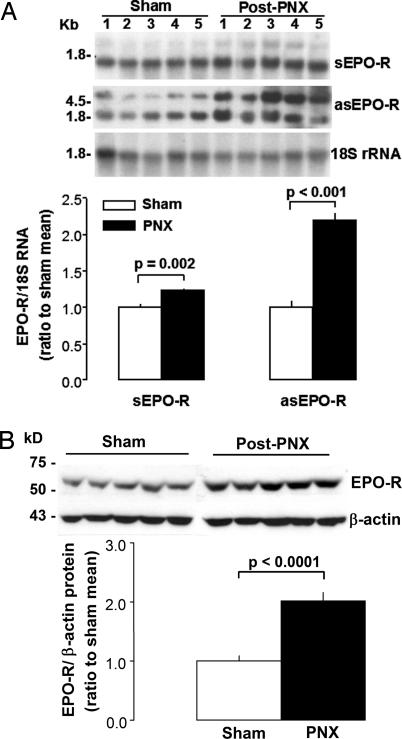
In the actively growing remaining left lung of dogs after right PNX, sEPO-R and asEPO-R transcripts and EPO-R protein levels were 23% (P = 0.002), 119% (P < 0.001), and 169% (P < 0.0001), respectively, higher than in the corresponding lobe of matched control animals after sham PNX (Fig. 2).
Fig. 2.
Expression of sEPO-R and asEPO-R transcripts and EPO-R protein in the remaining canine lung following right PNX or Sham PNX (n = 5 each). (A) RNA blots of sEPO-R and asEPO-R transcripts, with 18S rRNA as loading control. Individual sEPO-R/18S or asEPO-R/18S signals were expressed as a ratio to the mean value in Sham group. Triplicate assays used separate tissue samples from each animal. (B) Immunoblot of EPO-R protein. Mean ± SEM, unpaired t test.
Structure of asEPO-R Transcript.
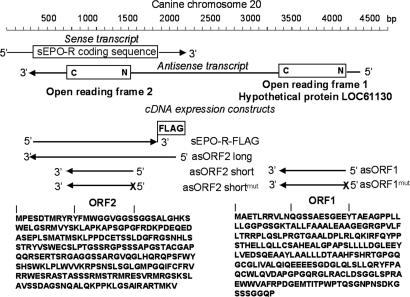
We amplified asEPO-R cDNA using the published sEPO-R sequence (GenBank accession No. AY908987) from canine lung mRNA with strand-specific RT-PCR. The 5′ and 3′ ends of asEPO-R cDNA were obtained by RACE PCR [supporting information (SI) Fig. S1]. The asEPO-R and sEPO-R transcripts are entirely complementary within the EPO-R-coding region (Fig. 3). The full-length cDNA of asEPO-R is ≈4.1 kb. The 5′ end contains an ORF (ORF1) that encodes a hypothetical 248-amino acid protein (LOC61130, GenBank accession no. XM_848773). Another ORF (ORF2; 248 aa) lies within the coding region of sEPO-R.
Fig. 3.
Schematic diagram of sEPO-R and asEPO-R transcripts from canine lung. The asEPO-R transcript contains two putative ORFs (ORF1 and ORF2, 248 aa each) with hypothetical predicted polypeptides shown (LOC61130 GenBank accession no. XM_84877). Below the native transcripts are cDNA constructs used for expression studies. sEPO-R-FLAG represents sense cDNA with C-terminal FLAG epitope tag. asORF2-long represents antisense cDNA with ≈300 bp, followed by the full ORF2 coding region. asORF2-short represents antisense cDNA spanning the ORF2 putative coding region. asORF2-shortmut represents asORF2-short with mutated start codon (ATG to ACG) denoted by “X.” asORF1 represents antisense cDNA spanning the ORF1 putative coding region. asORF1mut represents asORF1 with mutated start codon (ATG to ACG) denoted by “X.”
Localization.
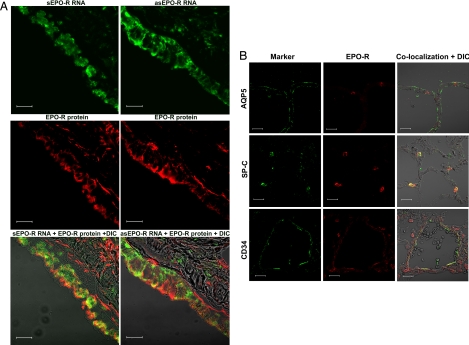
We colocalized sEPO-R or asEPO-R transcripts with EPO-R protein on the same lung sections by in situ hybridization and fluorescent immunohistochemistry. sEPO-R and asEPO-R transcripts each colocalized with EPO-R protein to the same bronchiolar epithelial cells (Fig. 4A). EPO-R protein colocalized with surfactant-associated protein-C (SP-C), a type 2 alveolar epithelial cell marker, and with CD34, an alveolar endothelial cell marker, but not with aquaporin-5 (AQP5), a type 1 epithelial cell marker (Fig. 4B). To exclude nonspecific annealing, we cloned a 412-bp cDNA fragment of canine surfactant-associated protein-A and generated cRNA probes for sense and antisense transcripts (sSP-A and asSP-A, respectively). The sSP-A transcripts were abundant within alveolar septa (positive control), but no hybridization signals were detected for asSP-A transcripts (negative control) (Fig. S2) or random RNA (data not shown).
Fig. 4.
Immunolocalization. (A) (Top) sEPO-R (Left) and asEPO-R (Right) transcripts were detected by in situ hybridization (green) using antisense and sense cRNA probes, respectively, in bronchiolar epithelial cells on OCT-embedded lung sections. (Middle) The same sections were labeled for EPO-R protein (red) by immunofluorescence. (Bottom) Superimposed images with differential interference contrast (DIC). (Scale bar, 20 μm.) (B) Immunofluorescent localization of EPO-R protein (red, Center) with an alveolar cell marker (green, Left): AQP5 (Top), SP-C (Middle), and CD34 (Bottom), on the same sections. Images were superimposed with DIC (Right). EPO-R colocalized with SP-C and CD-34 (yellow), but not with AQP5. (Scale bar, 20 μm.)
Effect of asEPO-R RNA in Vitro.
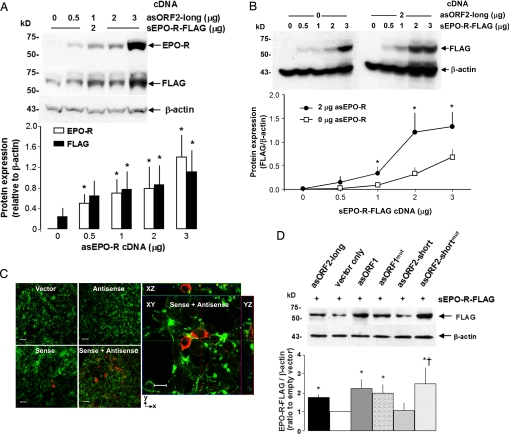
To examine whether asEPO-R mRNA expression increases EPO-R protein level, we expressed sEPO-R and asEPO-R transcripts in HEK-293 cells and studied their effect on EPO-R protein expression. The sEPO-R cDNA covered the entire coding region (508 aa) with a C-terminal FLAG epitope tag, whereas the asEPO-R cDNA started 314 bp 3′- from the stop codon of sEPO-R and covered the entire coding region of sEPO-R (Fig. 3; asORF2-long). At a constant level of sEPO-R cDNA, EPO-R protein level detected both by EPO-R and FLAG antibodies increased as more asORF2-long was added (Fig. 5A). In the absence of asORF2-long, adding sEPO-R cDNA to the cells increased EPO-R protein expression in a dose-dependent fashion. In the presence of asORF2-long, each dose of sEPO-R cDNA resulted in higher EPO-R protein expression compared to the corresponding level without asORF2-long (Fig. 5B). These results demonstrate the interaction between sense–antisense EPO-R transcripts in stimulating EPO-R protein expression. Under confocal microscopy, cotransfection of asORF2-long and sEPO-R cDNA increased the number of cells expressing EPO-R protein compared to transfection with sEPO-R DNA alone (Fig. 5C). No EPO-R expression was found in cells transfected with random DNA or asEPO-R DNA only. Transfected EPO-R partly localized to the cell membrane (see ZX- and ZY-sections), suggesting targeting to the cell surface.
Fig. 5.
Effect of asEPO-R cDNA transfection on EPO-R protein expression in cultured HEK-293 cells. (A) Cells were cotransfected with sEPO-R-FLAG cDNA (2 μg) and variable amounts of asORF2-long cDNA and immunoblotted for EPO-R, FLAG, and β-actin. *, P < 0.05 vs. 0 μg of asEPO-R cDNA. (B) Cells were cotransfected with asORF2-long cDNA (0–2 μg) and variable amounts of sEPO-R-FLAG cDNA and immunoblotted for FLAG and β-actin. *, P < 0.05, 2 μg vs. 0 μg asORF2-long at the same sEPO-R-FLAG cDNA level. For A and B, three independent experiments were performed (mean ± SD, one-way ANOVA). (C) Immunocytochemistry of transfected cells shows increased EPO-R production in response to sEPO-R-FLAG and asORF2-long cotransfection. (Left) Whole-cell images of transfection with vector only, asORF2-long only, sEPO-R-FLAG only, or sEPO-R-FLAG + asORF2-long cDNAs, stained with anti-FLAG antibody for EPO-R-FLAG (red) and phalloidin for β-actin (green). (Scale bars, 20 μm.) (Right) Confocal optical section of cells cotransfected with sEPO-R-FLAG + asORF2-long cDNAs, stained for EPO-R(FLAG) and β-actin. (Scale bar, 10 μm.) (D) Cultured cells were cotransfected with sEPO-R-FLAG cDNA (2 μg) and different antisense cDNAs (2 μg): asORF2-long, antisense cDNA with ≈300 bp + ORF2 coding region; asORF2-short, antisense cDNA spanning the putative ORF2 coding region; asORF2-shortmut, asORF2-short with mutated start codon; asORF1, antisense cDNA spanning the putative ORF1 coding region; asORF1mut, asORF1 with mutated start codon. EPO-R-FLAG expression was detected by immunoblot with anti-FLAG normalized to β-actin. A typical blot (Upper) and average data from four independent experiments (Lower) are shown. The mean EPO-R-FLAG/β-actin expression with vector only was arbitrarily set at 100%. Mean ± SD, one-way ANOVA: P < 0.05 * vs. empty vector † vs. corresponding wild-type antisense cDNA.
Effects of Putative Coding Regions of asEPO-R.
The asORF2-long cDNA spans ≈300 bp upstream of the putative coding sequence, whereas asORF2-short spans only the ORF coding region (Fig. 3). In some constructs the putative ATG start codon was mutated (asORF1mut, asORF2-shortmut, respectively). Cultured HEK293 cells were cotransfected with each asEPO-R construct at a constant level of sEPO-R-FLAG cDNA. Control cells were cotransfected with asORF2-long cDNA (positive control) or empty vector (negative control) (Fig. 5D). Cells cotransfected with intact asORF1 cDNA showed higher EPO-R protein expression compared to control cells. The increase was not eliminated by mutation (asORF1-shortmut), suggesting that translation of ORF1 is probably not critical for the observed synergism, because asORF1 with intact translation performs marginally better than translation-defective asORF1mut.
Results with asORF2 are more complex. Cotransfection with intact asORF2-short cDNA had minimal effect, whereas asORF2-long clearly increased EPO-R protein expression (Fig. 5D). There may be critical sequences 5′ to ORF2 that potently stimulate EPO-R protein expression. Mutation (ORF2-shortmut) significantly increased EPO-R protein expression, suggesting multifaceted regulation of sEPO-R transcripts by its antisense transcript. One possible model compatible with these data is that the asORF2 transcript positively regulates, whereas the hypothetical asORF2 protein negatively regulates, EPO-R protein expression. The opposing action can explain the minimal net change of EPO-R protein level when both asORF2-short transcript and protein are expressed. When translation of asORF2 is abolished by the ATG mutation, the positive effect of asORF2 transcript acts unopposed to increase EPO-R protein expression.
Discussion
Summary.
This is the first report of the existence and function of naturally occurring asEPO-R transcript in the lung. We found abundant asEPO-R transcript in multiple organs, and showed that sEPO-R and asEPO-R transcripts coexist in the same cells with EPO-R protein, which was localized to the alveolar type 2 epithelium and endothelium. We observed that in vivo up-regulation of asEPO-R transcripts was concurrent with increased sEPO-R transcript and with increased EPO-R protein expression in a physiological model of post-PNX compensatory lung growth. Finally, we demonstrated sense–antisense transcript synergism in vitro (i.e., asEPO-R transcript enhances EPO-R protein expression induced by sEPO-R transcript). Two putative ORFs within the asEPO-R transcript were identified and appear to regulate sEPO-R expression via different mechanisms.
EPO Signaling and Lung Growth.
Both EPO and EPO-R are widely expressed in embryonic and adult tissue, and both possess pleiotropic paracrine/autocrine function related to development, growth, and cytoprotection. Endogenous EPO signaling enhances vascular growth and protects against ischemic and hypoxic injury (5). EPO is as potent as VEGF in inducing angiogenesis in vitro (12). In mice with attenuated alveolar development due to inhibited angiogenesis, exogenous EPO maintains alveolar volume, gas exchange surface area, and endothelial cell volume (13). EPO recruits endothelial progenitor cells for pulmonary vascular remodeling and prevents hypoxia-induced pulmonary hypertension (7). During hyperoxic lung injury, EPO treatment improves alveolar morphology and vascularity and reduces fibrosis, suggesting protection against bronchopulmonary dysplasia (14).
Lung tissue EPO-R protein expression is elevated during postnatal growth and maturation (9); the normally high EPO-R expression is further enhanced in the post-PNX remaining lung during active compensatory alveolar growth (10), in which generation of additional gas exchange tissue and surface is accompanied by elevated expression of hypoxia-induced factor-1α (HIF-1α), a transcriptional regulator of EPO, as well as indices of cell proliferation and apoptosis (9, 15). The major stimulus for compensatory growth appears to be expansion of the remaining lung. Preventing post-PNX lung expansion impairs structural and functional compensation (16, 17), whereas induction of lung expansion up-regulates mRNA and protein expression of HIF-1α, EPO-R, and VEGF (11) and improves function (16). Cumulative data support the notion that HIF-EPO–VEGF signaling mediates compensatory lung growth, and that both sense and antisense EPO-R transcripts up-regulate post-PNX EPO-R expression in synergy.
Sense–Antisense Transcript Interactions.
Endogenous naturally occurring antisense RNA has been detected in viruses (18), prokaryotes (19), and many eukaryotic genes (20). Sense–antisense transcript pairs can be partially (21) or completely (22) complementary in sequence and expressed in either a reciprocal or concordant pattern (23). At least 22–29% of transcripts in the human and mouse genome have complementary counterparts (20, 24). In the lung, the major asEPO-R transcript is fully complementary to that of sEPO-R in the coding region, and is transcribed off the opposite DNA strands in the same cells (Fig. 4A).
Sense–antisense transcript pairs are more conserved in evolution and are concurrently or inversely expressed in a frequency higher than that expected from chance alone (25), but concrete examples of in vivo regulation are limited. Physiologic regulation of natural antisense transcripts was described in circadian rhythm (22) and craniofacial bone development (21). HIF-1α also has naturally occurring antisense transcripts that correspond to the 3′ UTR of the sense message (26). In cultured cells, chronic hypoxia decreases sense but increases antisense HIF-1α transcript, but corresponding data in intact tissue is lacking (27), and the function of antisense HIF-1α is unknown. In addition to concurrent up-regulation of natural sEPO-R and asEPO-R transcripts during postnatal lung growth in vivo, we show that at a fixed sEPO-R transcript level, increasing asEPO-R transcript increases EPO-R protein in vitro, strongly suggesting that asEPO-R transcripts enhance EPO-R protein expression in vivo.
Antisense transcripts can influence gene expression via different mechanisms, including interfering with sense strand transcription (28), silencing by methylation (29), masking of splice sites (30), RNA-duplex-dependent RNA editing and splicing (31), prolongation of mRNA half-life (32), or alteration of translational efficiency (33). Antisense transcripts can potentially be translated into “antisense polypeptides” that regulate sense transcript or sense polypeptide by affecting protein stability, folding, targeting, and half-life. Sense–antisense peptide interactions have been shown in synthetic peptides and in model simulation (34, 35). The asEPO-R transcript harbors two long ORFs that code for two hypothetical proteins. The fact that these ORFs regulate EPO-R protein production in different ways supports the existence of multiple interacting regulatory mechanisms.
Materials and Methods
Animals and Tissue Sampling.
The canine PNX model is well established (10, 15). Litter-matched male foxhounds (2.5 months of age) underwent right PNX or thoracotomy without lung resection (sham) (n = 5 each). Three weeks later, the left upper lobe was removed via left thoracotomy. Multiple tissue samples were taken from standardized peripheral and central regions, snap-frozen in liquid nitrogen, and stored (−70 °C). Replicate assays used separate tissue samples from different regions.
RNA Isolation.
Total RNA was isolated from ≈300 mg of tissue by using acid guanidinium thiocyanate–phenol–chloroform extraction. Integrity was validated by ethidium bromide RNA gel, mRNA was purified (PolyATract; Promega), and genomic DNA was removed (DNA-free; Ambion).
Strand-Specific RT-PCR, Cloning, and Sequencing.
Primers were based on a full-length canine EPO-R cDNA we cloned (GenBank accession no. AY908987) (Table S1): RT1 and RT2 were used for first-strand cDNA from antisense transcript, and RT3 and RT4 for first-strand cDNA from sense transcript (Fig. S1). Standard PCR on sense and antisense cDNA templates used a primers from different regions (Table S1, Fig. S1), and products were cloned and sequenced. The 3′ and 5′ ends of asEPO-R cDNA were obtained by RACE (BD Biosciences Clontech). PCR fragments were assembled by Omiga ver 1.1 (Oxford Molecular Group).
RNA Blot and in Situ Hybridization.
RNA blot (15) used 32P-UTP uniformly labeled single-stranded riboprobes (primers F4 and R4) of the EPO-R coding region (Fig. 3, Table S1). PCR products (432 bp) cloned in pBluescript (Stratagene) were linearized (BamH I or KpnI), and sense and antisense cRNA were transcribed in vitro with T3 and T7 RNA polymerase, respectively (Strip-EZ RNA kit, Ambion). Hybridization was done in Quick-Hyb (Stratagene) containing 32P-riboprobe (≈108 cpm/μg cRNA, 68°C for 1 h) as described (11).
For in situ hybridization, a 432-bp cDNA of canine EPO-R (primers F4 and R4) was cloned (pGEM-T vector; Promega) and linearized (SpeI or NcoI), and cRNA was transcribed with T7-RNA or SP6 polymerase to generate digoxigenin-labeled sense and antisense RNA probes, respectively (DIG RNA Labeling Kit; Roche Applied Science). For a lung-specific control gene, a 412 bp cDNA fragment of canine SP-A was amplified (primers 5′-ACTCCACGACTTTAGACATC-3′ and 5′-GAACTCACAGATGGTCAGTC-3′), cloned, and used to generate sense and antisense SP-A cRNA probes.
Fluorescence in situ RNA hybridization was performed in OCT-embedded frozen normal canine lung. Four-micrometer sections were rehydrated (RNase-free PBS), digested (RNase-free proteinase K; Invitrogen), immersed (0.25% acetic anhydride and 0.1 M triethanolamine), and hybridized [50% deionized formamide, 0.25 mg/ml yeast tRNA (Sigma-Aldrich), 5× Denhardt's solution (Sigma-Aldrich), 5× SSC, 0.5 mg/ml salmon sperm DNA, 200 ng/ml of the sense or antisense EPO-R probe (16 h, 50 °C)]. Slides were washed (5× SSC, 2× SSC with 50% formamide, 2× SSC, 0.2× SSC, 0.1× SSC) and blocked [100 mM Tris·HCl (pH 7.5), 150 mM NaCl, 0.1% Tween-20, 1% blocking agent (Roche Applied Science), and 1 μg/ml normal mouse IgG], and bound digoxigenin-labeled cRNA probes were detected by horseradish peroxidase-conjugated anti-digoxigenin antibody (1:600, Cat. no. 6212; Abcam) with amplification by TSA-direct deposition of fluorescein (1:50; NEL 741; PerkinElmer). For EPO-R protein colocalization, slides were blocked (1.5% BSA, 10% goat serum), incubated with rabbit polyclonal anti-EPO-R antibody (8 mg/ml, M20;Santa Cruz Biotechnology), rinsed, incubated with Texas red-coupled secondary antibody (T6391;Molecular Probes), dehydrated, mounted with Vectashield (Vector Laboratories), and visualized by fluorescent microscopy.
Immunofluorescent Histochemistry and Cytochemistry.
Cryosections (4 μm) were prepared from fixed lung (4% paraformaldehyde), rinsed, embedded in OCT, and stored (−80°C). Deparaffinized sections were pretreated with 10 mM sodium citrate buffer for antigen unmasking (pH 6.0, 95 °C, 10 min), rinsed (0.1% TritonX-100 in PBS), blocked (1.5% BSA, 10% goat serum in PBS buffer), incubated with rabbit polyclonal anti-EPO-R (1 mg/ml, M20, Santa Cruz Biotechnology), rinsed, and incubated with Texas red-coupled secondary antibody (T6391; Molecular Probes). Colocalization was performed by using mouse monoclonal anti-CD34, goat polyclonal anti-AQP5, and goat polyclonal anti-SP-C (all from Santa Cruz Biotechnology). Bound antibody was detected with Alexa Fluor 488-coupled goat anti-mouse or Alexa Fluor 488-coupled donkey anti-goat secondary antibody (Molecular Probes) in a Zeiss LSM-510 laser scanning confocal microscope (excitation = 543 and 488 nm for Texas red and Alexa Fluor 488, respectively).
Constructs and Cell Transfection.
Canine cDNA was obtained from normal lung mRNA by RT-PCR (primers F1 and R5 for sEPOR; F1 and RT3 for asEPO-R) (Table S1 and Fig. S1) and cloned (sEPOR in p3xFLAG-CMV-14, Sigma-Aldrich; asEPO-R-ORF2-long in pcDNA3.1, Invitrogen). Specific sequences of asEPO-R transcript (Fig. 3) were inserted in p3xFLAG-CMV-14 plasmid (Sigma-Aldrich), and corresponding constructs with mutated start codon (ATG to ACG) were generated by site-directed mutagenesis. Transfection was performed with Lipofectamine 2000 (Invitrogen) at 75% confluence, as reported (15). Empty plasmid equalized total DNA per transfection. Cells were harvested after 48 h, rinsed, and lysed on ice with standard radioimmunoprecipitation assay (RIPA) with protease inhibitor mixture (Roche Diagnostics). Lysates were centrifuged (16,000 g × 20 min). Total protein concentration in the supernatant was measured by the Bradford method, and used for immunoblot.
Immunoblot.
Lung tissue was minced on ice, transferred to RIPA buffer, homogenized (Pro 200; PRO Scientific), sonicated (Sonifier 450; Branson Ultrasonics), and cleared by centrifugation (16,000 × g for 20min). Equal amounts of protein were separated by 7.5% SDS/PAGE reducing gel (BioRad), transferred onto Immobilon-P (Millipore), and incubated with rabbit polyclonal anti-EPO-R (1 μg/ml, M20; Santa Cruz Biotechnology) and secondary donkey anti-rabbit IgG conjugated to horseradish peroxidase (Amersham Biosciences). Labeling was visualized by chemiluminescence (ECL; Amersham Biosciences) and quantified by densitometry. Membranes were stripped and reprobed with mouse anti-FLAG (1:10,000, F3165; Sigma-Aldrich) or monoclonal mouse anti-β-actin (1 μg/ml, A1978; Sigma-Aldrich).
Supplementary Material
Acknowledgments.
We thank Dr. Alex Bobulescu for helpful discussion and the staff of the Animal Resources Center for veterinary assistance. This work was supported by National Institutes of Health Grants R01 HL040070, HL054060, HL045716, HL062873, DK048482, and DK020543; the Simmons Family Foundation; and University of Texas Southwestern O'Brien Kidney Research Core Center Grant P30 DK079328.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802467105/DCSupplemental.
References
- 1.Jelkmann W. Erythropoietin after a century of research: Younger than ever. Eur J Haematol. 2007;78:183–205. doi: 10.1111/j.1600-0609.2007.00818.x. [DOI] [PubMed] [Google Scholar]
- 2.Juul SE, Yachnis AT, Christensen RD. Tissue distribution of erythropoietin and erythropoietin receptor in the developing human fetus. Early Hum Dev. 1998;52:235–249. doi: 10.1016/s0378-3782(98)00030-9. [DOI] [PubMed] [Google Scholar]
- 3.Kertesz N, et al. The role of erythropoietin in regulating angiogenesis. Dev Biol. 2004;276:101–110. doi: 10.1016/j.ydbio.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 4.Cai Z, et al. Hearts from rodents exposed to intermittent hypoxia or erythropoietin are protected against ischemia-reperfusion injury. Circulation. 2003;108:79–85. doi: 10.1161/01.CIR.0000078635.89229.8A. [DOI] [PubMed] [Google Scholar]
- 5.Junk AK, et al. Erythropoietin administration protects retinal neurons from acute ischemia-reperfusion injury. Proc Natl Acad Sci USA. 2002;99:10659–10664. doi: 10.1073/pnas.152321399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu X, et al. The human erythropoietin receptor gene rescues erythropoiesis and developmental defects in the erythropoietin receptor null mouse. Blood. 2001;98:475–477. doi: 10.1182/blood.v98.2.475. [DOI] [PubMed] [Google Scholar]
- 7.Satoh K, et al. Important role of endogenous erythropoietin system in recruitment of endothelial progenitor cells in hypoxia-induced pulmonary hypertension in mice. Circulation. 2006;113:1442–1450. doi: 10.1161/CIRCULATIONAHA.105.583732. [DOI] [PubMed] [Google Scholar]
- 8.Chin K, et al. Regulation of transcription of the human erythropoietin receptor gene by proteins binding to GATA-1 and Sp1 motifs. Nucleic Acids Res. 1995;23:3041–3049. doi: 10.1093/nar/23.15.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster DJ, Moe OW, Hsia CCW. Upregulation of erythropoietin receptor during postnatal and postpneumonectomy lung growth. Am J Physiol. 2004;287:L1107–L1115. doi: 10.1152/ajplung.00119.2004. [DOI] [PubMed] [Google Scholar]
- 10.Takeda S, et al. Compensatory alveolar growth normalizes gas exchange function in immature dogs after pneumonectomy. J Appl Physiol. 1999;86:1301–1310. doi: 10.1152/jappl.1999.86.4.1301. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, et al. Post-pneumonectomy lung expansion elicits hypoxia inducible factor-1alpha signaling. Am J Physiol. 2007;293:L497–L504. doi: 10.1152/ajplung.00393.2006. [DOI] [PubMed] [Google Scholar]
- 12.Jaquet K, et al. Erythropoietin and VEGF exhibit equal angiogenic potential. Microvasc Res. 2002;64:326–333. doi: 10.1006/mvre.2002.2426. [DOI] [PubMed] [Google Scholar]
- 13.Cho SJ, et al. Retinoic acid and erythropoietin maintain alveolar development in mice treated with an angiogenesis inhibitor. Am J Respir Cell Mol Biol. 2005;33:622–628. doi: 10.1165/rcmb.2005-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozer EA, et al. Effects of erythropoietin on hyperoxic lung injury in neonatal rats. Pediatr Res. 2005;58:38–41. doi: 10.1203/01.PDR.0000163391.75389.52. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, et al. Regulated expression of hypoxia-inducible factors during postnatal and postpneumonectomy lung growth. Am J Physiol. 2006;290:L880–L889. doi: 10.1152/ajplung.00213.2005. [DOI] [PubMed] [Google Scholar]
- 16.Wu EY, et al. Preventing mediastinal shift after pneumonectomy does not abolish physiologic compensation. J Appl Physiol. 2000;89:182–191. doi: 10.1152/jappl.2000.89.1.182. [DOI] [PubMed] [Google Scholar]
- 17.Hsia CCW, et al. Preventing mediastinal shift after pneumonectomy impairs regenerative alveolar tissue growth. Am J Physiol. 2001;281:L1279–L1287. doi: 10.1152/ajplung.2001.281.5.L1279. [DOI] [PubMed] [Google Scholar]
- 18.Barrell BG, Air GM, Hutchison CA., III Overlapping genes in bacteriophage phiX174. Nature. 1976;264:34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- 19.Tomizawa J, et al. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci USA. 1981;78:1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 21.Blin-Wakkach C, et al. Endogenous Msx1 antisense transcript: In vivo and in vitro evidences, structure, and potential involvement in skeleton development in mammals. Proc Natl Acad Sci USA. 2001;98:7336–7341. doi: 10.1073/pnas.131497098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer C, et al. Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature. 2003;421:948–952. doi: 10.1038/nature01427. [DOI] [PubMed] [Google Scholar]
- 23.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 24.Sun M, et al. Evidence for variation in abundance of antisense transcripts between multicellular animals but no relationship between antisense transcriptionand organismic complexity. Genome Res. 2006;16:922–933. doi: 10.1101/gr.5210006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, et al. Human antisense genes have unusually short introns: Evidence for selection for rapid transcription. Trends Genet. 2005;21:203–207. doi: 10.1016/j.tig.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Rossignol F, Vache C, Clottes E. Natural antisense transcripts of hypoxia-inducible factor 1alpha are detected in different normal and tumour human tissues. Gene. 2002;299:135–140. doi: 10.1016/s0378-1119(02)01049-1. [DOI] [PubMed] [Google Scholar]
- 27.Uchida T, et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: Implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–14878. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 28.Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imamura T, et al. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem Biophys Res Commun. 2004;322:593–600. doi: 10.1016/j.bbrc.2004.07.159. [DOI] [PubMed] [Google Scholar]
- 30.Hastings ML, et al. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: Alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 1997;25:4296–430. doi: 10.1093/nar/25.21.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borsani O, et al. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CY, Shyu AB. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 33.Werner A, et al. Regulation of the NPT gene by a naturally occurring antisense transcript. Cell Biochem Biophys. 2002;36:241–252. doi: 10.1385/CBB:36:2-3:241. [DOI] [PubMed] [Google Scholar]
- 34.Blalock JE, Smith EM. Hydropathic anti-complementarity of amino acids based on the genetic code. Biochem Biophys Res Commun. 1984;121:203–207. doi: 10.1016/0006-291x(84)90707-1. [DOI] [PubMed] [Google Scholar]
- 35.Siemion IZ, Cebrat M, Kluczyk A. The problem of amino acid complementarity and antisense peptides. Curr Protein Pept Sci. 2004;5:507–527. doi: 10.2174/1389203043379413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.