Abstract
The trp gene of Drosophila encodes a subunit of a class of Ca2+-selective light-activated channels that carry the bulk of the phototransduction current. Transient receptor potential (TRP) homologs have been identified throughout animal phylogeny. In vertebrates, TRP-related channels have been suggested to mediate “store-operated Ca2+ entry,” which is important in Ca2+ homeostasis in a wide variety of cell types. However, the mechanisms of activation and regulation of the TRP channel are not known. Here, we report on the Drosophila inaF gene, which encodes a highly eye-enriched protein, INAF, that appears to be required for TRP channel function. A null mutation in this gene significantly reduces the amount of the TRP protein and, in addition, specifically affects the TRP channel function so as to nearly shut down its activity. The inaF mutation also dramatically suppresses the severe degeneration caused by a constitutively active mutation in the trp gene. Although the reduction in the amount of the TRP protein may contribute to these phenotypes, several lines of evidence support the view that inaF mutations also more directly affect the TRP channel function, suggesting that the INAF protein may have a regulatory role in the channel function.
Phototransduction in Drosophila utilizes a phospholipase C (PLC)-mediated signaling cascade (1–3) and activates two classes of light-sensitive channels (4–8), a Ca2+-selective class and a nonselective cation class. A growing body of evidence suggests that the Ca2+-selective channels are formed by the protein product of the trp gene (5, 9). Moreover, the protein products of the trp gene and the trpl gene (10), transient receptor potential (TRP) and TRP-like (TRPL), appear to account for all light-activated channel activities in Drosophila photoreceptors (4, 6), suggesting that these two proteins either constitute both classes of light-sensitive channels in Drosophila or are required for their activation.
In a wide variety of vertebrate cells, activation of PLC-mediated signaling results in the influx of Ca2+ across the plasma membrane, triggered by the depletion of Ca2+ in intracellular stores. This process, known as store-operated Ca2+ entry or capacitative Ca2+ entry (11), is found ubiquitously and is implicated in such processes as T cell activation, mitogenesis, and osteoclast function (reviewed in ref. 12). Proteins related to TRP have been identified in a variety of organisms, both vertebrates and invertebrates; in mammals, a body of evidence suggests that at least some store-operated Ca2+ channels are composed of TRP-related proteins (reviewed in refs. 13 and 14).
The mechanisms of activation and regulation of TRP channels are not known, although a recent report suggests that polyunsaturated fatty acids may play a role (15). Elucidation of these mechanisms would be greatly facilitated by the identification of other proteins important in TRP function. One possible strategy for identifying such proteins is to examine a class of mutants with electrophysiological phenotypes similar to those of trp, known as ina (inactivation but no afterpotential) (16), because in these mutants the TRP channel function is likely to be affected. Thus, for example, INAD, a PDZ domain protein required for the localization of the TRP protein to a multimolecular signaling complex (17–20), was identified from an ina-class mutant, InaD (16, 21). Here, we report on an ina-class mutant, inaF. The inaF mutants more closely resemble trp in electrophysiological phenotypes (see Fig. 1) than any other mutant identified to date. We show that the inaF gene encodes an eye-enriched protein that appears to play a critical role in TRP channel activity.
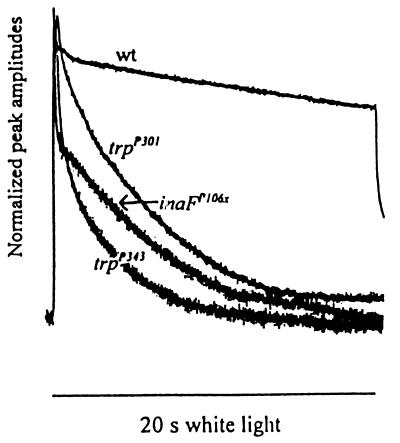
Figure 1.
Comparison of the photoreceptor potentials of inaF and trp mutants. The receptor potentials elicited from inaFP106x, trpP301, trpP343, and wild-type photoreceptors are presented normalized to their peak amplitudes. White light stimuli (20 s) were used.
Materials and Methods
Restriction Maps.
The P insertion site in inaFP105p was determined by sequencing a genomic fragment flanking the P element isolated from inaFP105p. Break points of deletions in inaFP106x and inaFP111x were identified in genomic Southern blot analyses by using fragments of an “eye-specific” clone, A23, as probes (see text; Fig. 2B).
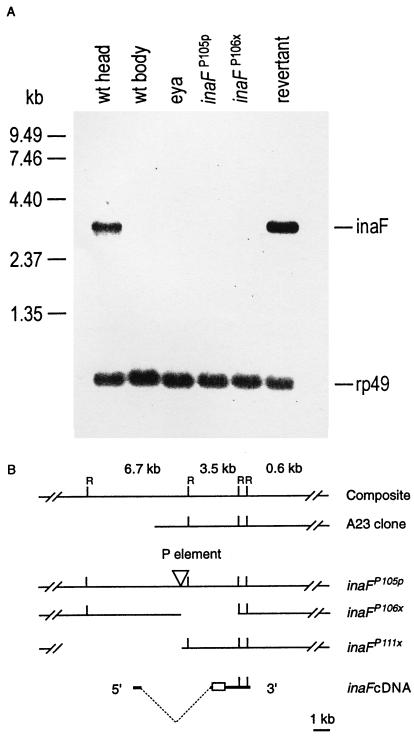
Figure 2.
(A) Northern blot analysis of inaF mutants and controls. The six lanes were loaded with poly(A)+ RNA isolated from wild-type heads, wild-type bodies, eyes absent (eya) mutant heads, inaFP105p heads, inaFP106x heads, and revertant heads (lanes 1–6, respectively). Approximately 1.5 μg of poly(A)+ RNA was loaded in each lane. The probes were prepared from a 3.5-kb EcoRI fragment of the A23 genomic clone (B). Probes prepared from the inaF cDNA gave the same results. A single 3.2-kb transcript is detected only in wild-type and revertant heads (lanes 1 and 6). A small amount (<5% of wild-type amount) of the same transcript becomes visible in eya and inaFP105p heads (lanes 3 and 4), but not in wild-type bodies or inaF106x heads (lanes 2 and 5), upon overexposure of the blot (data not shown). The uniformity of RNA loading was monitored by reprobing the blot with rp49 cDNA (Bottom). (B) Restriction maps of inaF cDNA and of the corresponding genomic region in the A23 clone and three inaF mutants. The inverted triangle in the inaFP105p map identifies the site of P element insertion. The empty spaces to the right and left of the P insertion site in the inaFP106x and inaFP111x maps, respectively, represent the deletions caused by imprecise excision of the P element. In the cDNA map, the broken dotted line indicates the extent of the intron, and the open rectangle identifies the ORF. A composite genomic map at the top shows EcoRI sites (R) and the sizes of EcoRI fragments.
Northern Blot Analysis.
Poly(A)+ RNA was isolated from heads or bodies of flies of various genotypes at 0–1 day posteclosion by using a poly(A)Ttract 1000 mRNA isolation kit (Promega). The probes were synthesized with random primers by using either the 3.5-kb EcoRI fragment of A23 clone or inaF cDNA as template.
Intracellular Recordings.
Intracellular recordings were performed as previously described (22) on 1-day-old flies. Both the recording and reference electrodes were introduced through a small cut made on the cornea and sealed with vacuum grease. The recording electrodes had 50- to 100-MΩ resistance when filled with 2 M KCl. All flies were marked with the mutation w (white) or cn bw (cinnabar brown) and were dark-adapted for 3 min before each stimulus. The light stimulus was from a 250-W tungsten halogen lamp, having unfiltered intensity of 800 μW/cm2 at the level of the fly. Signals were filtered out below 50 Hz and sampled at 2 kHz with an analog-to-digital converter (Digidata 1200A), and the data were acquired and analyzed in a computer with Axoscope (Axon Instruments, Foster City, CA).
Western Blot Analysis.
For analyses with anti-TRP, -INAD, and -PLC Abs, 12 heads were collected from 1-day-old adult flies that had been raised at 25°C in 12-hr-light/12-hr-dark illumination cycles. The heads were homogenized in 60 μl of SDS/PAGE sample buffer containing 50 mM DTT and proteinase inhibitors of recommended concentrations (Complete; Roche Molecular Biochemicals). The homogenate was boiled for 5 min and centrifuged (12,000 × g for 3 min), and 10 μl of the supernatant was loaded onto each lane of SDS/8% polyacrylamide gels. Analysis with anti-opsin (Rh1) Ab was carried out similarly with the following exceptions. Flies were raised and manipulated in complete darkness throughout, except that heads were collected in dim light; no proteinase inhibitors were used in the buffer; the homogenate was incubated at 37°C for 1 hr instead of boiling; and 12% gels were used for protein separation. To confirm uniformity of total protein loading in each lane, one of every few gels identically loaded with samples was stained with Coomassie blue. Western blots were generated by using a standard protocol. Each Ab was used at 1:3,000 dilution.
Immunofluorescence Confocal Microscopy.
Fly eyes were dissected and fixed in 4% paraformaldehyde (in PBS with 0.3% Triton X-100) for 1 hr. Nonspecific binding sites were blocked by incubation in 4% normal goat serum. The eyes were then incubated in primary Abs at 4°C overnight and in the fluorescein-conjugated goat anti-rabbit secondary Ab (Vector Laboratories) for 4 hr at 22°C. The anti-TRP Ab was used at 3% (vol/vol), and all the other Abs were used at 2% (vol/vol) in PBS containing 4% (vol/vol) normal goat serum. To optimize the quality of images, signals were obtained from optical sections of <1 μm thickness. To view TRP labeling in inaFP106x and trpP301, signals were summed from nine successive scannings of the same optical section at the same field and focal plane. The trpP301 samples were double-labeled with anti-TRP Ab and phalloidin-tetramethylrhodamine B isothiocyanate (Sigma), and the same section was viewed for both TRP and phalloidin labeling.
Deep Pseudopupil (dpp) Measurements.
The dpp (24) was examined as a function of age in flies of each genotype raised in 12-hr-light/12-hr-dark illumination cycles. A group of 40–50 flies of each genotype was examined on each day after eclosion to determine the fraction of flies that had not yet lost the dpp. The dpp was viewed in reflected light under a dissecting microscope. All flies were marked with the mutation w.
Results
The first inaF mutant, inaFP105p, was isolated in P-element-mediated mutagenesis (25). Deficiency mapping of the mutation and chromosomal in situ hybridization of a P element probe to polytene chromosomes placed the sites of both the mutation and P insertion to the same 10C2-E3 region of the X chromosome. Remobilization of the P insert in inaFP105p resulted in the isolation of >100 revertants with a wild-type phenotype, demonstrating that the P insertion is responsible for the inaFP105p phenotype. Also recovered in the remobilization crosses were many new inaF mutants, inaFP106x, inaFP107x, etc., resulting from imprecise excision events (“imprecise excision mutants”), in which some flanking DNA was excised together with the P element.
The photoreceptor potentials of inaF were studied in detail in comparison to those of strong trp mutants by intracellular recordings by using the mutants inaFP106x, trpP301, and trpP343. inaFP106x is a null inaF mutant generated by imprecise excision of the P insert. trpP301 and trpP343 are both protein-null in Western blot analysis (26); trpP343 is also functionally null (27), while trpP301 has some residual TRP activities (4, 6). Thus, the phenotypes of these null and near-null trp mutants are presumably the result of the absence or near-absence of the TRP protein.
Among the parameters of the photoreceptor potential examined were (i) the time course of decay of the potential during stimulus (Fig. 1); (ii) V–log I curves, which relate the response amplitudes with the log of stimulus intensity (data not shown); (iii) latency (data not shown); and (iv) recovery of response from a previous stimulation (see Fig. 4B, d and e). Although there were some subtle differences, the inaFP106x mutant closely resembled trpP343 and trpP301 in all these parameters. The effects of these mutations on parameter i are shown in Fig. 1, which displays the photoreceptor potentials elicited by a bright 20-s stimulus from the above three mutants and a wild-type control, normalized to peak amplitudes. As expected, the trp mutants are unable to sustain the response, and the response decays rapidly during stimulus (28, 29). The inaFP106x receptor potential also decays very rapidly and attains a steady-state value very similar to that of trpP343.
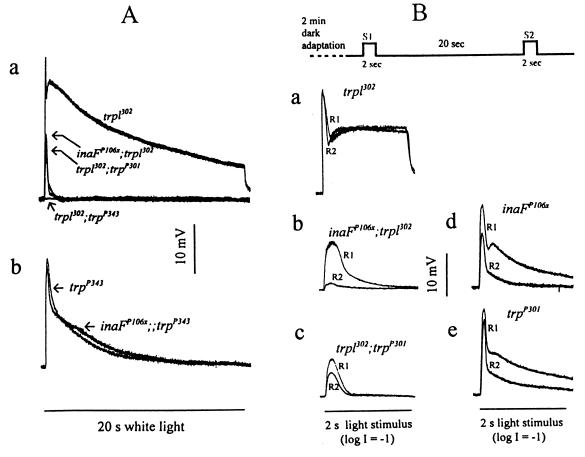
Figure 4.
Electrophysiological analysis of inaFP106x in comparison to trp mutants. (A) Effects of inaFP106x mutation on the TRP (a) and TRPL (b) channel responses. (a) The receptor potentials recorded from the double mutants inaFP106x;trpl302, trpl302;trpP301, and trpl302;trpP343 are compared with that of trpl302. The peak amplitudes of the inaFP106x;trpl302, trpl302;trpP301, and trpl302 responses are, respectively, 13.1 ± 3.3, 10.3 ± 1.9, and 27 ± 3.6 mV. (b) The receptor potential recorded from trpP343 is compared with that from the double mutant inaFP106x;;trpP343. The wild-type response is as shown in Fig. 1. White light stimuli (20 s) were used throughout. (B) Refractory properties of the residual TRP responses of the double mutants, inaFP106x;trpl302 and trpl302;trpP301. Two successive stimuli (S1 and S2) separated by a 20-s interval (top trace) are presented to trpl302 (a), inaFP106x;trpl302 (b), and trpl302;trpP301 (c) following 2-min dark adaptation, and the resulting responses (R1 and R2) for each genotype are compared by superimposing. Control responses obtained from inaFP106x and trpP301 by using the same protocol are shown in d and e. The stimuli are white light of 2-s duration filtered with a 1-log-unit neutral density filter.
The inaF gene was cloned by using a previously isolated “eye-specific” genomic clone, A23, mapping to the same region of the X chromosome as inaF. In Northern blot analysis, a 3.5-kb EcoRI fragment of the A23 clone (Fig. 2B) recognized a single 3.2-kb transcript that is reduced to a very low level in inaFP105p and absent in the imprecise excision mutants tested (Fig. 2A). The transcript is highly eye-enriched because it is detected in wild-type heads, but not in wild-type bodies or in heads of eyes absent (eya) mutants (Fig. 2A), which lack the compound eyes (30). The 3.5-kb fragment was used to isolate cDNA clones. One of the largest clones isolated (2.9 kb) was sequenced completely, and several others were sequenced partially. All cDNA clones analyzed belonged to the same class. In Northern blots, the 2.9-kb cDNA clone detected the same, single 3.2-kb eye-enriched transcript detected by the 3.5-kb genomic fragment (Fig. 2A). Rapid amplification of 5′ cDNA ends by PCR (5′RACE PCR) and end-sequencing of another cDNA clone extended the 5′ untranslated region of the 2.9-kb cDNA by 221 bp. The 2.9-kb clone ends in a short stretch of poly(A)+ tail preceded by a polyadenylation signal. The 2,905-bp sequence contained in the clone together with the 221-bp 5′ sequence obtained by the 5′ RACE thus represents a full-length cDNA. The cDNA contains a single long ORF that would encode a protein of 241 amino acids (accession no. AJ010868). The deduced protein contains no potential transmembrane segments and no obvious protein motifs and exhibits no significant homology to any known protein in the database.
To demonstrate definitively that the identified cDNA corresponds to the gene identified by inaF mutations, (i) the site of P insertion in inaFP105p was determined by sequencing the regions flanking the P insertion site, and (ii) the extent of deletions in two of the imprecise excision mutants, inaFP106x and inaFP111x, was determined by probing the genomic DNA blots of the mutants with various A23 fragments. Results showed that the P element in inaFP105p is inserted 1970 bp upstream of the proposed translation start site, within an intron that interrupts the 5′ untranslated region of the cDNA (Fig. 2B). Precise excision of this P element by remobilization fully restores the 3.2-kb eye-enriched transcript to wild-type levels (Fig. 2A). The imprecise excision mutant, inaFP106x, carries an ≈3.7-kb deletion that extends from the P insertion site in the 3′ direction to include the entire ORF and portions of both 3′ and 5′ untranslated sequences (Fig. 2B). The other excision mutant, inaFP111x, carries a deletion that extends more than 6.4 kb in the 5′ direction from the site of P insertion (Fig. 2B). If the gene corresponding to the cDNA is the inaF gene, these two deletion mutants are expected to be null mutants. Indeed, their electrophysiological phenotypes are stronger than that of inaFP105p, and inaFP106x has no detectable transcript in Northern blots, while some transcript can be detected in inaFP105p when the blots are overexposed (data not shown). Northern blot analysis was not carried out on inaFP111x because of its recessive female sterility. Thus, each of the three inaF mutants tested carries a disruption within the gene deduced from the cDNA and alters the amount of the only transcript recognized by the cDNA in an allele-dependent manner. Most importantly, remobilization of the P element reverts inaFP105p to wild type. We conclude that the cDNA we cloned does, indeed, correspond to the gene identified by inaF mutants.
Western blot analyses showed that the TRP protein is reduced to ≈15% and <10% of the wild-type level in inaFP105p and inaFP106x, respectively, at 1 day posteclosion (Fig. 3A). The reductions are not because of nonspecific reductions of retinal proteins. Other retinal proteins examined (INAD, PLCβ, and opsin) did not show similar reductions at this age (Fig. 3B), nor were there any signs of retinal degeneration in such young flies.
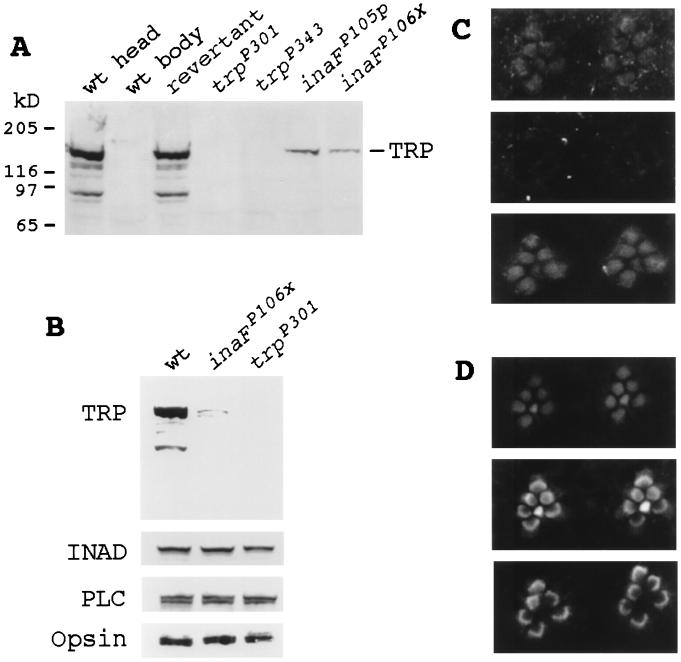
Figure 3.
Immunodetection of the TRP protein. (A) Western blot analyses of null and near-null inaF and trp mutants and wild-type and revertant controls. The seven lanes were loaded with total protein prepared from wild-type heads, wild-type bodies, revertant heads, trpP301 heads, trpP343 heads, inaFP105p heads, and inaFP106x heads (lanes 1 to 7, respectively). The blot was probed with a monoclonal anti-TRP Ab (23). (B) Western blot analysis with retinal protein controls. The three lanes were loaded with total protein prepared from wild-type heads, inaFP106x heads, and trpP301 heads, and the blot was probed with, from top to bottom, anti-TRP, anti-INAD, anti-PLCβ, and anti-opsin Abs, respectively. (C) Subcellular localization of the TRP protein by immunofluorescence confocal microscopy. (Top) Confocal micrograph of the rhabdomeres of the inaFP106x compound eye labeled with a TRP antiserum and visualized with an FITC-conjugated secondary Ab. TRP-specific staining is found only in rhabdomeres. (Middle) Confocal micrograph of trpP301 rhabdomeres, processed exactly the same as for inaFP106x (see Methods). There is no TRP-specific staining. (Bottom) Confocal micrograph of exactly the same field and focal plane of trpP301 rhabdomeres as Middle, but showing staining of filamentous actin in rhabdomeres. The preparation was double-labeled with a TRP antiserum and phalloidin (see Methods). (D) Subcellular localization of control retinal proteins in inaF by confocal microscopy. Confocal micrographs of inaFP106x retinas labeled with anti-INAD (Top), anti-PLCβ (Middle), and anti-opsin (Bottom) Abs and visualized with an FITC-conjugated secondary Ab.
However, the amount of reduction in TRP, even in null inaF mutants, does not approach that observed in trpP301 or trpP343 (Fig. 3A), and yet null inaF mutants closely resemble trpP301 and trpP343 in electrophysiological phenotypes. One possible explanation might be that most of the TRP protein detected in inaF mutants is not localized in the rhabdomeres and, therefore, is not functional. For example, the major class of Drosophila rhodopsin requires a cyclophilin, NINAA, as a chaperone for its transport to the rhabdomeres (31–33). The above hypothesis was tested by comparing the site of TRP localization in inaFP106x with that in trpP301 by immunofluorescence confocal microscopy (Fig. 3C). As expected, the amount of immunodetectable TRP protein was low in inaFP106x. However, all TRP immunostaining was found localized within the rhabdomeres (Fig. 3C, Top). There was no other site of TRP-specific immunostaining within inaFP106x photoreceptor cell bodies. By contrast, no TRP-specific immunostaining was evident in trpP301 (Fig. 3C, Middle). To be certain that the rhabdomeres are intact in trpP301, the same rhabdomeres were also stained with phalloidin to visualize filamentous actin in the rhabdomeres (Fig. 3C, Bottom). The absence of TRP-specific staining notwithstanding, the rhabdomeres were structurally intact in trpP301 (Fig. 3C, Bottom). Other retinal proteins important in phototransduction, such as INAD, PLCβ, and opsin, also localized properly to the rhabdomeres in inaFP106x (Fig. 3D), ruling out any nonspecific mislocalization of retinal proteins. We conclude that the phenotype of inaFP106x is as severe as those of trpP301, in spite of the fact that the former has considerably more TRP in the rhabdomeres than the latter.
Electrophysiological studies provided further insight into the nature of inaF mutations. We first considered the specificity of inaFP106x on TRP or TRPL channels. Its effect on TRP was examined by comparing the receptor potential recorded from trpl302, a null mutant of the trpl gene (4), with those recorded from the double mutants: inaFP106x;trpl302, trpl302;trpP301, and trpl302;trpP343 (Fig. 4A, a). Because TRP and TRPL account for all light-induced channel activities (4, 6), only the TRP channel activities would be present in the trpl302 mutant, and the responses of the double mutants would correspond to the effects of inaF or trp mutations on the isolated TRP channel activities. No response at all could be recorded from the trpl302;trpP343 double mutant. The residual responses recorded from inaFP106x;trpl302 and trpl302;trpP301 double mutants were very similar. Most of TRP channel responses have been abolished, and only small responses of very short durations remained (Fig. 4A, a). To test the effect of inaF on the TRPL channel, the receptor potentials of trpP343, which has only the TRPL channels remaining, were compared with those of the double mutant inaFP106x;;trpP343 (Fig. 4A, b). The inaFP106x mutation had little or no effect on the TRPL channel activity. Thus, the effects of inaFP106x are as severe as those of trpP301 and are specific to the TRP channel.
If the mechanism(s) responsible for the near-abolishment of the TRP activity is the same for both inaFP106x and trpP301 mutations, the residual TRP responses remaining in the two double mutants, inaFP106x;trpl302 and trpl302;trpP301, are expected to have the same properties because they are responses of approximately the same size generated by the same mechanism. These responses of the double mutants exhibit significant refractory properties. If two stimuli are presented in succession with a variable delay, the double mutants do not respond to the second stimulus (R2) at very short interstimulus intervals, but respond with an increasingly larger R2 as the interstimulus interval is increased. Under the stimulus conditions specified in Fig. 4B, the response to the second stimulus (R2) was still smaller than that to the first (R1) in both double mutants at an interstimulus interval of 20 s, whereas, in trpl302, which has a full complement of the TRP protein, R2 recovered to the size of R1 within a few seconds (shown at a 20-s interval in Fig. 4B, a). Significantly, the ratio of R2 to R1 is strikingly smaller in inaFP106x;trpl302 than in trpl302;trpP301 at this interstimulus interval (Fig. 4B, b and c), indicating that the refractory period is much longer for inaFP106x;trpl302, which has a significant amount of the TRP protein in the rhabdomeres, than trpl302;trpP301, which has no immunodetectable TRP protein. As controls, the same experiments were carried out with inaFP106x and trpP301 single mutants, i.e., without trpl302 to eliminate the TRPL channels (Fig. 4B, d and e). R2 was a significantly larger fraction of R1 in trpP301 than inaFP106x, indicating that the residual TRP response of trpP301 recovers faster than that of inaFP106x, regardless of whether the trpl302 mutation is present. As another control, Western blot analysis of the double mutants was carried out to show that the presence of trpl does not affect the amount of TRP reduction by inaF or trp (data not shown). Thus, the properties of the residual TRP responses in the two double mutants are not the same, indicating that inaF does not exert its effect simply by reducing the amount of TRP.
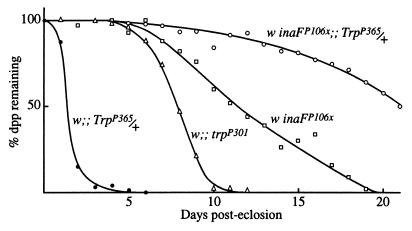
Another phenotype of inaF mutations is photoreceptor degeneration. Near-elimination of the TRP protein by the trpP301 mutation also causes photoreceptor degeneration. Fig. 5 compares the time course of degeneration in these mutants as determined from the disappearance of the dpp, the microscopically observed, superposed virtual image of a group of neighboring rhabdomere tips (24). As the photoreceptors degenerate, the superposition fails and the dpp disappears. TrpP365 is an identified trp allele that causes very rapid degeneration of photoreceptors through constitutive activity of the TRP channel (34). In TrpP365 homozygotes, no dpp is detectable from the time of eclosion, whereas in TrpP365 heterozygotes, the dpp disappears within 2–3 days after eclosion (Fig. 5). The inaFP106x mutation substantially rescues the degeneration phenotype of TrpP365 heterozygotes. In the double mutant, inaFP106x/inaFP106x;;TrpP365/+, not only is the dpp maintained longer than in TrpP365/+, but also longer than in inaFP106x or trpP301 (Fig. 5).
Figure 5.
Suppression of the degeneration phenotype of TrpP365 by inaF. The time course of disappearance of the dpp, used as a measure of photoreceptor degeneration time course, is compared among TrpP365/+, trpP301, inaFP106x, and the double mutant, inaFP106x/inaFP106x;;TrpP365/+. All flies were raised under 12-hr-light/12-hr-dark illumination conditions.
Discussion
Taken together, the above data suggest that INAF plays a critical role in TRP channel activity. In the absence of the INAF protein, the TRP channel response is nearly abolished (Fig. 4A). The pronounced reduction in the amount of the TRP protein by inaF mutations likely contributes to the inaF phenotypes. The following lines of evidence demonstrate, however, that inaF mutations have a more direct effect on TRP channel function, in addition to their indirect effect through a reduction in the TRP level. First of all, inaF null mutants, with a substantial amount of the TRP protein, have TRP responses that are nearly as severely affected as those of trpP301, which has no immunodetectable TRP protein (Figs. 1 and 4A). The argument that most of the detected TRP protein in inaFP106x mutants may not be localized in the rhabdomeres is untenable in light of the confocal microscopic evidence to the contrary (Fig. 3C). Second, the difference in the refractory periods of residual TRP responses in the double mutants, inaFP106x;trpl302 and trpl302;trpP301, cannot be explained by reductions in the TRP level. TRP responses obtained from trpl302, which has a full complement of the TRP protein, exhibit very short refractory periods. If inaF mutations exert their effects through a reduction in the amount of the TRP protein alone, one would expect the refractory period of the inaFP106x;trpl302 double mutant to be significantly shorter than that of trpl302;trpP301 because the former has considerably more TRP than the latter. Contrary to this expectation, the refractory period of the TRP response is significantly longer in inaFP106x;trpl302 than in trpl302;trpP301 (Fig. 4B, b and c). Finally, the near-rescue of the severe degeneration phenotype of TrpP365 heterozygotes by inaFP106x (Fig. 5) is difficult to explain if inaF acts solely by reducing the amount of the TRP protein. The reduction of the constitutively active TRP channels from 50% to <5% of the normal amount would have slowed down the time course of degeneration, but one would not have expected the resulting phenotype to be even more normal than those of inaFP106x or trpP301 (Fig. 5), neither of which has any constitutively active TRP channels.
Thus, inaF mutations, in addition to reducing the amount of the TRP protein, appear to affect the TRP channel function more directly. An attempt to demonstrate interaction between TRP and INAF proteins in vitro by the glutathione S-transferase–INAF fusion protein pulldown assay (35) was not successful. The reason could be that proper conditions for interaction were not met in the in vitro assay or that the target of INAF is not TRP but some other molecular component(s) involved in TRP channel-specific function because INAF need not necessarily interact with TRP to influence TRP channel function. The weight of evidence thus suggests that the INAF protein may have a modulatory role on the TRP channel function. It is possible that TRP-related channels of other species may also require corresponding INAF-related proteins for their function.
Acknowledgments
We thank John Bowman and Ann Pellegrino for technical assistance and Dr. Donald F. Ready for help with confocal microscopy. We also thank Dr. Seymour Benzer of California Institute of Technology, Dr. John A. Pollock of Carnegie Mellon University, Dr. Bih-Hwa Shieh of Vanderbilt University, Dr. Randall Shortridge of State University of New York at Buffalo, and Dr. Charles Zuker of University of California at San Diego for Abs. This work was supported by a grant from the National Eye Institute, National Institutes of Health (EY00033) (to W.L.P.). The departmental shared confocal microscope facility was supported by Grant BIR-9512962 from the National Science Foundation.
Abbreviations
- PLC
phospholipase C
- TRP
transient receptor potential
- TRPL
TRP-like
- dpp
deep pseudopupil
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ010868).
References
- 1.Selinger Z, Minke B. Cold Spring Harbor Symp Quant Biol. 1988;53:333–341. doi: 10.1101/sqb.1988.053.01.040. [DOI] [PubMed] [Google Scholar]
- 2.Bloomquist B B, Shortridge R D, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak W L. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- 3.Toyoshima S, Matsumoto N, Wang P, Inoue H, Yoshioka T, Hotta Y, Osawa T. J Biol Chem. 1990;265:14842–14848. [PubMed] [Google Scholar]
- 4.Niemeyer B A, Suzuki E, Scott K, Jalink K, Zuker C S. Cell. 1996;85:651–659. doi: 10.1016/s0092-8674(00)81232-5. [DOI] [PubMed] [Google Scholar]
- 5.Hardie R C, Minke B. Neuron. 1992;8:613–651. doi: 10.1016/0896-6273(92)90086-s. [DOI] [PubMed] [Google Scholar]
- 6.Reuss H, Mojet M H, Chyb S, Hardie R C. Neuron. 1997;19:1249–1259. doi: 10.1016/s0896-6273(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 7.Gillo B, Chorna I, Cohen H, Cook B, Manistersky I, Chorev M, Arnon A, Pollock J A, Selinger Z, Minke B. Proc Natl Acad Sci USA. 1996;93:14146–14151. doi: 10.1073/pnas.93.24.14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X-Z S, Li H-S, Guggino W B, Montell C. Cell. 1997;89:1155–1164. doi: 10.1016/s0092-8674(00)80302-5. [DOI] [PubMed] [Google Scholar]
- 9.Hardie R C. Curr Biol. 1996;6:1371–1373. doi: 10.1016/s0960-9822(96)00733-6. [DOI] [PubMed] [Google Scholar]
- 10.Phillips A M, Bull A, Kelly L E. Neuron. 1992;8:631–642. doi: 10.1016/0896-6273(92)90085-r. [DOI] [PubMed] [Google Scholar]
- 11.Putney J W J. Cell Calcium. 1990;11:611–624. doi: 10.1016/0143-4160(90)90016-n. [DOI] [PubMed] [Google Scholar]
- 12.Berridge M J. Biochem J. 1995;312:1–11. doi: 10.1042/bj3120001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birnbaumer L, Zhu X, Jiang M, Boulay G, Peyton M, Vannier B, Brown D, Platano D, Sadeghi H, Stefani E, Birnbaumer M. Proc Natl Acad Sci USA. 1996;93:15195–15202. doi: 10.1073/pnas.93.26.15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montell C. Mol Pharmacol. 1997;52:755–763. doi: 10.1124/mol.52.5.755. [DOI] [PubMed] [Google Scholar]
- 15.Chyb S, Raghu P, Hardie R C. Nature (London) 1999;397:255–259. doi: 10.1038/16703. [DOI] [PubMed] [Google Scholar]
- 16.Pak W L. In: Neurogenetics: Genetic Approaches to the Nervous System. Breakfield X, editor. New York: Elsevier/North–Holland; 1979. pp. 67–99. [Google Scholar]
- 17.Chevesich J, Kreuz A J, Montell C. Neuron. 1997;18:1069–1072. doi: 10.1016/s0896-6273(01)80049-0. [DOI] [PubMed] [Google Scholar]
- 18.Huber A, Sander P, Gobert A, Bahner M, Hermann R, Paulsen R. EMBO J. 1996;15:7036–7045. [PMC free article] [PubMed] [Google Scholar]
- 19.Shieh B-H, Zhu M-Y. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 20.Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker C S. Nature (London) 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 21.Shieh B-H, Niemeyer B. Neuron. 1995;14:201–210. doi: 10.1016/0896-6273(95)90255-4. [DOI] [PubMed] [Google Scholar]
- 22.Johnson E C, Pak W L. J Gen Physiol. 1986;88:651–673. doi: 10.1085/jgp.88.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollock J A, Assaf A, Peretz A, Nichols C D, Mojet M H, Hardie R C, Minke B. J Neurosci. 1995;15:3747–3760. doi: 10.1523/JNEUROSCI.15-05-03747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franceschini N. In: Information Processing in the Visual System of Arthropods. Wehner R, editor. New York: Springer; 1972. pp. 75–82. [Google Scholar]
- 25.Spradling A, Stern D, Kiss I, Roote J, Laverty T, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montell C, Rubin G M. Neuron. 1989;2:1313–1323. doi: 10.1016/0896-6273(89)90069-x. [DOI] [PubMed] [Google Scholar]
- 27.Scott K, Sun Y, Beckingham K, Zuker C S. Cell. 1997;91:375–383. doi: 10.1016/s0092-8674(00)80421-3. [DOI] [PubMed] [Google Scholar]
- 28.Cosens D J, Manning A. Nature (London) 1969;224:285–287. doi: 10.1038/224285a0. [DOI] [PubMed] [Google Scholar]
- 29.Minke B, Wu C-F, Pak W L. Nature (London) 1975;258:84–87. doi: 10.1038/258084a0. [DOI] [PubMed] [Google Scholar]
- 30.Sved J. Drosophila Inf Serv. 1986;63:169. [Google Scholar]
- 31.Schneuwly S, Shortridge R D, Larrivee D C, Ono T, Ozaki M, Pak W L. Proc Natl Acad Sci USA. 1989;86:5390–5394. doi: 10.1073/pnas.86.14.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shieh B-H, Stamnes M A, Seavello S, Harris G L, Zuker C S. Nature (London) 1989;338:67–70. doi: 10.1038/338067a0. [DOI] [PubMed] [Google Scholar]
- 33.Colley N J, Baker E K, Stamnes M A, Zuker C S. Cell. 1991;67:255–263. doi: 10.1016/0092-8674(91)90177-z. [DOI] [PubMed] [Google Scholar]
- 34.Yoon, J., Cohen-BenAmi, H., Hong, Y. S., Park, S., Strong, L. L. R., Bowman, J., Baek, K., Geng, C., Minke, B. & Pak, W. L. (1999) J. Neurosci., in press. [DOI] [PMC free article] [PubMed]
- 35.Brent R. Analysis of Protein Interactions. New York: Wiley; 1997. [Google Scholar]