Abstract
Glutaredoxins represent a ubiquitous family of proteins that catalyze the reduction of disulfide bonds in their substrate proteins by use of reduced glutathione. In an attempt to identify the full complement of glutaredoxins in baker's yeast, we found three so-far uncharacterized glutaredoxin-like proteins that we named Grx6, Grx7, and Grx8. Grx6 and Grx7 represent closely related monothiol glutaredoxins that are synthesized with N-terminal signal sequences. Both proteins are located in the cis-Golgi, thereby representing the first glutaredoxins found in a compartment of the secretory pathway. In contrast to formerly described monothiol glutaredoxins, Grx6 and Grx7, showed a high glutaredoxin activity in vitro. Grx6 and Grx7 overlap in their activity and deletion mutants lacking both proteins show growth defects and a strongly increased sensitivity toward oxidizing agents such as hydrogen peroxide or diamide. Our observations suggest that Grx6 and Grx7 do not play a general role in the oxidative folding of proteins in the early secretory pathway but rather counteract the oxidation of specific thiol groups in substrate proteins.
INTRODUCTION
The thiol groups of cysteine residues are of outstanding physiological relevance for many biological reactions as they are critical for the coordination of metal ions like zinc, copper, or iron. Furthermore, they are required for the binding of iron sulfur clusters and heme cofactors, the covalent attachment of prenyl anchors, and various types of cysteine modifications. Moreover, their unique ability to form intra- or intermolecular disulfide bridges by oxidation is of considerable significance both for protein folding processes and for the dynamic regulation of protein structures in response to cellular conditions (for review see Paget and Buttner, 2003; Barford, 2004; Lill and Mühlenhoff, 2006). Because these functions of cysteine residues strictly depend on their redox state, a plethora of cellular components is dedicated to the precise control of reduced and oxidized states of thiol groups.
In the cytosol, the nucleus and the matrix of mitochondria, most cysteine residues are maintained in a reduced state. This is achieved by high concentrations of reduced glutathione that were measured to be around 13 mM in the yeast cytosol (Ostergaard et al., 2004). The strong reducing capacity of glutathione is conferred to the bulk of cytosolic proteins by a group of enzymes referred to as glutaredoxins (Grant, 2001; Holmgren et al., 2005). Glutaredoxins are characterized by conserved thioredoxin-like structures with redox-active Cys-x-x-Cys (in the case of dithiol glutaredoxins) or Cys-x-x-Ser motifs (in the case of monothiol glutaredoxins) in the active center. Glutaredoxins represent a ubiquitous protein family with orthologues in the cytosol, in mitochondria and in the nucleus, whereas no glutaredoxins were found in compartments of the secretory pathway up to date.
In Saccharomyces cerevisiae five glutaredoxins have been described so far, referred to as Grx1 through Grx5 (see Figure 1; Grant, 2001; Herrero and de la Torre-Ruiz, 2007; Lillig and Holmgren, 2007). Grx1 and Grx2 are dithiol glutaredoxins of the cytosol, which counteract oxidative stress especially at higher temperatures. Both proteins share an identity of 64% but appear to interact with distinct populations of substrates so that deletion of each gene shows a characteristic phenotype (Luikenhuis et al., 1998). The Grx2 protein is expressed in two forms by the use of two alternative start codons (Pedrajas et al., 2002; Porras et al., 2006). The shorter form remains in the cytosol and the longer form is translocated into the matrix of mitochondria. The function of monothiol glutaredoxins is less clear because so far no glutaredoxin activity could be measured in vitro for these enzymes, which might point to a high substrate specificity of these components. The monothiol glutaredoxins Grx3 and Grx4 both have an additional N-terminal thioredoxin domain and were found to be predominantly located in the nucleus, where they appear to regulate transcription of a small set of substrate proteins (Lopreiato et al., 2004; Ojeda et al., 2006; Pujol-Carrion et al., 2006). Grx5 is targeted to the mitochondrial matrix, where it plays an ill-defined role in the synthesis or assembly of iron-sulfur clusters (Rodriguez-Manzaneque et al., 2002; Lill and Mühlenhoff, 2006).
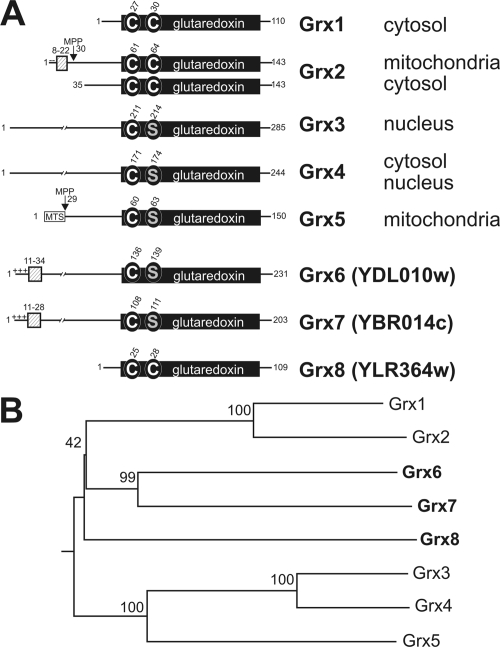
Figure 1.
(A) Schematic representation of glutaredoxin homologues of S. cerevisiae. The conserved glutaredoxin domain is depicted as black box, in which the conserved cysteine and serine residues are indicated. The numbers refer to positions of amino acid residues in the sequences. Hydrophobic sequences are indicated by hatched boxes and charged residues N-terminal to these sequences are indicated by minus and plus signs. MTS indicates the mitochondrial-targeting sequence of Grx5, the position at which proteins are cleaved by the mitochondrial-processing peptidase (MPP) is shown. The intracellular locations of the proteins are depicted on the right. (B) Phylogenetic analysis of the yeast glutaredoxin sequences. Bootstrap numbers are indicated. The tree was calculated using the DNAman software package (Lynnon, Quebec, Canada).
In contrast to the situation in the cytosol, thiol groups are typically converted to disulfide bonds in the lumen of the endoplasmic reticulum (ER). The oxidation system of the ER was studied to quite some detail: it is comprised of the soluble protein disulfide isomerase (Pdi1) and the membrane-associated sulfhydryl oxidase Ero1, which carry out the electron transfer between their substrate proteins and oxygen in a concerted and sequential manner (for review see (Sevier and Kaiser, 2002; Sitia and Braakman, 2003; Ellgaard, 2004; Tu and Weissman, 2004; Wilkinson and Gilbert, 2004). This system is very efficient and might oxidize most, if not all, cysteine residues on the polypeptides during or directly after their translocation into the ER lumen. The redox conditions in the other compartments of the secretory pathway, as in the Golgi apparatus, in endosomes or in lysosomes, are not known nor were any redox-active factors identified thus far in these compartments. The ratio of reduced to oxidized glutathione in microsomes is about 3:1 (Hwang et al., 1992; Bass et al., 2004), making the reduction of certain initially oxidized proteins possible, but there is no evidence that secreted proteins in general would not remain oxidized.
In this study, we describe two novel closely related glutaredoxins that we named Grx6 (YDL010w) and Grx7 (YBR014c). On the basis of their Cys-x-x-Ser motif in the active center, they belong to the monothiol glutaredoxins, although they differ considerably from so-far characterized representatives of this group, both by their primary sequence and by their much higher glutaredoxin activity in vitro. Interestingly, both proteins are located in the cis-Golgi, as indicated by immunofluorescence and cellular fractionation. Thus, they represent the first redox components of the secretory pathway downstream of the ER. We show Grx6 and Grx7 are critical for the resistance of cells to oxidative stress and discuss potential physiological roles of these proteins.
MATERIALS AND METHODS
Yeast Strains and Growth Media
All strains used in this study were isogenic to the wild-type strains W303–1A (Mat a ade2 his3 leu2 trp1 ura3; Myers et al., 1985) and YPH499 (Mat a ura3 lys2 ade2 trp1 his3 leu2; Sikorski and Hieter, 1989). For generation of deletion mutants of GRX6 (YDL010w), GRX7 (YBR014c), or both, the entire coding regions of the genes were replaced in YPH499 cells by HIS3 and kanamycin resistance cassettes, respectively. For generation of the GRX6-HA and GRX7-HA variants, the sequence encoding three hemagglutinin (HA) epitopes and a TRP1 marker gene was inserted downstream of the chromosomal GRX6 and GRX7 genes by use of the pYM22 template (PCR toolbox; Janke et al., 2004). This insertion resulted in the expression of the entire Grx6 and Grx7 proteins carrying three HA epitopes at their C terminus under control of their endogenous promoters. For overexpression of Ero1 and Pdi1, both genes were amplified by PCR and subcloned into NcoI and SacI or EcoRI and SacI sites of pYX132 (Novagen, Nottingham, United Kingdom), respectively. The resulting plasmids were transformed into the GRX6 GRX7 double-deletion strain. Yeast cultures were grown at 30°C in lactate medium, YP (1% yeast extract, 2% peptone) medium supplemented with 2% glucose (YPD), or 2% galactose, or in minimal medium supplemented with 20 μg/ml adenine, histidine, and tryptophan, and 30 μg/ml leucine and lysine (Sherman et al., 1986; Herrmann et al., 1994).
Expression of Grx6 and Grx7 in Escherichia coli
GRX6 and GRX7 were amplified from base pair 100 and the stop codon by PCR, digested with BamHI and SalI, and cloned into the E. coli expression vector pQE30 (Qiagen, Chatsworth, CA). A cysteine-to-serine mutant of Grx6 (Grx6S) was generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the supplier's instructions. N-terminally His-tagged variants of Grx6, Grx6S, and Grx7 all beginning with Val34 were expressed in E. coli (XL1-Blue). Freshly transformed bacteria were grown at 37°C to an OD600 of 0.5 in LB medium containing 100 μg/ml ampicillin. Expression was induced with 0.5 mM isopropyl-thiogalactopyranoside for 4 h. Cells from 100 ml culture were harvested by centrifugation and resuspended in 20 ml buffer containing 50 mM sodium phosphate, 300 mM NaCl, and 10 mM imidazole, pH 8.0. Cell walls were digested with lysozyme followed by sonication on ice. The suspension was clarified by centrifugation at 4°C (30 min, 10,000 × g) and loaded on a column containing 0.5 ml Ni-NTA. The recombinant protein was eluted with the same buffer containing 250 mM imidazole. PfGrx was purified as described (Rahlfs et al., 2001).
Translocation Assays
GRX6 and GRX7 genes were subcloned into pGEM4 expression plasmids (Promega, Madison, WI) and used for in vitro transcription/translation in the presence of [35S]methionine in reticulocyte lysate according to the protocol of the manufacturer (Promega, San Luis Obispo, CA). A microsome-containing membrane fraction was essentially purified as described (Wuestehube and Schekman, 1992) with the exception that zymolyase was used for preparation of spheroplasts and that the sucrose step gradient was omitted, resulting in a fraction containing both microsomes and mitochondria. Typically, 100 μg of this microsome-containing fraction was used per reaction in 0.6 M sorbitol, 0.1 mg/ml BSA, 2 mM potassium phosphate, 50 mM HEPES-KOH, pH 7.4, 2 mM ATP, 2 mM NADH, and 10 μM valinomycin, if not indicated otherwise. Import was stopped by a 10-fold dilution in ice-cold 0.6 M sorbitol, and 20 mM HEPES-KOH, pH 7.4, with 50 μg/ml proteinase K. Signals of radiolabeled proteins were detected by autoradiography on Biomax MR-1 films (Eastman Kodak, Rochester, NY).
Halo Assays
Wild-type or mutant cells were grown in liquid culture to midlog phase. Equal amounts of cells were spread onto YP plates containing 2% glucose as carbon source. Filter discs soaked with 10 μl of 9.8 M hydrogen peroxide or 500 mM diamide were placed onto the cell loan. Plates were incubated at 30°C for 2 d.
HEDS Assay
Steady-state kinetics of PfGrx, Grx6, Grx7, and Grx6S were monitored at 340-nm wavelength with a Jasco V-550 UV/Vis double-beam spectrophotometer essentially as described (Holmgren and Aslund, 1995). The consumption of NADPH in a coupled enzyme reaction was measured. Reactions were carried out in 0.1 M Tris/HCl, and 1 mM EDTA, pH 8.0, and contained 0.1 mM NADPH, 0.25 U/ml glutathione reductase, and 1 mM glutathione. The reaction was started with addition of hydroxyethyl disulfide (HEDS) to a final concentration of 736 μM. Measured activities in all assays were corrected by subtracting the absorbance of a reference cuvette containing all components excluding glutaredoxins and by subtracting the slope (ΔAbs/min) of the baseline.
Ribonuclease Refolding Assay
Renaturation of scrambled RNase A from bovine pancreas (Frickel et al., 2004) was measured spectrophotometrically by monitoring hydrolysis of its substrate cCMP at 296 nm. The samples contained 100 mM Tris, pH 8.0, 4.5 mM cCMP, 1 mM reduced glutathione, 0.2 mM oxidized glutathione, 1 mM EDTA, 25 μM scrambled RNase, 1 μM Pdi1, and 18 μM Grx7, if not indicated otherwise. The assay was carried out at 25°C.
Immunofluorescence
Cells were grown to early log phase in YP supplemented with 2% glucose and were analyzed by immunofluorescence as described (Chuang and Schekman, 1996). After fixation and conversion into spheroplasts, the cells were stained with Grx6-, Grx7-, HA- (Sigma) and green fluorescent protein (GFP; Torrey Pines)-specific antibodies. Cy3- and FITC-coupled secondary antibodies were used for visualization (Jackson ImmunoResearch Laboratories, West Grove, PA).
Subcellular Fractionation
Cells were grown to midlog phase, converted to spheroplasts, and resuspended in 3.5 ml 10 mM HEPES, pH 7.5, 12.5% sucrose, 1 mM EDTA, and 1 mM PMSF. The spheroplasts were disrupted by 10 strokes with a Dounce homogenizer. The cell debris was removed by spinning twice 5 min at 450 × g. The supernatant was loaded on a sucrose gradient consisting of 1 ml of each 22, 26, 30, 34, 38, 42, 50, 54, and 60% sucrose (wt/vol) in 10 mM HEPES, pH 7.5, and 1 mM MgCl2. The samples were spun 2.5 h at 4°C and 37,000 rpm in a TST41.14 rotor. Fractions (800 μl) were colleted from the top, and 5 μl/fraction was analyzed by Western blotting.
Northern Blot Analysis
Yeast RNA, 15–25 μg, was separated on an 1.2% agarose/formaldehyde gel. The RNA was transferred over night onto Hybond N+ membrane (GE Healthcare, Waukesha, WI) by capillary blot. The RNA was cross-linked to the membrane and hybridized with an HAC1 probe. To generate the probe an ∼1000-base pair HAC1 PCR product was labeled with alkaline phosphatase using the AlkPhos Direct labeling and detection kit (GE Healthcare). The Northern blots were developed according to the manufacturer's protocol.
RESULTS
Grx6 and Grx7, Two Novel Glutaredoxin-like Proteins
Recent studies indicated that a certain balance of oxidizing and reducing processes is critical for cellular compartments in which no thioredoxin- or glutaredoxin-like enzymes were identified so far, namely in the intermembrane space of mitochondria and in the secretory pathway (Mesecke et al., 2005; Sevier et al., 2007). To identify the full set of glutaredoxins present in S. cerevisiae, BLAST searches were performed of the yeast genome. These revealed the presence of three uncharacterized potential gene products with glutaredoxin-like domains, YDL010w, YBR014c, and YLR364w (Figure 1A), which we named Grx6, Grx7, and Grx8, respectively. Grx8 represents a presumably cytosolic dithiol glutaredoxin of 109-amino acid residues. It was identified as a gene strongly induced by the exposure to arsenic which causes glutathione-depletion in yeast (Haugen et al., 2004), consistent with a role of Grx8 in the protection against oxidative stress. Grx6 and Grx7 comprise 231 and 203 amino acid residues, respectively, and show significant sequence identity to each other over their entire sequence (45%), but lower identity to other glutaredoxins (between 14 and 21%). Both are predicted monothiol glutaredoxins although a phylogenetic analysis suggested a relationship to the yeast dithiol glutaredoxins Grx1 and Grx2 (Figure 1B). Interestingly, the N termini of Grx6 and Grx7 similarly show a short sequence with three positively charged residues followed by a hydrophobic domain of 13 and 19 residues, respectively. Prediction programs for subcellular localization like TargetP (Emanuelsson et al., 2007) indicate for Grx6 and Grx7 the presence of signal peptides with high scores (0.938 and 0.875) and of mitochondrial targeting sequences with lower scores (0.083 and 0.12).
Grx6 and Grx7 Localize to the cis-Golgi
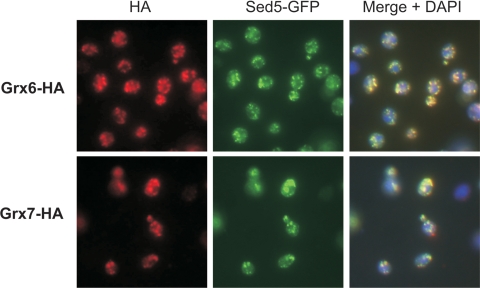
To analyze the subcellular localization of Grx6 and Grx7 experimentally, we constructed yeast strains in which either Grx6 or Grx7 were expressed with C-terminal HA epitopes by chromosomal integration of PCR-amplified cassettes. Immunofluorescence revealed in both cases a similar pattern of punctate staining throughout the cell (Figure 2). This staining did not match the pattern expected for a mitochondrial or perinuclear/ER distribution but rather was similar to a staining of Golgi or endosomes. To identify the Grx6/Grx7-containing compartment, we performed double-labeling experiments by using markers for the cis- and the trans-Golgi, Sed5-GFP and Sec7-GFP, respectively. As shown in Figure 2, both Grx6-HA and Grx7-HA colocalized very well with Sed5-GFP, a marker of the cis-Golgi (Banfield et al., 1994). No colocalization of Grx6 or Grx7 with Sec7-GFP was observed (our unpublished observations).
Figure 2.
Grx6 and Grx7 localize to the cis-Golgi by immunofluorescence. A GFP-tagged version of cis-Golgi marker Sed5 was expressed in Grx6-HA and Grx7-HA cells. The cells were grown in YPD and processed for immunofluorescence using mouse anti-HA and rabbit anti-GFP antibodies. The anti-mouse antibody was coupled to CY3 and the anti-rabbit to FITC. Overlay images of the glutaredoxin and the GFP signals are shown on the right. The DNA was visualized with DAPI.
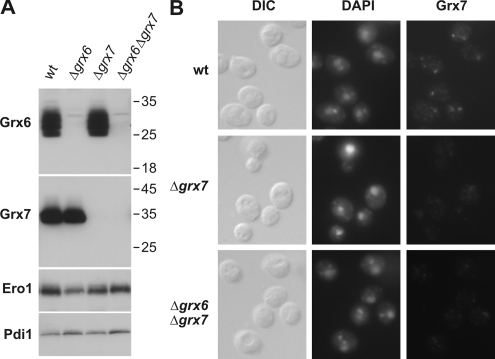
To exclude a mislocalization of the tagged glutaredoxins, we generated antibodies against Grx6 and Grx7, which reacted in a specific manner with both proteins (Figure 3A). Immunofluorescence with the Grx7-specific antibodies revealed a punctate staining in the immunofluorescence (Figure 3B), consistent with a localization of the protein in the Golgi.
Figure 3.
Immunofluorescence with Grx7-specific antibodies reveals a punctate staining consistent with the localization in the Golgi apparatus. (A) Protein extracts (100 μg) of the strains indicated were analyzed by Western blotting using antibodies against Grx6, Grx7, Ero1, and Pdi1. Positions of molecular-weight markers are indicated. (B) The strains indicated were processed for immunofluorescence using Grx7-specific antibodies. The DNA was visualized with DAPI.
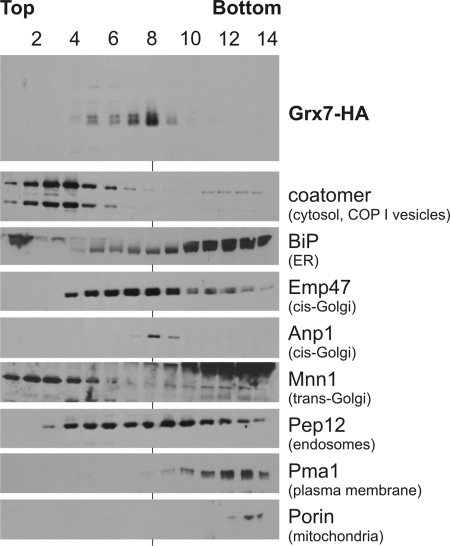
As a second independent approach to asses the intracellular localization of the glutaredoxin proteins, we fractionated yeast cells by sucrose gradient centrifugation. Grx7-HA comigrated with the cis-Golgi marker Anp1 on these gradients confirming its localization in the cis-Golgi (Figure 4). Cofractionation with mitochondria or other compartments was not observed.
Figure 4.
Grx7 fractionates with proteins of the cis-Golgi on sucrose gradients. Cells expressing Grx7-HA were converted into spheroplasts. The spheroplasts were lysed, and the lysate was separated on a sucrose step gradient. Fourteen fractions were collected from the top and analyzed by Western blotting using different marker proteins and Grx7-HA. Grx7-HA cofractionated with the cis-Golgi markers Emp47 and Anp1. Grx7-HA was absent from fractions containing ER (BiP), medium/late Golgi (Mnn1), mitochondria (porin), or plasma membrane (Pma1).
Grx6 and Grx7 Are Orientated toward the Lumen of the Golgi
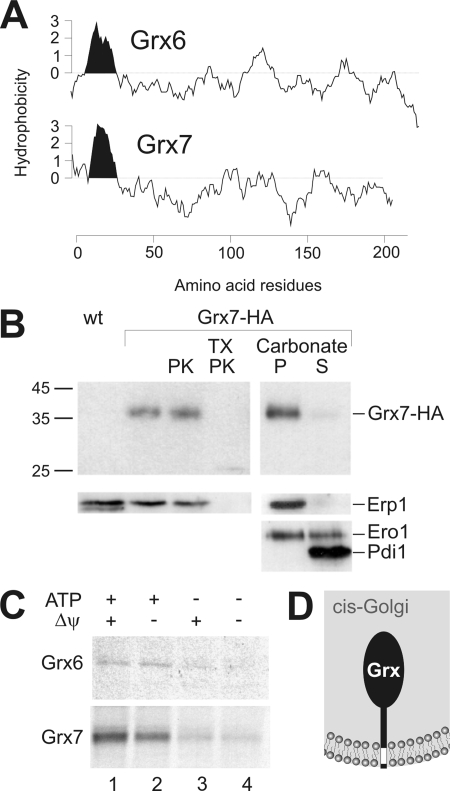
Topology prediction suggested the existence of transmembrane segments close to the N termini of Grx6 and Grx7 (Figure 5A). To assess their orientation in the membrane, we isolated a Golgi-containing microsome fraction from Grx7-HA cells. Treatment of these organelles with proteinase K did not lead to the degradation of Grx7 (Figure 5B). This was not due to an intrinsic protease resistance of the protein because Grx7 was efficiently degraded by added proteases when membranes were solubilized with detergents. From this we conclude that Grx7 is exposed to the Golgi lumen. Fractionation of the samples with carbonate indicated a tight association of Grx7 with membranes. Under the conditions of the experiments, integral membrane proteins like Erp1 (Marzioch et al., 1999) are present in the pellet, whereas membrane-associated proteins such as Ero1 distribute between the supernatant and the membrane pellet.
Figure 5.
Grx7 is a membrane-anchored protein exposing its soluble domain to the Golgi lumen. (A) Hydrophobicity profiles of Grx6 and Grx7 were generated using the DNAman software. N-terminal hydrophobic regions of the proteins are indicated in black. (B) Crude microsome fractions of a wild-type (wt) and Grx7-HA cells were isolated. 100 μg of the Grx7-HA fraction were incubated in the presence or absence of 100 μg/ml proteinase K (PK) or 0.2% Triton X-100 (TX) for 30 min on ice. For the extraction of membrane proteins, 100 μg of membranes were incubated with unbuffered sodium carbonate on ice. Membrane-containing (P) and soluble fractions (S) were separated by centrifugation. Proteins were precipitated with trichloroacetic acid and applied to SDS-PAGE and Western blotting. Controls show the integral membrane protein Erp1, the membrane-associated protein Ero1, and the soluble protein Pdi1. (C) Radiolabeled Grx6 or Grx7 were incubated with a crude microsome fraction for 20 min at 30°C. During this reaction, 2 mM ATP (lanes 1 and 2), 2 mM NADH (lanes 1 and 3), or 10 μM valinomycin (lanes 2 and 4) were present. To remove nonimported material after the translocation reaction, the samples were treated with proteinase K. Microsomes were reisolated and subjected to SDS-PAGE and autoradiography. (D) Scheme of the Grx7 topology concluded from our results.
The localization of Grx6 and Grx7 in the lumen of a compartment of the secretory pathway is further supported by in vitro translocation experiments (Figure 5C). Incubation of Grx6 and Grx7 with crude microsome fractions, which also contain mitochondria, led to translocation of both proteins into a protease-protected location. Efficient translocation depended on the presence of ATP but was not sensitive to valinomycin, an uncoupler of the mitochondrial membrane potential (Figure 5C). Moreover, Grx7 could not be imported into a microsome-free mitochondrial fraction (Supplementary Data, Figure S1).
In summary, we conclude that Grx6 and Grx7 are luminal proteins of the cis-Golgi that are tightly anchored into the Golgi membrane, presumably by their N-terminal transmembrane domains (Figure 5D).
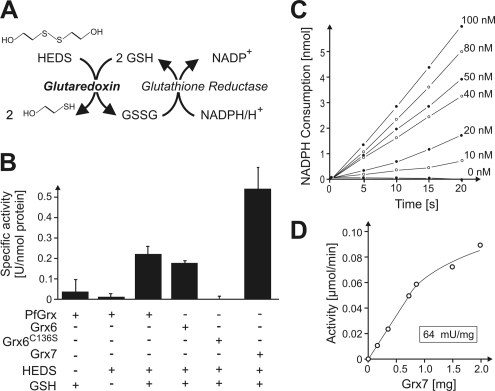
Grx6 and Grx7 Show Glutaredoxin Activity In Vitro
The sequences of Grx6 and Grx7 show characteristic monothiol glutaredoxin domains (Herrero and de la Torre-Ruiz, 2007). Unlike for dithiol glutaredoxins, glutaredoxin activity could not be experimentally attributed to any of the so-far characterized monothiol glutaredoxins by using standard assays (Tamarit et al., 2003; Deponte et al., 2005), and their molecular activity therefore remains unclear. To test whether Grx6 and Grx7 exhibit glutaredoxin activity in the standard HEDS assay (Holmgren and Aslund, 1995), we expressed the proteins recombinantly in bacteria and purified them by affinity chromatography. The HEDS assay couples glutathione-dependent glutaredoxin activity to the glutathione reductase–mediated consumption of NADPH, which can be easily monitored in a photospectrometer (Figure 6A). The method was successfully applied to analyze activity of the dithiol glutaredoxin PfGrx of Plasmodium falciparum (Rahlfs et al., 2001), which we used for control. As shown in Figure 6B, purified PfGrx showed a high glutaredoxin activity of 0.21 U/nmol enzyme. The activity of Grx6 (0.18 U/nmol) was similar to that of PfGrx, whereas Grx7 was ∼2.5 times more active (0.51 U/nmol). As control, we expressed a variant of Grx6 in which the single conserved cysteine residue was exchanged for a serine residue (Grx6C136S mutant). This mutant protein did not show any activity in the assay (Figure 6B). The measured activities correlated well with the amounts of added glutaredoxin protein (see Figure 6, C and D, for Grx7). From this result, we conclude that Grx6 and Grx7 both exhibit significant glutaredoxin activity and catalyze the reduction of oxidized substrates by use of reduced glutathione.
Figure 6.
Grx6 and Grx7 show glutaredoxin activity in vitro. (A) Scheme of the glutaredoxin activity assay using HEDS as electron acceptor (Holmgren and Aslund, 1995). (B) Recombinant PfGrx, Grx6, Grx6C136S, and Grx7 were expressed and purified to homogeneity from E. coli cells. Their activity was monitored at 340 nm by oxidation of NADPH in a coupled enzyme assay. Glutaredoxin proteins were present in the reactions at a final concentration of 52 nM. For control, reactions with PfGrx were carried out in the absence of glutathione (GSH) or HEDS (left columns). All reactions were carried out in a two-beam photometer in which a glutaredoxin-free sample served as reference. (C) The NADPH consumption in the HEDS assay over time was analyzed for different concentrations of purified Grx7. (D) The activity of Grx7 in the experiment shown in C was plotted against the Grx7 concentration showing a linear correlation up to 1 mg Grx7 in the assay.
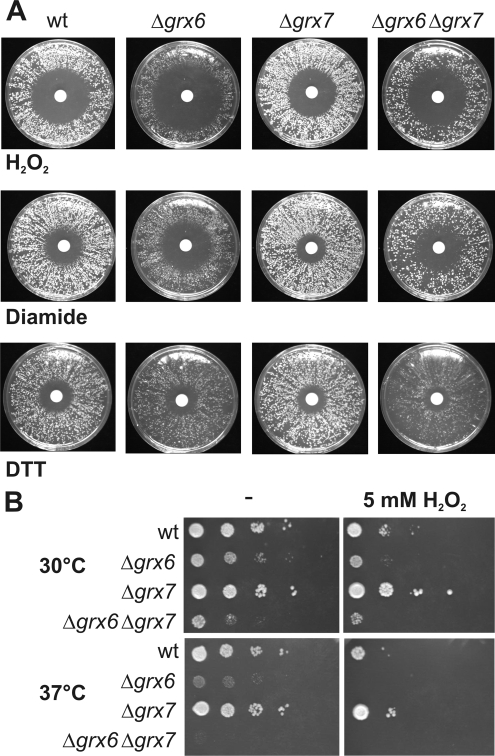
Deletion of Grx6 and Grx7 Lead to Synthetic Growth Defects
Glutaredoxins play a critical role in the redox regulation of the cell. Therefore we tested the growth phenotype of cells lacking Grx6, Grx7, or both using filter disk assays. The Δgrx6 deletion mutant showed a significantly increased sensitivity toward hydrogen peroxide or diamide (Figure 7, A and B). In contrast, the Δgrx7 mutant was hardly affected and grew as well as wild-type cells. A more important role of Grx6 is also reflected by its higher abundance in yeast cells: Quantitative Western blotting showed that Grx6 and Grx7 are both expressed and present at concentrations of 40 and 3 μg/100 mg cellular protein, respectively (our unpublished observations).
Figure 7.
The deletion of Grx6 and Grx7 leads to increased sensitivity toward oxidizing reagents. (A) The indicated strains were grown in liquid medium over night. Equal amounts of cells were plated on rich media plates containing 2% glucose (YPD). Filter disks were placed in the middle of each plate soaked with 10 μl of either 500 mM diamide, 9.8 M hydrogen peroxide, or 3 M DTT. Plates were incubated at 30°C for 2 d. (B) The indicated strains were grown to midlog phase. Serial dilutions of the culture were dropped on YPD plates lacking or containing 5 mM hydrogen peroxide and incubated for 3 d at 30°C.
The Δgrx6 and Δgrx7 mutants were not hypersensitive to dithiothreitol (DTT) but rather slightly less sensitive than the wild type. Moreover, the loss of Grx6 caused a temperature-dependent growth defect that was even more pronounced in double mutants lacking both Grx6 and Grx7 (Figure 7B). These results indicate a cooperative function of both glutaredoxins that might counteract the deleterious effects of oxidative agents in the secretory pathway.
Loss of Grx6 and Grx7 Does Not Induce the Unfolded Protein Response Pathway
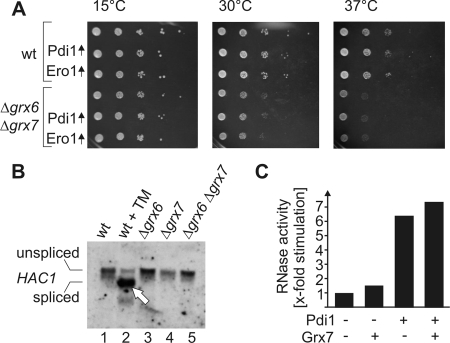
Because the activity of a reducing enzyme (like glutaredoxin) may counteract the activity of oxidizing components (such as Ero1 and Pdi1), we asked whether the overexpression of Ero1 or Pdi1 increases the growth defect observed in Δgrx6Δgrx7 cells (Figure 8A). The glutaredoxin-deficient strain showed reduced growth rates that were most pronounced at higher temperatures. However, the overexpression of Pdi1 or Ero1 in these cells did not influence growth. This suggests that Grx6 and Grx7 are not primarily required to counteract the activities of Ero1 and Pdi1.
Figure 8.
Relevance of Grx6 and Grx7 for oxidative protein folding in the secretory pathway. (A) The indicated strains were grown to midlog phase. Serial dilutions of the cultures were dropped on SD plates lacking tryptophan and incubated at 15, 30, or 37°C for 7, 3, or 6 d, respectively. (B) RNA was isolated from strains lacking Grx6, Grx7, or both and the corresponding wild type. For control, one wild-type culture was treated with 10 μg/ml tunicamycin (TM) for 30 min before RNA was extracted. The splicing of HAC1 mRNA was analyzed by Northern blotting. On induction of the UPR pathway, HAC1 mRNA is spliced leading to its increased mobility in the gel (arrowhead). (C) Scrambled RNase was incubated with 1 mM reduced glutathione and 0.2 mM oxidized glutathione in the presence or absence of 1 μM purified Pdi1 and/or 18 μM purified Grx7. The RNase activity was photometrically measured and is depicted in comparison of the spontaneous RNase refolding which was set as one.
The formation of disulfide bridges strongly contributes to the stable folding of proteins in compartments of the secretory pathway. Deficiencies in the components that facilitate the oxidative protein folding induce a signaling cascade known as the unfolded protein response (UPR) pathway (for review see Ellgaard and Helenius, 2003; Bernales et al., 2006). Oxidative stress is thereby sensed in the ER and transduced via a transmembrane receptor into the cytosol, where it leads to the splicing of HAC1 mRNA. Hac1 is the transcription factor that regulates UPR-controlled genes. To identify a potential role of Grx6 and Grx7 in protein folding in the secretory pathways, we tested whether the deletion of Grx6 and Grx7 leads to the splicing of HAC1 mRNA (Figure 8B). Although the addition of the glycosylation inhibitor tunicamycin induced the UPR pathway (Figure 8B, lane 2, arrowhead), no splicing of the HAC1 mRNA was observed in Δgrx6, Δgrx7, and Δgrx6Δgrx7 cells. Consistent with this observation, Δgrx6Δgrx7 mutants did not show increased expression levels of Pdi1 and Ero1 (Figure 3A). Moreover, no accumulation of aggregates was found in Golgi fractions of these mutants (our unpublished observations).
To investigate whether Grx6 and Grx7 play a role in the oxidative folding of proteins, we used an in vitro assay that monitors the refolding of oxidized scrambled RNAse. In contrast to Pdi1, Grx7 did not significantly accelerate the refolding of RNase over the spontaneous rate of refolding (Figure 8C). In summary, our results suggest that Grx6 and Grx7 are not involved in the general folding of proteins in the secretory pathway nor are they simple antagonists of Ero1 or Pdi1 but rather perform a distinct function that is especially important during oxidative stress conditions.
DISCUSSION
The glutaredoxin family comprises a large group of related enzymes that are ubiquitously present in different compartments of prokaryotic and eukaryotic cells. Here we identified three novel members of this family in baker's yeast in a comprehensive BLAST search, suggesting that the yeast genomes encodes for eight glutaredoxins in total. Two of the novel glutaredoxins, Grx6 and Grx7, are structurally closely related to each other. In the course of evolution, the genome of S. cerevisiae was duplicated (Wolfe and Shields, 1997). This event presumably also caused the duplication of an ancestor of Grx6 and Grx7, as only one Grx6/Grx7-like protein is encoded in fungi in which the genome had not been duplicated during evolution like Ashbya gossypii (Dietrich et al., 2004).
Both Grx6 and Grx7 contain N-terminal signal sequences and are translocated into purified microsomes in an ATP-dependent manner. Immunofluorescence and cellular subfractionation indicated their presence in the lumen of the cis-Golgi. To our knowledge, redox-active enzymes were not identified so far in a compartment of the secretory pathway beyond the ER.
Cysteine residues in proteins transported along the secretory pathway of eukaryotic cells are typically oxidized during their insertion into the ER and are believed to stay in an oxidized state. It was therefore unexpected to find glutaredoxin enzymes in the cis-Golgi where they might counteract the activity of the oxidizing machinery of the ER. The discovery of glutaredoxins in the secretory pathway shows that secreted proteins are not only exposed to significant amounts of reduced glutathione (Hwang et al., 1992) but also to specific enzymes that use the glutathione pool to catalyze the reduction of disulfide bridges. Because it is well established that secreted proteins are maintained in an oxidized state, it can be excluded that Grx6 and Grx7 antagonize protein oxidation in general. In consistence, we found no genetic interaction of the glutaredoxins with the oxidation machinery. In the absence of Grx6 and Grx7 we did not observe an increased aggregation of proteins in the Golgi or changes in the pattern of secreted proteins (our unpublished observations). Moreover, Δgrx6Δgrx7 mutant cells did not induce the UPR pathway, whereas mutants in Pdi1 or Ero1 lead to a strong UPR response (Pollard et al., 1998; Norgaard et al., 2003).
If not the general reduction of disulfide bonds, what might then be the specific function of Grx6 and Grx7? We regard it as very likely that these enzymes are critical for the reduction of specific cysteine residues in certain substrate proteins in the secretory pathway that require reduced thiol residues for their functionality. Candidates for these proteins might be enzymes with thiol groups in their active center (like cysteine proteases), proteins using thiol groups as ligands for cofactors like metals, heme groups or iron sulfur clusters, or components in which cysteine residues need to be posttranslationally modified. Biochemical analyses of Grx6 and Grx7 indeed suggest a role of these proteins in the formation of metal-containing reaction centers (Mesecke et al., 2008). In all these cases, the reduction of potential disulfide bridges will be critical, especially under oxidizing growth conditions, explaining the observed sensitivity of Δgrx6Δgrx7 mutant cells to hydrogen peroxide and diamide. It will be interesting to trace these specific substrate proteins in the future.
Originally, the ER was regarded as an oxidizing environment that favors the formation of disulfide bonds simply by the presence of considerable concentrations of oxidized glutathione (Hwang et al., 1992). Over the last decade it became clear, however, that proteins are oxidized by a dedicated set of components, namely Ero1 and Pdi1. These components contain intramolecular disulfide bridges with high transfer potentials, and transport the electrons from their substrates to molecular oxygen (Sevier et al., 2001; Tu and Weissman, 2002). Although the concentration of reduced glutathione in microsomes is lower than in the cytosol, the “chemical” environment in the secretory pathway is still reducing, which makes a glutaredoxin-catalyzed reduction of substrates possible.
We measured a significant reducing activity with purified Grx6 and Grx7. This is in striking contrast to observations with other monothiol glutaredoxins for which no activity in standard HEDS assays could be detected (Tamarit et al., 2003; Deponte et al., 2005). Also structurally, Grx6 and Grx7 are clearly distinct from other monothiol glutaredoxins in yeast and more closely related to the dithiol glutaredoxins Grx1 and Grx2 (Figure 1B). The reaction mechanism by which monothiol glutaredoxins catalyze the reduction of their substrates is still not clear. One plausible model suggests that monothiol glutaredoxins initially form mixed disulfides with their substrates. These complexes then need to be released in a reaction in which electrons are transferred from reduced glutathione to the intermolecular disulfide bond between the substrate and the enzyme. The possibility to analyze the activity of Grx6 and Grx7 in vitro should allow the biochemical dissection of their reaction cycle in order to unravel the working mode of this ubiquitous class of enzymes in the future.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Soledad Funes and Karl Bihlmaier for help with some experiments and Walter Neupert for constant support. We thank Chris Kaiser and Carolyn Sevier (both of the Massachusetts Institute of Technology, Cambridge, MA) for antibodies against Ero1, and Jakob Winther (University of Copenhagen, Denmark) for Pdi1 serum. We thank Katja Becker (University of Giessen, Germany) for providing the pQE30 plasmid encoding PfGrx. This work was supported by grants to J.M.H from the Deutsche Forschungsgemeinschaft (He2803/4-1) and the Stiftung für Innovation in Rheinland-Pfalz, to N.M. by the Bayerische Eliteförderung and to A.S. by the University of Basel.
Abbreviations used:
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HA
hemagglutinin
- HEDS
hydroxyethyl disulfide
- UPR
unfolded protein response.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0896) on April 9, 2008.
REFERENCES
- Banfield D. K., Lewis M. J., Rabouille C., Warren G., Pelham H. R. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J. Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr. Opin. Struct. Biol. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Bass R., Ruddock L. W., Klappa P., Freedman R. B. A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J. Biol. Chem. 2004;279:5257–5262. doi: 10.1074/jbc.M304951200. [DOI] [PubMed] [Google Scholar]
- Bernales S., Papa F. R., Walter P. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- Chuang J. S., Schekman R. W. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deponte M., Becker K., Rahlfs S. Plasmodium falciparum glutaredoxin-like proteins. Biol. Chem. 2005;386:33–40. doi: 10.1515/BC.2005.005. [DOI] [PubMed] [Google Scholar]
- Dietrich F. S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Ellgaard L. Catalysis of disulphide bond formation in the endoplasmic reticulum. Biochem. Soc. Trans. 2004;32:663–667. doi: 10.1042/BST0320663. [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Frickel E. M., Frei P., Bouvier M., Stafford W. F., Helenius A., Glockshuber R., Ellgaard L. ERp57 is a multifunctional thiol-disulfide oxidoreductase. J. Biol. Chem. 2004;279:18277–18287. doi: 10.1074/jbc.M314089200. [DOI] [PubMed] [Google Scholar]
- Grant C. M. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 2001;39:533–541. doi: 10.1046/j.1365-2958.2001.02283.x. [DOI] [PubMed] [Google Scholar]
- Haugen A. C., Kelley R., Collins J. B., Tucker C. J., Deng C., Afshari C. A., Brown J. M., Ideker T., Van Houten B. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 2004;5:R95. doi: 10.1186/gb-2004-5-12-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., de la Torre-Ruiz M. A. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol. Life Sci. 2007;64:1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J. M., Fölsch H., Neupert W., Stuart R. A. Isolation of yeast mitochondria and study of mitochondrial protein translation. In: Celis J. E., editor. Cell Biology: A Laboratory Handbook. vol. 1. San Diego: Academic Press; 1994. pp. 538–544. [Google Scholar]
- Holmgren A., Aslund F. Glutaredoxin. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- Holmgren A., Johansson C., Berndt C., Lonn M. E., Hudemann C., Lillig C. H. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem. Soc. Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- Hwang C., Sinskey A. J., Lodish H. F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Lill R., Mühlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu. Rev. Cell Dev. Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- Lillig C. H., Holmgren A. Thioredoxin and related molecules—from biology to health and disease. Antioxid. Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- Lopreiato R., Facchin S., Sartori G., Arrigoni G., Casonato S., Ruzzene M., Pinna L. A., Carignani G. Analysis of the interaction between piD261/Bud32, an evolutionarily conserved protein kinase of Saccharomyces cerevisiae, and the Grx4 glutaredoxin. Biochem. J. 2004;377:395–405. doi: 10.1042/BJ20030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikenhuis S., Perrone G., Dawes I. W., Grant C. M. The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol. Biol. Cell. 1998;9:1081–1091. doi: 10.1091/mbc.9.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch M., Henthorn D. C., Herrmann J. M., Wilson R., Thomas D. Y., Bergeron J. J., Solari R. C., Rowley A. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecke N., Mittler S., Eckers E., Herrmann J. M., Deponte M. Two novel monothiol glutaredoxins from Saccharomyces cerevisiae provide further insight into iron-sulfur cluster binding, oligomerization, and enzymatic activity of glutaredoxins. Biochemistry. 2008;47:1452–1463. doi: 10.1021/bi7017865. [DOI] [PubMed] [Google Scholar]
- Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Pape L. K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgaard P., Tachibana C., Bruun A. W., Winther J. R. Gene regulation in response to protein disulphide isomerase deficiency. Yeast. 2003;20:645–652. doi: 10.1002/yea.978. [DOI] [PubMed] [Google Scholar]
- Ojeda L., Keller G., Mühlenhoff U., Rutherford J. C., Lill R., Winge D. R. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- Ostergaard H., Tachibana C., Winther J. R. Monitoring disulfide bond formation in the eukaryotic cytosol. J. Cell Biol. 2004;166:337–345. doi: 10.1083/jcb.200402120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget M. S., Buttner M. J. Thiol-based regulatory switches. Annu. Rev. Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- Pedrajas J. R., Porras P., Martinez-Galisteo E., Padilla C. A., Miranda-Vizuete A., Barcena J. A. Two isoforms of Saccharomyces cerevisiae glutaredoxin 2 are expressed in vivo and localize to different subcellular compartments. Biochem. J. 2002;364:617–623. doi: 10.1042/BJ20020570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M. G., Travers K. J., Weissman J. S. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- Porras P., Padilla C. A., Krayl M., Voos W., Barcena J. A. One single in-frame AUG codon is responsible for a diversity of subcellular localizations of glutaredoxin 2 in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:16551–16562. doi: 10.1074/jbc.M600790200. [DOI] [PubMed] [Google Scholar]
- Pujol-Carrion N., Belli G., Herrero E., Nogues A., de la Torre-Ruiz M. A. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J. Cell Sci. 2006;119:4554–4564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- Rahlfs S., Fischer M., Becker K. Plasmodium falciparum possesses a classical glutaredoxin and a second, glutaredoxin-like protein with a PICOT homology domain. J. Biol. Chem. 2001;276:37133–37140. doi: 10.1074/jbc.M105524200. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manzaneque M. T., Tamarit J., Belli G., Ros J., Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier C. S., Cuozzo J. W., Vala A., Aslund F., Kaiser C. A. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat. Cell Biol. 2001;3:874–882. doi: 10.1038/ncb1001-874. [DOI] [PubMed] [Google Scholar]
- Sevier C. S., Kaiser C. A. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- Sevier C. S., Qu H., Heldman N., Gross E., Fass D., Kaiser C. A. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell. 2007;129:333–344. doi: 10.1016/j.cell.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. Methods in Yeast Genetics: A Laboratory Course. New York: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia R., Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- Tamarit J., Belli G., Cabiscol E., Herrero E., Ros J. Biochemical characterization of yeast mitochondrial Grx5 monothiol glutaredoxin. J. Biol. Chem. 2003;278:25745–25751. doi: 10.1074/jbc.M303477200. [DOI] [PubMed] [Google Scholar]
- Tu B. P., Weissman J. S. The FAD- and O2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol. Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- Tu B. P., Weissman J. S. Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B., Gilbert H. F. Protein disulfide isomerase. Biochim. Biophys. Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Shields D. C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Wuestehube L. J., Schekman R. W. Reconstitution of transport from endoplasmic reticulum to Golgi complex using endoplasmic reticulum-enriched membrane fraction from yeast. Methods Enzymol. 1992;219:124–136. doi: 10.1016/0076-6879(92)19015-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.