Abstract
CRM1 exports proteins that carry a short leucine-rich peptide signal, the nuclear export signal (NES), from the nucleus. Regular NESs must have low affinity for CRM1 to function optimally. We previously generated artificial NESs with higher affinities for CRM1, termed supraphysiological NESs. Here we identify a supraphysiological NES in an endogenous protein, the NS2 protein of parvovirus Minute Virus of Mice (MVM). NS2 interacts with CRM1 without the requirement of RanGTP, whereas addition of RanGTP renders the complex highly stable. Mutation of a single hydrophobic residue that inactivates regular NESs lowers the affinity of the NS2 NES for CRM1 from supraphysiological to regular. Mutant MVM harboring this regular NES is compromised in viral nuclear export and productivity. In virus-infected mouse fibroblasts we observe colocalization of NS2, CRM1 and mature virions, which is dependent on the supraphysiological NS2 NES. We conclude that supraphysiological NESs exist in nature and that the supraphysiological NS2 NES has a critical role in active nuclear export of mature MVM particles before cell lysis.
INTRODUCTION
CRM1 is a general purpose nuclear export receptor, which transports a wide range of proteins and RNAs out of the nucleus. Many cargoes that associate with CRM1 do so via a short leucine-rich nuclear export signal (NES), either directly or indirectly (reviewed in Fornerod and Ohno, 2002). The direction of nuclear export is imposed by RanGTP. This is because NESs can only bind CRM1 stably in the presence of RanGTP, which is present in high concentration in the nucleus and not in the cytoplasm. Nuclear export complexes dissociate in the cytoplasm through the combined action of RanGAP and RanBP1 (reviewed in Fornerod and Ohno, 2002). The translocation through the NPC is dependent on CRM1/nucleoporin interactions and in its simplest mode has little or no directionality (Becskei and Mattaj, 2003). Indeed, export can be reversed if NES/CRM1 association is allowed to occur in the cytoplasm (Nachury and Weis, 1999). Therefore, a key factor for the export system to work efficiently is that the affinity of the NES for CRM1 in the absence of RanGTP should be low enough to prevent CRM1/cargo complexes from forming in the cytoplasm.
Recently, we generated a series of NES variants through reiterative screening of a phage library that display ∼100 fold greater affinity for CRM1 than regular NESs (Engelsma et al., 2004). We termed these artificially generated NES variants supraphysiological NESs or supraNESs. These NESs bind CRM1 stably in the cytoplasm in the absence of RanGTP, thereby functioning as competitive inhibitors of CRM1 function. Another distinct feature of these supraNESs is their prominent localization at the nuclear envelope (NE), largely dependent on nucleoporin Nup358. This characteristic NE localization likely reveals a dissociation step in nuclear export in vivo (Engelsma et al., 2004). So far, supraNESs have not been encountered in proteins evolved by natural selection.
Minute Virus of Mice (MVM) is an autonomous mouse parvovirus with a 5-kb single-stranded DNA genome consisting of two overlapping transcriptional units. Early transcription drives the expression of two nonstructural proteins NS1 and NS2. At later time points the structural proteins VP1 and VP2 are expressed. In the nucleus, VP proteins form an icosahedral capsid with a diameter close to 25 nm that encapsulates the viral DNA. Parvoviruses assemble in the nucleus and have evolved mechanisms for nuclear export independent of cell lysis, conceivably in order to gain systemic infection (discussed in Maroto et al., 2004). Parvovirus particles are small enough to pass through the NPC, which has been thought to have a maximal inner diameter of ∼39 nm (Pante and Kann, 2002). However, only DNA-filled viral capsids are efficiently exported from the nucleus. This requires the N-terminus of VP2 (2Nt), which is exposed from the viral capsid when an ssDNA molecule is present within (Maroto et al., 2004). MVM virions have two different ways of nuclear exit. In transformed human fibroblastst, the capsids can exit in an NS2 and CRM1 independent way (Maroto et al., 2004). In mouse fibroblasts, export of MVM virions is inhibited by the CRM1 inhibitor leptomycin B (LMB; Maroto et al., 2004), indicating involvement of CRM1. Nuclear export of MVM in these cells is also dependent on NS2, which is the only known CRM1 substrate encoded by MVM (Bodendorf et al., 1999; Eichwald et al., 2002). Consistently, viruses with a mutated NS2 NES fail to be exported to the cytoplasm of mouse fibroblasts and display strongly reduced fitness in culture (Eichwald et al., 2002). Thus, NS2-dependent nuclear export of mature viral particles appears, at least in their natural host, to be CRM1-mediated and a critical step in MVM infection cycle (see below).
Interestingly, CRM1 has been shown to relocalize to the cytoplasm in cells infected with an immunosuppressive strain of MVM (MVMi; Lopez-Bueno et al., 2004). In particular, cytoplasmic sequestration of CRM1 was prominent in viruses selected in severe combined immunodeficiency (SCID) mice that carried point mutations close to the NS2 NES and showed enhanced CRM1 binding in cell lysates (Lopez-Bueno et al., 2004). These findings led us to investigate whether NS2 may in fact contain a supraNES. We first analyzed the localization of NS2 in more detail and found that it was present not only in the cytoplasm, but accumulated at the nuclear periphery as well. Study of the behavior and biochemical characteristics of the NS2 NES indeed identified NS2 as a supraNES-bearing protein. We find that the NS2 supraNES is critical for MVM nuclear export in mouse fibroblasts. Collectively, our findings demonstrate that supraNESs are observed in nature and have been coopted by the MVM virus to promote its life cycle and pathogenicity.
MATERIALS AND METHODS
Antibodies
Antibodies were diluted in PBS/1% milk and used as follows; rabbit anti-serum against NS2 (Faisst et al., 1995) was used 1:200, rabbit anti-serum against CRM1 (Fornerod et al., 1997b) 1:250, mAb raised against an epitope of the MVM capsid surface (B7a; Lopez-Bueno et al., 2003) 1:200, rabbit anti-serum 2Nt raised against the N-terminus of VP2 (Maroto et al., 2004)1:200, guinea-pig anti-serum Nup358 (Walther et al., 2002) 1:100, and rabbit antibody NFκB p65 (sc-372) 1:500.
Plasmids and Plasmid Construction
pdBMVp and pdBMV-NES22 (Eichwald et al., 2002), Nmd3-GFP (Thomas and Kutay, 2003), and pS and pSNup358 (Bernad et al., 2004) were described previously. To construct pX-NS2P, a NcoI/BamHI fragment containing the MVMp NS2P cDNA (nt 260 to nt 2692 minus nt 515-1989 and nt 2281–2376) was excised from pTM1-NS2P (kindly provided by P. Tattersall, Yale University, New Haven, CT) and was first subcloned into pGBT9 (Clontech, Palo Alto, CA), in which an NcoI site was previously inserted in the multiple cloning site using the following EcoRI linker: 5′ AATTCACCATGGTTAAC 3′ and 5′ AATTGTTAACCATGGTG, to generate pGBT9-NS2P and then into pALTER-1 (Promega, Madison, WI) as a EcoRI/BamHI fragment to generate pALTER-NS2P. The NS2P cDNA-containing EcoRI/HindIII fragment was further excised from pALTER-NS2P and inserted into the EcoRI/HindIII site of pX (a vector kindly provided by G. Superti-Furga, EMBL, Heidelberg, Germany). To construct pX-NS2_NES8, the so-called “NES6” and “NES8” primers with the respective sequences: 5′-AGTGGATGAAGCGACCAAAAAGGCCGGCACGCTC-3′ and 5′-CCAAAAAGGCCGGCACGGCCACCGCTCACGACACCGAAAAG-3′ were subsequently introduced into pALTER-NS2P using the Altered SitesR in vitro Mutagenesis System (Promega) according to the manufacturer's instructions. The resulting pALTER-NS2P_NES6 and pALTER-NS2P_NES8 were further sequenced to check for the presence of the mutations within the NS2 NES site. The EcoRI/HindIII fragment, containing the NS2P mutated cDNA, was further excised from pALTER-NS2-NES8 and inserted into the EcoRI/HindIII site of pX-NS2P. To construct green fluorescent protein (GFP)-NS2 and pRSET-NS2, a BamHI/HindIII fragment excised from pX-NS2 (NS2p isoform, obtained from Peter Tattersall, Yale University, New Haven, CT) was inserted into the BamHI/HindIII sites of pEGFP-C1 or pRSET-A. To construct GFP-NS2AAAA and pRSET-NS2AAAA, pX-NS2-NES8 was digested with XmnI and the insert containing the mutated NES was inserted in XmnI-digested GFP-NS2 and pRSET-NS2. For GFP-NS2-NES, GFP-NS2-NESL89A, GFP-NS2-NESI91A, GFP-NS2-NESL89A/I91A, and GFP-PKIαL44A, oligos (only Fw are shown) containing HindIII and BamHI overhangs (not shown) with the following respective sequences were annealed: 5′-TCGGTGGATGAAATGACCAAAAAGTTCGGCACGCTCACCATTCACGACG-3′, 5′-TCGGTGGATGAAATGACCAAAAAGTTCGGCACGGCCACCATTCACGACG-3′, 5′-TCGGTGGATGAAATGACCAAAAAGTTCGGCACGCTCACCGCTCACGACG-3′, 5′-TCGGTGGATGAAATGACCAAAAAGTTCGGCACGGCCACCGCTCACGACG-3′,5′-AGCTTACTTGGCCCTGAAGCTGGCCGGCGCCGATATCG-3′. These oligos were subcloned into HindIII/BamHI digested pEGFP-C1 (Clontech). For pRev-S4-GFP, 5′-TTGGGGCTGTTGATTTGTTGGCTCGTCTGTTTTCTGCTCTGAGTGTTAGTGGGGCCGCA-3′ oligos with a BamHI and PinAI overhangs were subcloned into pRev(1.4)-GFP (Henderson and Eleftheriou, 2000). The eGFP-NES insert containing the protein kinase A inhibitor NES was amplified by PCR from pBSSK (Roth et al., 2003) using the forward primer containing a HindIII site 5′-CGATAAGCTTGATGGTGAGCAAGGGCGAGGAG-3′ and a reverse primer, 5′-AGCTGGAGCTCCACCGCGGTG-3′, and subcloned into pCDNA3 (Stratagene, La Jolla, CA) by digestion with HindIII and NotI.
Recombinant Protein Expression and Purification
Expression of GFP3-S1, Rna1p, CRM1, and GFP-PKI was described previously (Engelsma et al., 2004). Native His6-tagged NS2 and NSAAAA (∼10% purity; used for RanGAP assays) were purified using Ni-NTA agarose (Qiagen, Chatsworth. CA). Highly pure His6-tagged NS2 protein (>95% purity, used for Biocore assays) was isolated from inclusion bodies in 8 M urea purified in Ni-NTA and refolded by stepwise dialysis to 20 mM HEPES, pH 7.9, 200 mM NaCl, 1 mM DTT, and 8.7% glycerol. Refolded protein was subsequently eluted as a single peak below 30 kDa from a HiLoad S75 16/60 gel filtration column (GE Healthcare, Waukesha, WI). Eluted protein was concentrated using Centricon concentrator (10-kDa cutoff) to 0.8 mg/ml.
CRM1 RanGAP Assays
GAP assay were performed as described previously (Askjaer et al., 1999). The following sequences of peptides were used: NS2-NES, CVDEMTKKFGTLTIHDTEK; NS2-NES22, CVDEMTKKFGTQSIHDTEK; and REVNES, CLQLPPLERLTL.
Cell Culture and Transfections
MCF7, low passage HeLa (ATCC, Manassas, VA; CCL-2), L929, and A9 cells were cultured in DMEM, containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and antibiotics. MCF7, HeLa, and L929 cells were transfected using Fugene 6 (Roche, Indianapolis, IN), and A9 cells were transfected with Lipofectamine (Invitrogen). Images were taken using Leica TCS-NT2, SP2, and AOBS confocal microscopes (Deerfield, IL) and analyzed using Image J software (http://rsb.info.nih.gov/ij/). pSuper constructs were transfected for 72 h. Only cells with reduced Nup358 levels were scored for the presence of GFP protein at the NE. 20–40 cells per conditions were scored. LMB, 50 nM, was used when indicated. GFP-NS2-NES22 transfection was recorded live using a temperature-controlled chamber build onto the Leica AOBS microscope.
MVM Infections and Plaque Assay
Infections and virus production was performed as described previously (Eichwald et al., 2002). Plaque assays were performed as described previously (Lopez-Bueno et al., 2004).
Immunofluorescence Stainings and Image Analysis
Stainings were done as described previously (Bernad et al., 2004). Pre-extraction was performed with 0.5% Triton X-100 in PBS for 1 min on ice, after which the coverslips were fixed in 3.7% formalin in PBS for 10′. Images were acquired on a Leica AOBS confocal microscope. Image analysis was done in Image J, a program, developed at the U.S. National Institutes of Health. For nucleocytoplasmic distributions, log2 nuclear to cytoplasmic ratios were calculated. Log transformation was applied to obtain equal representation of all ratios (log ratios showed normal distributions, whereas ratios usually did not). Colocalization analysis was done using the Colocalization Test plugin developed by Wayne Rasband (Research Services Branch, National Institute of Mental Health, Bethesda, MD) and Tony Collins (Wright Cell Imaging Facility, University Health Network, Toronto, Canada), available at http://www.uhnresearch.ca/facilities/wcif. Pearson's correlation coefficients (R) of intranuclear regions were calculated as well as those of randomized images. Correlation coefficients were squared to obtain a linear approximation to explained variance. Statistical analyses were done in R (www.r-project.org).
Surface Plasmon Resonance Spectroscopy
Surface plasmon resonance (SPR) spectroscopy was performed at 25°C on a Biacore T100 (GE Healthcare). CRM1 and NS2 were immobilized on a CM5 chip with amine coupling in 10 mM sodium acetate at pH 5.5 (CRM1) or pH4.5 (NS2). Approximately 17,000 response units (RUs) of CRM1 and 1800 response units (RUs) of NS2 were immobilized on the chip. For binding measurements, NS2 (18 μM), CRM1 (0.64 μM), or PKI NES peptide (50 μM) in the presence or absence of 30 μM (CRM1 chip) or 50 μM (NS2 chip) RanGTP were streamed over the chip in 20 mM HEPES-KOH pH 7.9, 200 mM sodium chloride, 8.7% glycerol at 0.03 ml/min. Simultaneously, an empty flow cell was used as reference. Biacore T100 evaluation software was used for analysis of the data.
RESULTS
Parvovirus NS2 Is Localized at Nucleoporin Nup358
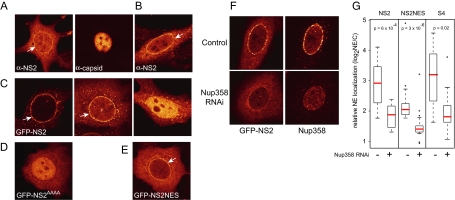
The interaction between CRM1 and NS2 is directly mediated by an NES in NS2 (Askjaer et al., 1999; Miller and Pintel, 2002; Ohshima et al., 1999). Recently, expression of NS2 was shown to increase the cytoplasmic fraction of CRM1 (Lopez-Bueno et al., 2004), resembling the effect of supraNES proteins (Engelsma et al., 2004). As supraNES-bearing proteins are expected to reside prominently at the NE, we first studied the localization of NS2. Immunostaining of NS2 in A9 cells expressing the MVM genome showed that NS2 localized in part to the NE (Figure 1A, specificity of the antibody for this method is shown in Supplementary Figure 1). We also observed this NE localization in cells expressing untagged or GFP-tagged NS2 (Figure 1, B and C). GFP-NS2 accumulated in the nucleus of cells expressing higher NS2 levels, possibly because supraNESs inhibit their own export at higher concentrations (Figure 1C, right). To address whether the NE localization was carried out by the NES of NS2, consensus hydrophobic residues were mutated into alanines (NS2AAAA). Expression of this construct resulted in a complete loss of nuclear rim staining (Figure 1D). In addition, the isolated NS2 NES peptide fused to GFP (NS2NES-GFP) was localized at the NE (Figure 1E). Therefore, the NS2 NES is required and sufficient for NS2 NE localization.
Figure 1.
NS2 localizes to the NE via Nup358. (A) A9 cells transfected with MVM expression plasmid pdBMVp for 24 h and stained with anti-NS2 and anti-capsid antibodies. (B) HeLa cells expressing untagged NS2 were stained after 24 h with anti-NS2 antibodies. MCF7 cells were transfected for 24 h with (C) NS2-GFP, (D) NS2AAAA–GFP, or (E) NS2-NES-GFP. White arrows indicate NE localization. (F and G) MCF7 cells were transfected with pSuper-Nup358 and Nup358 levels were determined after 72 h using Nup358 antibodies. (F) An example of GFP-NS2 localization in control cells or cells depleted of Nup358. (G) Boxplots showing relative amounts of NE-located GFP-tagged protein in control cells or Nup358 knockdown cells were determined by dividing the NE pixel intensity by the cytoplasmic pixel intensity, measured at three sites of the NE. Medians are indicated in red. p values were calculated using Mann-Whitney tests.
Because supraNES/CRM1 complexes arrest at Nup358 (Engelsma et al., 2004), we tested whether NS2 also localized to this cytoplasmic nucleoporin. On knockdown in MCF7 cells of Nup358 levels by expression of shRNAs targeting Nup358 (Engelsma et al., 2004) GFP-NS2 was depleted from the nuclear rim (Figure 1, F and G). NS2NES-GFP and the synthetic GFP-S4 supraNES, a variant of the previously described S1 supraNES (Engelsma et al., 2004), were similarly depleted from the nuclear rim, indicating that Nup358 mediates nuclear rim localization of NS2 (Figure 1G).
NS2 Relocalizes CRM1 to the Cytoplasm and Interferes with the CRM1-Export Pathway
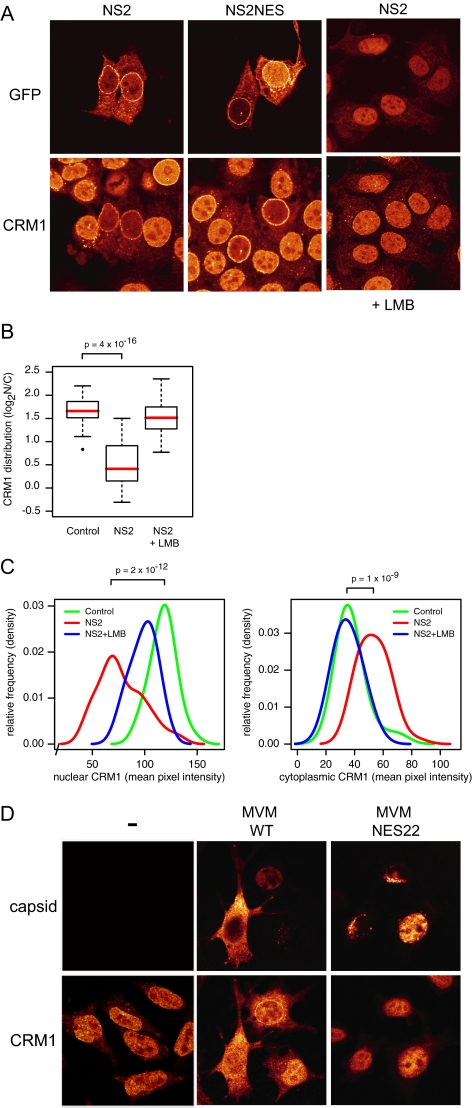
CRM1 is relocalized to the cytoplasm in response to the expression of supraNES variants (Engelsma et al., 2004). As human cells infected with MVMi similarly relocalize CRM1 to the cytoplasm (Lopez-Bueno et al., 2004), we wondered whether CRM1 relocalization was mediated by the NS2 NES. Indeed, NS2-GFP expression depleted CRM1 from the nucleus and relocalized it to the cytoplasm (Figure 2A, quantified in B and C). Expression of the isolated NS2-NES fused to GFP resulted in a similar CRM1 relocalization (Figure 2A), indicating that it is NES mediated. On addition of LMB, NS2 accumulates in the nucleus, and CRM1 relocalization was largely abolished (Figure 2, A–C). In cells infected with MVM, CRM1 was also increased in the cytoplasm (Figure 2D). Furthermore, MVM carrying a previously described NS2 NES mutant, NES22, which carries point mutations in the NS2 NES (Eichwald et al., 2002), did not relocalize CRM1 to the cytoplasm (Figure 2D).
Figure 2.
NS2 relocalizes CRM1 from the nucleus to the cytoplasm (A) MCF7 cells expressing NS2-GFP (left and right) or NS2-NES-GFP (middle). When indicated, cells were treated with LMB for 3 h. Cells were stained with anti-CRM1 antibodies. (B) Boxplot summarizing quantifications (n = 36 each) of CRM1 subcellular distribution exemplified in A. Medians are indicated in red. (C) Distribution of nuclear (left) and cytoplasmic (right) CRM1 intensities, showing both an NS2-dependent nuclear decrease and a cytoplasmic increase, which is LMB-sensitive. Densities were calculated using a bandwidth of 8. p values according to Mann-Whitney tests (D) A9 cells mock-infected (−) or infected with MVMp (wt) or MVMp-NES22 (NES22) for 48 h and stained for both capsid and CRM1.
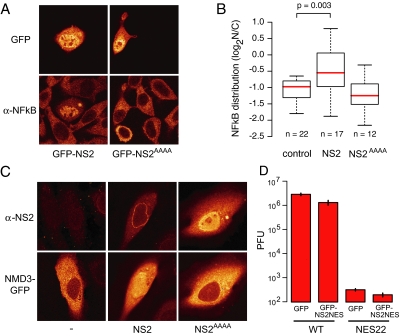
Previously, we have shown that the expression of a supraNES leads to inhibition of its own export by compromising CRM1 function. We therefore investigated if NS2 could also block CRM1-mediated export. This was tested for the NFκB protein, whose cytoplasmic localization is mediated by CRM1 (Tam et al., 2000). As shown in Figure 3A, expression of GFP-NS2, but not the NES-dead variant GFP-NS2AAAA, led to the accumulation of endogenous NFκB in the nucleus. A quantification of this experiment is presented in Figure 3B. Significant nuclear accumulation of NFκB only occurred in cells expressing relatively high amounts of NS2 (Supplementary Figure 2). We also tested whether export of an exogenously expressed protein was hampered. We therefore studied the localization of the 60S preribosomal export adaptor Nmd3 (Ho et al., 2000; Thomas and Kutay, 2003). Export of GFP-Nmd3 was also inhibited in cells expressing NS2, but not in cells expressing NS2AAAA (Figure 3C).
Figure 3.
NS2 interferes with the CRM1 export pathway. (A) NS2 inhibits the cytoplasmic localization of NFκB. L929 cells were transfected with GFP-NS2 or GFP-NS2AAAA and stained for NFκB. (B) Boxplots summarizing quantifications of NFκB subcellular distribution shown in (A). p value from Mann-Whitney test. (C) NS2 inhibits export of Nmd3. HeLa cells expressing Nmd3-GFP alone (right) Nmd3-GFP and untagged NS2 (middle), or Nmd3-GFP and untagged NS2AAAA (right). NS2 was detected with NS2 antibodies. (D) Exogenously expressed supraNES cannot rescue the defect in viral propagation of MVMNES22. A9 cells (n = 105) were transfected with either wild-type pdBMVp or pdBMV-NES22 (NES22) and cotransfected with either GFP or GFP-NS2-NES. After 48 h the extracellular virus pool was titered by a plaque assay using NB324K cells. Y-axis shows the plaque forming units (PFU). Infection with MVM-NES22 yielded microplaques. Error bars, SEs from independent experiments.
Some viruses inhibit nuclear transport pathways of the host cell in order to enhance export of their own cargoes (Fontoura et al., 2005). The previously described MVM NES22 mutant is an MVM strain that contains a mutated supraNES. This viral strain is severely deficient in viral propagation. We investigated whether the viral propagation of this mutant virus could be rescued by expressing a supraNES exogenously, leading to CRM1 inhibition. A9 cells were transfected with pdBMVp or pdBMV-NES22 viral DNA in combination with NS2NES-GFP or GFP. Viral particles present in the supernatant were harvested 48 h after transfection and titered by plaque assays using human transformed cells (NB324K). As shown before, NES22 yielded less plaque forming units as compared with wild-type virus (Eichwald et al., 2002). Coexpression of a supraNES, however, did not rescue the NS2 NES mutated virus (Figure 3C), but instead lead to a significant decrease. This result suggests that inhibition of the CRM1 pathway by the supraNES is not the its main function and is consistent with earlier reports that virus nuclear export requires intact CRM1 function (Maroto et al., 2004).
Intranuclear Colocalization of CRM1 with Mature Virions Is Dependent on the NS2 supraNES
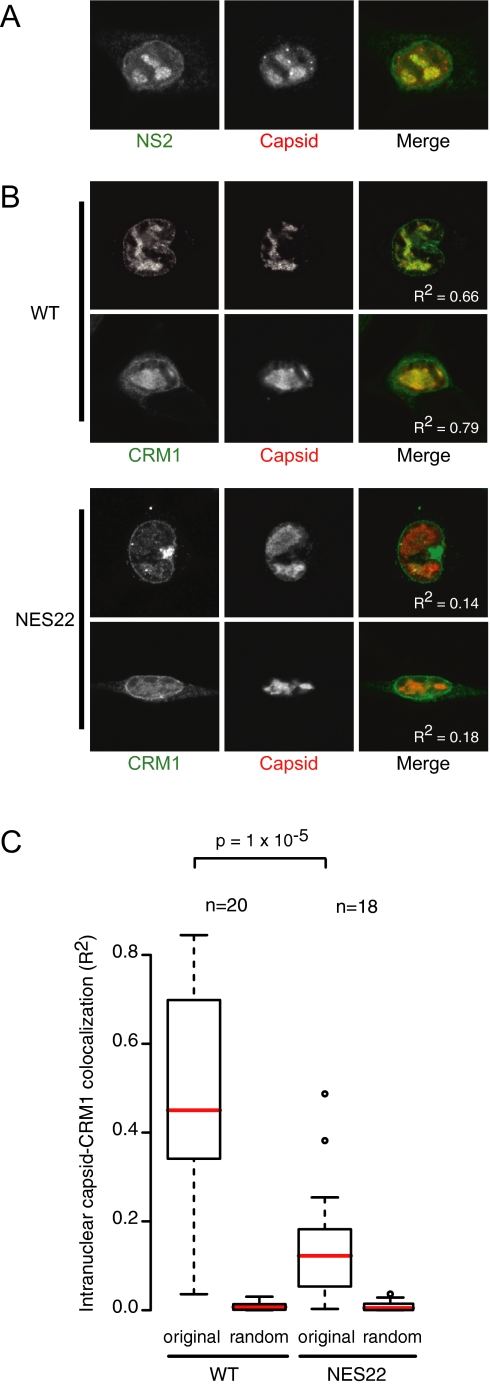
Maturation of MVM coincides with the externalization of the N-terminus of VP2 (2Nt) when capsid is filled with DNA (Maroto et al., 2004). This viral maturation occurs in large nuclear structures, termed Smn-associated autonomous parvovirus-associated replication bodies, or SAABs, which appear late in infection (Young et al., 2002b). Interestingly, NS2 has also been detected in SAABs (Young et al., 2002a,b), suggesting a direct role in late stages of viral maturation. Mature MVM virions can be visualized by antibodies directed against the exposed 2Nt signal of VP2 (Maroto et al., 2004), while both mature and immature virions are detected by an anti-capsid antibody (Lopez-Bueno et al., 2004). As shown in Supplementary Figure 3, 2Nt staining was indeed detected primarily in SAABs. For optimal detection of SAABs using the anti-capsid antibody, mouse cells were briefly pre-extracted by triton permeabilization before fixation (Supplementary Figure 3). Consistent with earlier observations, NS2 is present in SAABs, colocalizing with capsid (Figure 4A). This colocalization is independent of the wild-type NS2 NES as NS2 and capsid similarly colocalize in wild-type virus and NES22 mutant virus–expressing cells (data not shown). CRM1, however, colocalizes with SAABs to a far greater degree in cells expressing the wild-type MVM genome compared with cells expressing an NES22 mutant genome (Figure 4, B and C). These data suggest an interaction between CRM1 and the viral capsid in mouse fibroblasts, which is mediated by the NS2 NES.
Figure 4.
Intranuclear colocalization of CRM1 with mature virions is dependent on the NS2 supraNES. A9 cells transfected with pdBMVp (WT) or pdBMV-NES22 (NES22) for 48 h. (A) NS2 and capsid colocalize in SAABs. (B) CRM1 and capsid colocalize in SAABs in a supraNES-dependent manner. Colocalization values (R2) are displayed in the merged images. (C) Boxplots summarizing quantification of intranuclear colocalization of CRM1 with capsid. Pixel correlations (R) were calculated in the intranuclear region (excluding membrane invaginations) of CRM1 with original (original) or means of 25 randomized (random) capsid images using ImageJ. Correlations were squared to provide a linear approximation to colocalization. p values according to Mann-Whitney test.
NS2 Has Supraphysiological Affinity for CRM1 and Binds Stably without RanGTP
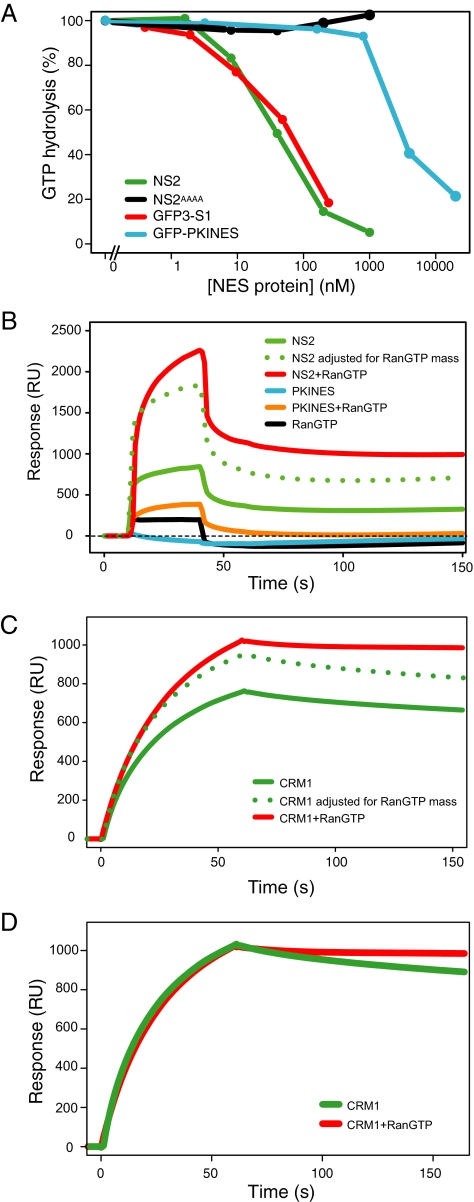
By definition, supraNESs should display supraphysiological affinity for CRM1. A quantitative method to test this is the CRM1 RanGTPase assay. This in vitro assay is based on the inability of RanGAP to hydrolyze RanGTP when present in transport receptor/RanGTP export complexes (Bischoff and Gorlich, 1997; Lounsbury and Macara, 1997). RanGTP is hydrolyzed in the absence of NES cargo, but with increasing NES concentrations export complexes form and RanGTP is protected from hydrolysis. High-affinity NESs will engage more RanGTP to CRM1 and increase RanGTP protection from hydrolysis (Kutay et al., 1997; Askjaer et al., 1999; Paraskeva et al., 1999). As shown in Figure 5A, recombinant NS2 bound CRM1 with ∼ 100-fold greater affinity as compared with the affinity of the reference GFP-PKI-NES. As a control for supraphysiological affinity, GFP3-S1 was used, which possesed a similar high-affinity as NS2 (Figure 5A), As expected, NS2AAAA did not bind CRM1.
Figure 5.
NS2 binds stably to CRM1 in a RanGTP-independent manner. (A) Full-length NS2 protein binds CRM1 with supraphysiological affinities similar to the supraNES S1. The affinity for CRM1 of NS2, NS2AAAA, the regular PKI-NES, and the supraNES S1 was measured using the CRM1 GTPase assay. Complexes formed with Ran[γ-GT32P], purified recombinant NES-bearing proteins (x-axis) and CRM1 were subjected to RanGAP, and hydrolyzed 32P was measured (y-axis). 100% hydrolysis was recorded when CRM1-Ran[γ-GT32P] samples without cargo were subjected to RanGAP-stimulated hydrolysis. Full-length NS2 (green) and GFP3-S1 (red) were used as reference for supraphysiological affinity; GFP-PKI-NES (blue) was used as control for regular affinity, and full-length NS2AAAA protein (black) was used as negative control. (B–D) Surface plasmon resonance (SPR) analysis of the NS2/CRM1 interaction. (B) NS2 (green) or PKI NES (blue), alone or in combination with RanGTP (NS2 plotted in red and PKI NES plotted in orange) were streamed over chip-immobilized CRM1 for 40s. Resonance response (in RU) was measured to 150 s. The dotted line denotes the theoretical response of an NS2/RanGTP complex based on the NS2 response adjusted for the additional mass of Ran. RanGTP (black) alone displays minor affinity for CRM1. (C) CRM1 (green) or CRM1+RanGTP (red) was streamed over chip-immobilized NS2 for 60 s. Resonance response was measured to 150 s. The dotted line denotes the theoretical response of a CRM1/RanGTP complex based on the CRM1 response adjusted for the additional mass of Ran. (D) Superimposed CRM1 and CRM1+RanGTP response curves indicate a greater stability of NS2/CRM1/RanGTP (red) over NS2/CRM1 (green).
To test RanGTP requirement of the NS2/CRM1 interaction, we used surface plasmon resonance (SPR) spectroscopy using a Biacore setup. We initially chip-immobilized CRM1 and measured surface resonance upon addition of a recombinant NS2 or PKI NES stream. The plasmon resonance response is proportional to the total mass of bound molecules (1000 response units ∼ 1 ng of protein). As shown in Figure 5B, significant response was detected upon addition of NS2, whereas no response was detected with the PKI NES alone. The observed increase of signal upon addition of recombinant RanGTP was close the calculated increase in mass by RanGTP joining the NS2/CRM1 complex, which is calculated as the ratio between NS2 + RanGTP (58 kDa) and NS2 alone (27 kDa). This suggests that the increased resonance response can be largely attributed to RanGTP joining the complex, without further stimulation of the NS2/CRM1 interaction per se. In contrast, RanGTP increased the response of the PKI NES substantially above the level that can be attributed to RanGTP alone. Termination of the NS2 stream allows one to assess the stability of the CRM1/NS2 interaction. Strikingly, both in presence or absence of RanGTP, part of the complex is rapidly dissociated, whereas a significant fraction of NS2 remains stably bound to CRM1. By contrast, all CRM1/PKI/RanGTP complexes are rapidly dissociated (Figure 5B).
CRM1 is a large suprahelical molecule that likely carries out large-scale conformational changes upon optimal binding (Petosa et al., 2004). We therefore wondered whether the two pools of NS2, one stably bound and one rapidly dissociating, might represent pools of CRM1 differently cross-linked on the chip. We therefore reversed the experimental setup by chip-immobilizing NS2 and binding CRM1 in the presence or absence of RanGTP. As shown in Figure 5C, a significant resonance response was detected upon addition of CRM1 alone, whereas addition of RanGTP yielded an increase of 1.3-fold. This again can be largely attributed to RanGTP joining the complex, because the mass ratio of CRM1 together with RanGTP (157 kDa) and CRM1 alone (125 kDa) equals 1.26. Interestingly, upon termination of the CRM1 or CRM1/RanGTP stream, most of the complex decayed very slowly. Plotting the two response curves together (Figure 5D) suggests that the binary complex decays faster than the trimeric complex, indicating that the presence of RanGTP stabilizes the CRM1/NS2 interaction, but has little effect on the initial binding.
The NS2 supraNES Is Required for Viral Nuclear Export
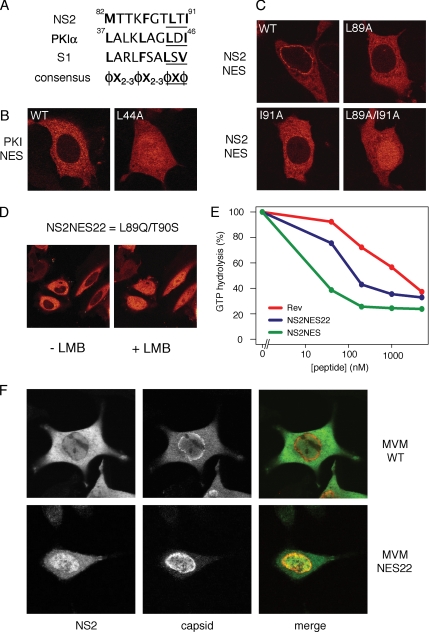
Next, we wanted to evaluate the relevance of the supraphysiological NES observed in NS2 for the viral life cycle. To do so, we compared wild-type MVM virus to an MVM virus carrying an NES of regular strength substituted in place of its supraNES. In regular strength NESs such as the protein kinase inhibitor α (PKIα) NES, mutation of one of two core hydrophobic residues into alanine renders the NES inactive and deficient in nuclear export (Figure 6, A and B; Wen et al., 1994). In contrast, substitution of either of the core hydrophobic amino acids (L89 or I91) of the NS2 NES into alanines converts the NS2 supraNES to a regular NES (Figure 6C). Mutation of both hydrophobic residues is required to completely inactivate the supraNES (Figure 6C).
Figure 6.
The NS2 supraNES is functionally required for viral nuclear export and infectivity. (A) Comparison of NES sequences of NS2, PKIα, and S1. Hydrophobic residues are marked bold. The core regions of the NESs are underlined. φ: L, I, F, M, or V; X: any other residue. (B and C) MCF7 cells were transfected with GFP-tagged NESs for 24 h. (B) Export of the PKIα-NES is inhibited upon mutation of one hydrophobic residue. (C) A single mutation in the NS2 supraNES downgrades the supraNES to a regular NES. To inactivate the NES activity an additional hydrophobic residues needs to be mutated. (D) NES22 behaves like a regular NES. Cells were transfected with GFP-NES22 and imaged using live confocal microscopy. + LMB: cells were treated with LMB for 10 min. (E) NES22 retains CRM1 binding activity NES. Affinity of NES22 was tested in GAP assay as described in Figure 2A. Synthetic peptides were used. NES22 (blue), supraNES (green), and RevNES (red). (F) NES22 viruses fail to be exported and the NS2-NES22 protein is retained in the nucleoplasm. Mouse fibroblasts expressing the WT or NES22 genome for 42 h were stained with anti-NS2 and anti-capsid antibodies.
Interestingly, MVM NS2 NES22 harbors mutations equivalent to those of the L98A mutant, namely L89Q/T90S (T90S likely is a conservative mutation). This NES22 mutant NES has been considered to be defective in nuclear export (Eichwald et al., 2002). On the basis of our NS2 point mutation analysis, we set out to test whether the NES22 mutant is in fact a regular strength NES. Indeed, an NS2-NES22-GFP fusion protein was efficiently exported from the nucleus in HeLa cells (Figure 6D). In addition, the NS2-NES22 NES retained significant affinity toward CRM1 in vitro, albeit substantially lower than the wild-type NS2 NES (Figure 6E). We therefore conclude that the L89Q/T90S mutation in NES22 decreases, but does not abolish CRM1 interaction, yet compromises its supraphysiological capacities. Interestingly, in mouse cells infected with MVM-NES22 virions, the NS2 protein is localized mainly to the nucleus, despite its active NES (Eichwald et al., 2002). Moreover, the MVM-NES22 virus particles are export deficient (Eichwald et al., 2002). To integrate these two observations, we expressed the MVM wild-type or NES22 genome in A9 mouse fibroblasts and monitored NS2 and viral nuclear export 42 h after transfection. As shown in Figure 6F, wild-type virus is efficiently exported from the nucleus and NS2 is predominantly cytoplasmic. In contrast, NES22 virus fails to be exported and NS2 is predominantly in the nucleoplasm. Because the NS2-NES22-GFP fusion protein alone is efficiently exported, these data indicate that in the context of mature viral particle export, the NS2-NES22 protein is retained in the nucleus by the virus and is unable to promote virus export. Measurement of viral production showed that the NS2-NES22 virus is severely hampered in viral propagation (Eichwald et al., 2002). We therefore conclude that the supraphysiological properties of the NS2 NES are required for nuclear export of the mature virus and crucial to the MVM life cycle in natural mouse fibroblast host cells.
DISCUSSION
Supraphysiological NESs were initially identified using a random peptide screen (Engelsma et al., 2004) and given their cytotoxicity, it remained unclear whether this type of NES might be encountered in nature. In this study we identify an NES with extraordinary CRM1-binding characteristics in a viral protein. In retrospect, previous work on the NS2/CRM1 interaction might already have hinted at the unusual characteristics of the NS2/CRM1 interaction, because this interaction was readily detected in whole cell lysates (Eichwald et al., 2002; Miller and Pintel, 2002) even in the absence of nonhydrolyzable forms of RanGTP, which are usually required (Fornerod et al., 1997a). The NS2 NES has been a most useful nuclear transport research reagent (e.g., Daelemans et al., 2002; Petosa et al., 2004), presumably because of its robust CRM1 interaction. The robustness of the NS2/CRM1 interaction may lay the foundation to further address structural and biophysical aspects of CRM1/cargo interactions.
Infectivity of the MVM virus in the host cell requires the supraphysiological characteristics of the NS2 NES. This raises the question why these supraphysiological characteristics have been coopted by MVM to enhance its viral life cycle. It is conceivable that inhibiting CRM1 provides the virus with a cytotoxic mechanism, accelerating cell death and thereby enhancing viral fitness. However, expression of a supraNES did not rescue mutant NES22 infectivity (Figure 3C), consistent with the requirement for intact CRM1 function in virus nuclear export during the viral life cycle (Eichwald et al., 2002; Maroto et al., 2004). Also, inhibition of CRM1 function only occurred at high NS2 expression levels and did not correlate with Nup358 association of NS2 per se. For these reasons it is likely that both the Nup358 interaction and CRM1 relocalization activities of NS2 are effects of its strong CRM1 interaction and do not necessarily imply a primary inhibiting role. Nevertheless, inhibition of endogenous NESs may disrupt trafficking of proteins involved in the antiviral response (e.g., NFκB), enhancing viral fitness on an organismal level. Considering the interphase function of CRM1, NS2 might act as a nuclear export factor for mature parvovirus capsids. Indeed, nuclear egress of MVM is inhibited upon LMB treatment (Miller and Pintel, 2002; Maroto et al., 2004) and NS2 supraNES mutants are defective in nuclear export of MVM viral particles. Furthermore, we observe colocalization of CRM1, NS2, and mature virions in nuclei of virus-infected cells (Figure 4), and NS2 can be retained in the nucleus by mature virus (Figure 6). These data demonstrate the requirement for the supraphysiological properties of the NS2 NES in mediating capsid nuclear export. We have not been able to demonstrate a direct interaction between NS2 and the viral capsid and/or individual capsid proteins in vitro. It is possible, however, that the interaction is highly regulated, complicating the in vitro detection of the export complex. In case of other large CRM1 cargoes, notably the 60S/Nmd3/CRM1 complex, it has similarly proven difficult to demonstrate a direct interaction. Interestingly, Nmd3, the proposed nuclear export adaptor for 60S preribosomal proteins, has recently been reported to behave in a manner reminiscent of supraNESs (West et al., 2007). A mutant of Nmd3 unable to interact with the 60S ribosomal protein localized to the nuclear periphery in a CRM1-dependent manner and stably interacted with nucleoporins. The exact requirements of this interaction remain to be determined. Because of the similarities between 60S preribosomes and parvovirus capsids both in size and abundance, it is tempting to speculate that these large cargoes require an adaptor with high-affinity interactions for CRM1 in order to be exported.
In summary, we identified a supraphysiological NES in a viral protein that is required for viral nuclear export in natural host cells. Our data show that in spite of their cytotoxicity, suprahysiological NESs have evolved in nature and are coopted to enable nuclear export of specific CRM1 cargoes like MVM viral particles. Supraphysiological export signals may serve as a paradigm for adapting cellular transport to cargo-specific needs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ursula Bodendorf (Deutsches Krebsforschungszentrum Heidelberg) for constructing pX-NS2P and pX-NS2-NES8, Frauke Melchior for Nup358 antibodies, Lauran Oomen and Lenny Brocks for assistance with confocal microscopy, Marnix Jansen, Bernike Kalverda, Stijn Heessen, and Nikos Xylourgidis for helpful discussions and comments on the manuscript. N.V. was in part supported by an EMBO short-term postdoctoral fellowship.
Abbreviations used:
- 2Nt
amino-terminus of VP2
- GFP
green fluorescent protein
- LMB
leptomycin B
- MVM
minute virus of mice
- NE
nuclear envelope
- NES
nuclear export signal
- NPC
nuclear pore complex
- NS2
non-structural protein 2
- SAABs
Smn-associated autonomous parvovirus-associated replication bodies
- supraNES
supraphysiological NES
- VP
viral protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0009) on April 2, 2008.
REFERENCES
- Askjaer P., et al. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A., Mattaj I. W. The strategy for coupling the RanGTP gradient to nuclear protein export. Proc. Natl. Acad. Sci USA. 2003;100:1717–1722. doi: 10.1073/pnas.252766999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R., Van Der Velde H., Fornerod M., Pickersgill H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 2004;24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F. R., Gorlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bodendorf U., Cziepluch C., Jauniaux J. C., Rommelaere J., Salome N. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J. Virol. 1999;73:7769–7779. doi: 10.1128/jvi.73.9.7769-7779.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daelemans D., Afonina E., Nilsson J., Werner G., Kjems J., De Clercq E., Pavlakis G. N., Vandamme A. M. A synthetic HIV-1 Rev inhibitor interfering with the CRM1-mediated nuclear export. Proc. Natl. Acad. Sci. USA. 2002;99:14440–14445. doi: 10.1073/pnas.212285299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichwald V., Daeffler L., Klein M., Rommelaere J., Salome N. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J. Virol. 2002;76:10307–10319. doi: 10.1128/JVI.76.20.10307-10319.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsma D., Bernad R., Calafat J., Fornerod M. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 2004;23:3643–3652. doi: 10.1038/sj.emboj.7600370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisst S., Faisst S. R., Dupressoir T., Plaza S., Pijol A., Jauniaux J. C., Rhode S. L., Rommelaere J. Isolation of a fully infectious variant of parvovirus H-1 supplanting the standard strain in human cells. J. Virol. 1995;69:4538–4543. doi: 10.1128/jvi.69.7.4538-4543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura B. M., Faria P. A., Nussenzveig D. R. Viral interactions with the nuclear transport machinery: discovering and disrupting pathways. IUBMB Life. 2005;57:65–72. doi: 10.1080/15216540500078608. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M. Exportin-mediated nuclear export of proteins and ribonucleoproteins. Results Probl. Cell Differ. 2002;35:67–91. doi: 10.1007/978-3-540-44603-3_4. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I. W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997a;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fornerod M., van-Deursen J., van-Baal S., Reynolds A., Davis D., Murti K. G., Fransen J., Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997b;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B. R., Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- Ho J. H., Kallstrom G., Johnson A. W. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Bischoff F. R., Kostka S., Kraft R., Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Lopez-Bueno A., Mateu M. G., Almendral J. M. High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J. Virol. 2003;77:2701–2708. doi: 10.1128/JVI.77.4.2701-2708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bueno A., Valle N., Gallego J. M., Perez J., Almendral J. M. Enhanced cytoplasmic sequestration of the nuclear export receptor CRM1 by NS2 mutations developed in the host regulates parvovirus fitness. J. Virol. 2004;78:10674–10684. doi: 10.1128/JVI.78.19.10674-10684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury K. M., Macara I. G. Ran-binding protein 1 (RanBP1). forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP). inhibition by karyopherin beta. J. Biol. Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- Maroto B., Valle N., Saffrich R., Almendral J. M. Nuclear export of the nonenveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J. Virol. 2004;78:10685–10694. doi: 10.1128/JVI.78.19.10685-10694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. L., Pintel D. J. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 2002;76:3257–3266. doi: 10.1128/JVI.76.7.3257-3266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M. V., Weis K. The direction of transport through the nuclear pore can be inverted. Proc. Natl. Acad. Sci. USA. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T., Nakajima T., Oishi T., Imamoto N., Yoneda Y., Fukamizu A., Yagami K. CRM1 mediates nuclear export of nonstructural protein 2 from parvovirus minute virus of mice. Biochem. Biophys. Res. Commun. 1999;264:144–150. doi: 10.1006/bbrc.1999.1478. [DOI] [PubMed] [Google Scholar]
- Pante N., Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskeva E., Izaurralde E., Bischoff F. R., Huber J., Kutay U., Hartmann E., Lührmann R., Görlich D. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 1999;145:255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petosa C., Schoehn G., Askjaer P., Bauer U., Moulin M., Steuerwald U., Soler-Lopez M., Baudin F., Mattaj I. W., Muller C. W. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol. Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Roth P., Xylourgidis N., Sabri N., Uv A., Fornerod M., Samakovlis C. The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J. Cell Biol. 2003;163:701–706. doi: 10.1083/jcb.200304046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W. F., Lee L. H., Davis L., Sen R. Cytoplasmic sequestration of rel proteins by IkappaBalpha requires CRM1-dependent nuclear export [In Process Citation] Mol. Cell. Biol. 2000;20:2269–2284. doi: 10.1128/mcb.20.6.2269-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F., Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 2003;116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- Walther T. C., Pickersgill H. S., Cordes V. C., Goldberg M. W., Allen T. D., Mattaj I. W., Fornerod M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 2002;158:63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Harootunian A. T., Adams S. R., Feramisco J., Tsien R. Y., Meinkoth J. L., Taylor S. S. Heat-stable inhibitors of cAMP-dependent protein kinase carry a nuclear export signal. J. Biol. Chem. 1994;269:32214–32220. [PubMed] [Google Scholar]
- West M., Hedges J. B., Lo K. Y., Johnson A. W. Novel interaction of the 60S ribosomal subunit export adapter Nmd3 at the nuclear pore complex. J. Biol. Chem., 2007;282:14028–14037. doi: 10.1074/jbc.M700256200. [DOI] [PubMed] [Google Scholar]
- Young P. J., Jensen K. T., Burger L. R., Pintel D. J., Lorson C. L. Minute virus of mice NS1 interacts with the SMN protein, and they colocalize in novel nuclear bodies induced by parvovirus infection. J. Virol. 2002a;76:3892–3904. doi: 10.1128/JVI.76.8.3892-3904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P. J., Jensen K. T., Burger L. R., Pintel D. J., Lorson C. L. Minute virus of mice small nonstructural protein NS2 interacts and colocalizes with the Smn protein. J. Virol. 2002b;76:6364–6369. doi: 10.1128/JVI.76.12.6364-6369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.