Abstract
The bacterium Bacillus subtilis undergoes endospore formation in response to starvation. σ factors play a key role in spatiotemporal regulation of gene expression during development. Activation of σ factors is coordinated by signal transduction between the forespore and the mother cell. σE is produced as pro-σE, which is activated in the mother cell by cleavage in response to a signal from the forespore. We report that expression of SpoIIR, a putative signaling protein normally made in the forespore, and SpoIIGA, a putative protease, is necessary and sufficient for accurate, rapid, and abundant processing of pro-σE to σE in Escherichia coli. Modeling and mutational analyses provide evidence that SpoIIGA is a novel type of aspartic protease whose C-terminal half forms a dimer similar to the human immunodeficiency virus type 1 protease. Previous studies suggest that the N-terminal half of SpoIIGA is membrane-embedded. We found that SpoIIGA expressed in E. coli is membrane-associated and that after detergent treatment SpoIIGA was self-associated. Also, SpoIIGA interacts with SpoIIR. The results support a model in which SpoIIGA forms inactive dimers or oligomers, and interaction of SpoIIR with the N-terminal domain of SpoIIGA on one side of a membrane causes a conformational change that allows formation of active aspartic protease dimer in the C-terminal domain on the other side of the membrane, where it cleaves pro-σE.
Understanding how gene expression is controlled temporally and spatially in living organisms is a fundamental challenge. The sporulation process of the Gram-positive bacterium Bacillus subtilis provides an attractive model to address this challenge. Sporulation is initiated in response to nutrient limitation, and it involves a highly ordered program of gene expression and morphological change (reviewed in Ref. 1). The first morphological change of sporulation is the appearance of an asymmetrically positioned septum that divides the cell into a larger mother cell (MC)3 compartment and a smaller forespore (FS) compartment (Fig. 1). The first compartment-specific transcription factor to become active is σF in the FS (reviewed in Refs. 2, 3). This subunit of RNA polymerase (RNAP) directs transcription of 47 genes (4, 5), including spoIIR, whose product signals activation of σE in the MC (6–8). σE RNAP transcribes over 270 genes (4, 9–11). The products of certain genes under σF control in the FS and other genes under σE control in the MC bring about further morphological change (reviewed in Ref. 1). The mother cell membrane (MCM) (Fig. 1) migrates around the forespore membrane (FSM) during a phagocytic-like process called engulfment. The completion of engulfment involves fusion of the MCM to pinch off the FS within the MC, triggering activation of σG in the FS, which signals activation of σK in the MC. The products of genes under the control of σG and σK bring about maturation of the spore and its release upon lysis of the MC. Hence, much of the temporal and spatial regulation of gene expression during B. subtilis sporulation is tied to two morphological events, formation of the asymmetrically positioned septum and completion of engulfment, and in each case a FS σ (σF or σG) is activated, and this leads to activation of a MC σ (σE or σK).
FIGURE 1.
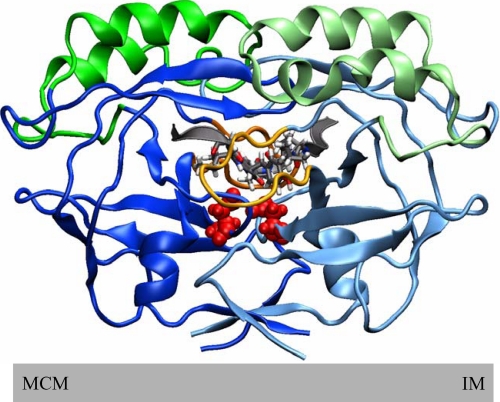
Model of pro-σE processing during sporulation of B. subtilis and in E. coli. Left, asymmetric septation during B. subtilis sporulation generates a smaller forespore (FS) compartment and a larger mother cell (MC) compartment. Middle, expanded view of the septum depicts proteins involved in pro-σE processing. SpoIIR is produced in the FS and is believed to cross the forespore membrane (FSM) and activate pro-σE processing. SpoIIGA and pro-σE are expressed predominantly in the MC. SpoIIGA is predicted to have an N-terminal domain with five transmembrane segments embedded in the mother cell membrane (MCM) and a C-terminal domain in the MC cytoplasm. Pro-σE associates with the MCM via its N-terminal pro-sequence and is believed to be cleaved (arrowhead) by SpoIIGA, releasing active σE into the MC. Right, SpoIIR, SpoIIGA, and pro-σE are produced under T7 RNA polymerase (T7 RNAP) control in E. coli. Middle, expanded view of the E. coli outer membrane (OM), periplasm, and inner membrane (IM). SpoIIR is expected to be secreted to the periplasm where it could interact with the IM-embedded N-terminal domain of SpoIIGA, stimulating the C-terminal domain of SpoIIGAs to cleave IM-associated pro-σE, releasing σE to the cytoplasm. See the text for references.
Here we focus on activation of σE in the MC in response to the SpoIIR signal protein made under σF control in the FS (Fig. 1). σE is synthesized as an inactive precursor, pro-σE, and is activated by cleavage of 27 residues from its N-terminal end (12, 13). The gene (spoIIGB or sigE) encoding pro-σE is cotranscribed in an operon with spoIIGA (14), whose product is necessary for processing of pro-σE to σE (15, 16). Coexpression of pro-σE and SpoIIGA in growing B. subtilis was sufficient for weak expression of a σE-dependent gene, suggesting that SpoIIGA might be a protease that processes pro-σE, and it was noted that SpoIIGA contains a DSG sequence matching the DS/TG sequence important in aspartic proteases (16). However, this hypothesis was not tested, and another group noted sequences in SpoIIGA that suggest it might be a serine protease (17). Several lines of evidence suggested that a gene under σF control in the FS is normally required for pro-σE processing in the MC (16, 18). That gene was identified as spoIIR (6, 7). SpoIIR has a putative N-terminal signal sequence and is believed to be secreted across the FSM (Fig. 1). Consistent with this model, wild-type B. subtilis (but not a spoIIR mutant) engineered to produce σF during growth secreted a factor (presumably SpoIIR) capable of stimulating processing of pro-σE to σE in B. subtilis protoplasts engineered to express SpoIIGA and pro-σE, and SpoIIR partially purified after overexpression in E. coli exhibited this activity (8). Therefore, it was proposed that during sporulation σF RNAP transcribes spoIIR; the SpoIIR synthesized in the FS is secreted across the FSM into the septal space, and SpoIIR activates membrane-embedded SpoIIGA to cleave pro-σE to σE (6–8).
Additional support for the model depicted in Fig. 1 comes from studies of SpoIIGA and pro-σE. A SpoIIGA-LacZ fusion protein appears to be membrane-associated (19), and SpoIIGA-GFP fusion proteins localize to the asymmetrically positioned septum during sporulation (20). The N-terminal half of SpoIIGA is predicted to contain five membrane-spanning segments (16), and the topology shown in Fig. 1 is supported by analysis of phoA and lacZ fusions in Escherichia coli (21). The first 55 residues of pro-σE are sufficient to direct a pro-σE-GFP fusion protein to the asymmetrically positioned septum during sporulation (22), and the ability to associate with the membrane is essential for pro-σE processing (23). The pro-sequence of pro-σE is predicted to form an amphipathic α-helix, but it is not known whether the pro-sequence interacts directly with the membrane surface (as depicted in Fig. 1) or with an integral membrane protein, although SpoIIGA is not necessary for the membrane association of pro-σE (24). The first 29 residues of pro-σE are sufficient for processing (25). Substituting lysine for glutamate at residue 25 of pro-σE impairs processing and the formation of heat-resistant spores (26). A suppressor mutation that restores sporulation and processing was identified in spoIIGA (27). The mutation changed proline to leucine at residue 259 in the predicted C-terminal cytoplasmic domain of SpoIIGA. This P259L mutant form of SpoIIGA also processed wild-type pro-σE and two mutant forms of pro-σE (that were not processed by wild-type SpoIIGA) in addition to the E25K form, indicating that SpoIIGA P259L has broadened specificity for pro-σE processing. Although consistent with the notion that SpoIIGA is the protease that processes pro-σE, the suppression was not allele-specific, so it remained possible that SpoIIGA modifies the activity of another protein that directly cleaves pro-σE (27).
Fig. 1 depicts SpoIIGA and pro-σE associated with the MCM, but the spoIIG operon from which these proteins are expressed is induced prior to formation of the asymmetrically positioned septum during sporulation (28). Therefore, SpoIIGA and pro-σE are presumably also associated with the FSM, yet σE fails to accumulate in the FS. This is not because SpoIIR acts vectorially (i.e. can only stimulate processing in the MC after being secreted from the FS) because coexpression of SpoIIR with SpoIIGA and pro-σE in growing B. subtilis resulted in efficient processing (6), and this depended on translocation of SpoIIR across the membrane but not its release from the membrane (21). Rather, it appears that σE accumulates only in the MC because SpoIIGA and pro-σE are expressed more highly in the MC than in the FS after formation of the asymmetrically positioned septum (29) and because any σE made in the FS is rapidly degraded (29–32).
The observation that SpoIIR can stimulate pro-σE processing in the same cell in which it is synthesized (provided SpoIIGA is also coexpressed) (6, 21) suggested that it might be possible to reconstitute pro-σE processing in a heterologous host, unless an unidentified B. subtilis protein was necessary. We report that coexpression of SpoIIR with SpoIIGA and pro-σE in E. coli is sufficient for accurate, rapid, and abundant processing of pro-σE to σE. We have used the E. coli system to test a model for SpoIIGA function. Our results provide strong evidence that SpoIIGA is an aspartic protease and that its C-terminal cytoplasmic domain forms a dimer similar to the HIV-1 protease and related retroviral proteases. Because SpoIIR was found to interact with SpoIIGA in E. coli, we propose that SpoIIR secreted to the periplasm interacts with extracellular parts of the N-terminal domain of SpoIIGA, causing a conformational change that activates its C-terminal aspartic protease domain to cleave pro-σE, releasing σE from the inner membrane (IM) into the cytoplasm (Fig. 1).
EXPERIMENTAL PROCEDURES
Plasmids—Some of the plasmids used in this study are described briefly in Table 1. All of the plasmids are described fully in supplemental Table S1, and primers used in their construction are listed in supplemental Table S2. All cloned PCR products and all genes subjected to mutagenesis (QuikChange kit, Stratagene) were sequenced to confirm that no undesired mutations were present.
TABLE 1.
Select plasmids used in this study
| Plasmid | Resistancea | Descriptionb |
|---|---|---|
| pID2 | Ap | PT7-spoIIGA-gfp-H6 |
| pID15 | Km | PT7-spoIIR-F2-rbs-sigE-H6 |
| pID16 | Km | PT7-spoIIGA-gfp-H6-rbs-sigE-H6 |
| pID17 | Ap | PT7-spoIIR-H6 |
| pID44 | Km | PT7-spoIIR-F2-rbs-sigE(M171)-H6 |
| pID48 | Ap | PT7-spoIIGA-gfp-F2 |
| pID49 | Km | PT7-spoIIR-F2-(STOP)-rbs-sigE-H6 |
| pID50 | Km | PT7-spoIIR-F2-(STOP)-rbs-sigE(M17I)-H6 |
| pID87 | Ap | PT7-spoIIGA-gfp-F2-rbs-spoIIGA-gfp-H6 |
| pID88 | Km | PT7-spoIIR-F2 |
The abbreviations used are as follows: Ap, ampicillin; Km, kanamycin
PT7 indicates the T7 RNA polymerase promoter and translation initiation sequence; H6 indicates six histidine residues; rbs indicates a ribosome-binding site; F2 indicates two FLAG epitopes. Details of plasmid construction are in supplemental Tables S1 and S2
Cotransformation—Plasmids bearing different antibiotic resistance genes were cotransformed into E. coli strain BL21(DE3) (Novagen) as described previously (33), except ampicillin was used at 50 μg/ml.
Induction of Gene Expression—E. coli BL21(DE3) can be induced to synthesize T7 RNAP by addition of isopropyl β-d-thiogalactopyranoside (IPTG). This strain, bearing plasmids with genes fused to the T7 RNAP promoter, was grown in Luria-Bertani medium containing 50 μg/ml kanamycin sulfate and/or 50 μg/ml ampicillin overnight at 37 °C with shaking. One hundred μl of culture was transferred to 10 ml of fresh Luria-Bertani medium with antibiotics, and incubation was continued at 37 °C with shaking at 350 rpm until the culture reached 80 Klett units. To induce gene expression, IPTG (0.5 mm) was added, and incubation with shaking was continued for 2 h unless noted otherwise.
Western Blot Analysis—E. coli cells from cultures (1 ml) induced as described above were collected by centrifugation (12,000 × g). Samples were prepared by resuspending cells in 100 μl of sample buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 10% (v/v) glycerol, 0.015% bromphenol blue) and boiling for 3 min. Samples were loaded onto SDS-12% polyacrylamide gels for separation of proteins and Western blot analysis as described previously (34). Monoclonal antibody against σE (a gift from W. Haldenwang) was used at 1:1,000 dilution to detect pro-σE and σE, with horseradish peroxidase-conjugated goat anti-mouse IgG (Promega) at 1:1,000 dilution serving as the secondary antibodies. Horseradish peroxidase-conjugated antibodies to pentahistidine (Qiagen) were used at 1:5,000 dilution to detect SpoIIGA-GFP-H6. Horseradish peroxidase-conjugated antibodies to FLAG (Sigma) were used at 1:5,000 dilution to detect SpoIIR-F2 and SpoIIGA-GFP-F2. For comparison with B. subtilis, sporulation of strain PY79 (35) was induced by resuspension of cells in Sterlini-Mandelstam medium as described previously (36). Cells were collected from 1 ml of culture by centrifugation (12,000 × g). Extracts were prepared by resuspending cells in 50 μl of lysis buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm EDTA, 1 mm Pefabloc (Roche Applied Science), 1 mg/ml lysozyme, 10 μg/ml DNase I) and incubating for 10 min at 37 °C, then adding 5 μl of 10% (w/v) SDS and 50 μl of 2× sample buffer and boiling for 3 min.
Purification of Histidine-tagged σE for N-terminal Sequencing—E. coli cells bearing pID15 and pID2 were induced as described above in a 2-liter culture, and cells were harvested by centrifugation (12,000 × g), washed in 25 ml of 10 mm Tris-HCl, pH 7.9, resuspended in 25 ml of buffer H (10 mm Tris-HCl, pH 7.9, 500 mm NaCl, 1 mm β-mercaptoethanol, 10 mm imidazole) containing protease inhibitor (Complete Mini EDTA-free, Roche Applied Science), then passed twice through a French pressure cell (American Instruments) at 14,000 pounds/inch2 (96 MPa). The lysate was centrifuged (12,000 × g) at 4 °C for 10 min, and the supernatant was centrifuged again. The supernatant was centrifuged (200,000 × g) at 4 °C for 1 h in an SW50.1 rotor (Beckman Instruments). To the 20-ml supernatant, 20 μl of Triton X-100 was added and mixed with 500 μl of nickel-nitrilotriacetic acid beads (Qiagen) that had been previously washed with 5 ml of buffer H three times. The mixture was rotated for 45 min and then poured into a Cell Thru 2-ml disposable column (Clontech). The beads were washed with 1 ml of buffer H containing protease inhibitor and 0.1% Triton X-100, 4 ml of buffer H containing protease inhibitor and 40 mm imidazole, 4 ml of buffer H containing protease inhibitor and 60 mm imidazole and eluted with 1 ml of elution buffer (10 mm Tris-HCl, pH 7.9, 50 mm NaCl, 1 mm β-mercaptoethanol, 250 mm imidazole). To the 1-ml eluted sample, 0.3 g of ammonium sulfate was added with mixing. The mixture was centrifuged (12,000 × g) for 10 min to collect precipitated proteins. The precipitate was resuspended in 20 μl of sample buffer and boiled for 3 min. Proteins in a 5-μl sample were separated on an SDS-12% polyacrylamide gel, electroblotted to Sequi-Blot polyvinylidene difluoride membranes (Bio-Rad), stained with Coomassie solution (0.1% Coomassie Brilliant Blue R-250, 1% acetic acid, 40% methanol), destained with 50% methanol, and the fastest migrating protein, σE-H6, was sequenced by Edman degradation at the Michigan State University Macromolecular Structure Facility.
Fractionation of Cellular Proteins—E. coli from cultures (100 ml) induced as described above were fractionated as described previously (33).
Modeling of the SpoIIGA C-terminal Domain—After identification of the HIV-1 retroviral protease structure as the most suitable template and alignment of the sequences, a model of the C-terminal domain of SpoIIGA was built as follows. First, the crystal structure of unliganded, tethered HIV-1 protease (Protein Data Bank code 1G6L) (37) was used to build a templated structure for the aligned sections. The short insertions at residues 173, 250, 268, and 283 were then modeled with the loop modeling protocol in MODELER (38). The longer insertion containing the three-helix bundle from residues 193 to 235 was modeled ab initio with a fragment modeling procedure based on lattice-based sampling (39) as implemented in the MMTSB Tool Set (40). A dimer was assembled based on HIV-1 protease. To refine the structure further, residues 22–33 of the pro-σE substrate, which encompass the proteolytic cleavage site (after residue 27), were modeled into the dimer in analogy to the substrate positioning in the crystal structure of an inactive variant of feline immunodeficiency virus protease (Protein Data Bank code 3FIV) (41). The model was then subjected to force field-based minimization over 1000 steps with distance-dependent dielectric and weak restraints on C-α atoms using the program CHARMM (42) and the CHARMM force field (43) followed by 10 ps of molecular dynamics with weakened restraints and Generalized Born with Molecular Volume implicit solvent (44) to allow further relaxation.
Determination of Relative Processing Efficiency—To quantify the effect of mutations in spoIIGA on pro-σE processing, the band intensities of pro-σE(M17I)-H6 and σE-H6 were measured using ImageJ 1.38 software (National Institutes of Health) analysis of Western blot images. Bands in the linear response range were quantified, and the range was established by analysis of Western blot images of samples containing different amounts (10% increments) of pro-σE(M17I)-H6 and σE-H6. The ratio of σE-H6 band intensity to that of pro-σE(M17I)-H6 for E. coli coexpressing a mutant SpoIIGA-GFP-F2 protein was normalized to the corresponding ratio for cells coexpressing wild-type SpoIIGA-GFP-F2 in the same experiment, yielding the relative processing efficiency. The relative processing efficiency for wild-type SpoIIGA-GFP-F2 was determined in the same way by normalizing the ratios from three independent cultures to that from a fourth independent culture in the same experiment.
Membrane Solubilization and Affinity Purification—E. coli from cultures (500 ml) induced as described above were harvested by centrifugation (12,000 × g) and resuspended in 6 ml of PBS, pH 7.2 (Invitrogen), containing 10 mm imidazole and then passed twice through a French pressure cell (American Instruments) at 14,000 pounds/inch2 (96 MPa). The lysate was centrifuged (12,000 × g) at 4 °C for 10 min, and the supernatant was centrifuged again. To the 5-ml supernatant, 250 μl of 20% (w/v) Sarkosyl (sodium dodecanoyl sarcosine; Anatrace) was added, and the sample was rotated at room temperature for 30 min and then centrifuged (200,000 × g) in an SW50.1 rotor (Beckman) at 4 °C for 1 h. A sample (50 μl) of the supernatant was taken before affinity purification. The rest of the supernatant was mixed with 500 μl of nickel-nitrilotriacetic acid beads (Qiagen) prepared by washing with 5 ml of PBS three times, and the mixture was rotated for 30 min and then poured into a Cell Thru 2-ml disposable column (Clontech). The beads were washed with 4 ml of PBS containing 10 mm imidazole and 1% Sarkosyl, and 4 ml of PBS containing 40 mm imidazole and 1% Sarkosyl, then eluted with 1 ml of PBS containing 250 mm imidazole and 1% Sarkosyl. Samples (50 μl) from before and after affinity Ni2+-affinity purification were mixed with 50 μl of 2× sample buffer and boiled for 3 min prior to Western blot analysis.
RESULTS
Processing of Pro-σE in a Heterologous Host—To establish the minimum requirements for pro-σE processing, we expressed B. subtilis proteins in E. coli. The putative protease SpoIIGA was C-terminally tagged with green fluorescent protein and His6, creating SpoIIGA-GFP-H6. The green fluorescent protein tag enhanced accumulation of SpoIIGA-GFP-H6 relative to SpoIIGA-H6 in E. coli engineered to express each protein from a T7 RNAP promoter, as determined by Western blot analysis of whole-cell extracts with penta-His antibodies (data not shown). Coexpression of SpoIIGA-GFP-H6 with its putative substrate, pro-σE-H6, resulted in three species (Fig. 2, lane 2). The one in the top panel of Fig. 2 is SpoIIGA-GFP-H6. The two in the bottom panel of Fig. 2 are pro-σE-H6 and a faster migrating species (labeled “Smaller pro-σE-H6”) that proved to result from translation initiation at an alternative start codon within sigE, the gene that encodes pro-σE (see below). The Smaller pro-σE-H6 was observed when only pro-σE-H6 was expressed (data not shown) or when pro-σE-H6 and SpoIIR-F2 (SpoIIR C-terminally tagged with 2-FLAG) were coexpressed (Fig. 2, lane 1). Coexpression of pro-σE-H6 and SpoIIR-F2 also resulted in a species that migrated slightly more slowly than pro-σE-H6 (Fig. 2, lane 1, bottom panel). This slower migrating species (Fig. 2, labeled “Larger pro-σE-H6”) was because of translation initiation at a start codon within the F2 tag, upstream of sigE (see below). When all three proteins (SpoIIGA-GFP-H6, pro-σE-H6, and SpoIIR-F2) were coexpressed from two different combinations of plasmids, a new species appeared in each case (Fig. 2, lanes 3 and 4, bottom panel). The new species (from cells containing the same plasmids as in Fig. 2, lane 3) was purified using metal-affinity chromatography followed by separation on an SDS-12% polyacrylamide gel, and after electroblotting and staining its N-terminal amino acid sequence was determined by Edman degradation to be YIGGS. This sequence is identical to the N-terminal amino acid sequence of σE purified from sporulating B. subtilis (13). We conclude that both SpoIIGA and SpoIIR are necessary for processing of pro-σE in E. coli, and that no other B. subtilis proteins are required for accurate processing in this heterologous host.
FIGURE 2.
SpoIIGA and SpoIIR are necessary and sufficient for processing of pro-σE in E. coli. Western blot analysis of SpoIIGA-GFP-H6, SpoIIR-F2, and pro-σE-H6/σE-H6 using antibodies against penta-His, FLAG, and σE, respectively. Cells were collected 2 h after IPTG induction. Samples were prepared from E. coli bearing pID15 to produce SpoIIR-F2 and pro-σE-H6 (lane 1, 0.5 μl), pID16 to produce SpoIIGA-GFP-H6 and pro-σE-H6 (lane 2, 2.5 μl), or pID15 and pID2 (lane 3, 5 μl), or pID16 and pID17 (lane 4, 5 μl) to produce all three proteins, and the indicated amounts of extract were loaded.
Improvements to the E. coli System for Studying Pro-σE Processing—The plasmid designed to coexpress pro-σE-H6 and SpoIIR-F2 produced two unexpected species that were detected with antibodies against σE (Fig. 2, lanes 1 and 3, bottom panel). This plasmid contains a T7 RNAP promoter followed by the gene for SpoIIR-F2 and then the gene for pro-σE-H6. The coding sequence for the F2 tag contains an out-of-frame ATG that is in-frame with the coding sequence for pro-σE-H6. We hypothesized that translation initiation at this alternative start codon produces the slower migrating species detected with σE antibodies (Fig. 3, lane 1). Consistent with this hypothesis, introduction of a stop codon in-frame with the ATG and downstream of it (between the genes for SpoIIR-F2 and pro-σE-H6) eliminated the slower migrating species (Fig. 3, lane 2).
FIGURE 3.
Improvement of the E. coli system for studying pro-σE processing. Western blot analysis of pro-σE-H6/σE-H6 using antibodies against σE. Cells were collected 2 h after IPTG induction. Samples were prepared from E. coli bearing pID15 to produce SpoIIR-F2 and pro-σE-H6 (lane 1), pID49 with a stop codon to eliminate larger pro-σE-H6 (lane 2), pID44 with a substitution that eliminates smaller pro-σE-H6 and produces pro-σE(M17I)-H6 (lane 3), or pID50 with both mutations (lane 4). All strains contained pID48 to produce SpoIIGA-GFP-F2, and 5 μl of sample was loaded in each lane.
We hypothesized that the unexpected species that migrates between pro-σE-H6 and σE-H6 (Fig. 3, lane 1) is because of translation initiation at one of four potential alternative start codons within sigE. We changed the nucleotide at the third position of each potential start codon. Only changing codon 17 eliminated the unexpected species (Fig. 3, lane 3). This change, from ATG to ATT, results in substitution of Ile for Met at position 17 of pro-σE-H6 (designated pro-σE(M17I)-H6), which does not interfere with processing (Fig. 3, lane 3).
To eliminate both unexpected species, the two mutations (i.e. the stop codon between the genes for SpoIIR-F2 and pro-σE-H6, and the M17I substitution in pro-σE-H6) were engineered into the same plasmid (Fig. 3, lane 4). The other improvement we made involved changing the C-terminal tag of SpoIIGA-GFP from H6 to F2. SpoIIGA-GFP-F2 was easier to detect by Western blotting (see below) than SpoIIGA-GFP-H6 (Fig. 2, lanes 2–4, top panel), and both proteins allowed a similar level of σE-H6 to accumulate (SpoIIGA-GFP-F2 was used in the experiment shown in Fig. 3; compare Fig. 2, lanes 3 and 4, bottom panel, in which SpoIIGA-GFP-H6 was used).
Therefore, subsequent experiments to study pro-σE processing in E. coli used a system in which SpoIIR-F2 and pro-σE(M17I)-H6 were expressed from one plasmid (pID50), eliminating the unexpected species, and SpoIIGA-GFP-F2 was expressed from another plasmid (pID48), facilitating detection of wild-type or mutant forms of the putative protease.
Characterization of the E. coli System—Expression of SpoIIGA-GFP-F2 from one plasmid inhibited expression of SpoIIR-F2 and pro-σE(M17I)-H6 from the other by about 10-fold. E. coli transformed with only the plasmid designed to express SpoIIR-F2 and pro-σE(M17I)-H6, began to accumulate both proteins by 10 min after induction of T7 RNAP, and both proteins reached a maximum level by 30 min (Fig. 4A, lanes 1–6). Cotransformation of E. coli with both plasmids (one expressing SpoIIR-F2 and pro-σE(M17I)-H6 and conferring kanamycin resistance, and the other expressing SpoIIGA-GFP-F2 and conferring ampicillin resistance), followed by growth in the presence of both antibiotics and induction of T7 RNAP, resulted in the rapid accumulation of all three proteins (Fig. 4A, lanes 7–12), but the maximum levels of SpoIIR-F2 and pro-σE(M17I)-H6 were about 10-fold less than in E. coli containing only the plasmid for expression of these two proteins (Fig. 4A, lanes 7–12 contain proteins from 10-fold more cells than lanes 1–6). The reduction in expression could be due, at least in part, to effects of one plasmid on the copy number of the other. In any case, the level of σE-H6 produced reached a maximum by 10–30 min post-induction and remained unchanged at least until 180 min, and the level of SpoIIGA-GFP-F2 showed a similar pattern (Fig. 4A, lanes 7–12). We chose 120 min to evaluate pro-σE(M17I)-H6 processing and the level of wild-type or mutant forms of SpoIIGA-GFP-F2 in subsequent experiments (see below). Under the conditions used in the experiment shown in Fig. 4A, pro-σE(M17I)-H6 was processed rapidly, making it difficult to observe time-dependent cleavage of pro-σE(M17I)-H6 to σE-H6. By decreasing aeration of the culture (i.e. shaking at 120 rpm rather than 350 rpm), it was possible to observe the level of pro-σE(M17I)-H6 to increase between 10 and 30 min after induction of T7 RNAP, and σE-H6 increased between 30 and 60 min after induction (supplemental Fig. 1, lanes 1–4). Addition of the translation inhibitor chloramphenicol to the culture 10 min after the induction of T7 RNAP blocked further accumulation of pro-σE(M17I)-H6 but did not block the appearance of σE-H6 by 60 min (supplemental Fig. 1, lanes 5–8), indicating that pro-σE(M17I)-H6 is cleaved to σE-H6 post-translationally.
FIGURE 4.
Characterization of the E. coli system. A, time courses of protein accumulation. Western blot analysis of SpoIIGA-GFP-F2 and SpoIIR-F2 using antibodies against FLAG, and of pro-σE(M17I)-H6/σE-H6 using antibodies against σE. Cells were collected at the indicated times after IPTG induction. Samples were prepared from E. coli bearing pID50 (lanes 1–6, 0.5 μl) or pID50 and pID48 (lanes 7–12, 5 μl). B, comparison of pro-σE processing in E. coli and in sporulating B. subtilis. Western blot analysis of pro-σE(M17I)-H6/σE-H6 (lane 1) and pro-σE/σE (lanes 2–6) using antibodies against σE. E. coli bearing pID50 and pID48 were collected 2 h after IPTG induction (lane 1). Proteins from an approximately equal number of B. subtilis cells (based on cell density measured in Klett units) were analyzed at the indicated times after the initiation of sporulation (lanes 2–6). C, fractionation of cell extracts. E. coli cells bearing pID50 and pID48 were induced with IPTG for 2 h. The whole-cell extract (lane 1), supernatant after low speed (12,000 × g) centrifugation (lane 2), and cytosolic (lane 3) and cell envelope (lane 4) fractions from the same number of cells were subjected to Western blot analysis with antibodies against FLAG (upper and middle) or σE (lower).
To compare processing of pro-σE in E. coli with that during sporulation of B. subtilis, proteins from an approximately equal number of IPTG-induced E. coli or sporulating B. subtilis cells were analyzed by Western blotting. Pro-σE(M17I)-H6 and σE-H6 accumulated to much higher levels in E. coli (Fig. 4B, lane 1) than did pro-σE and σE in sporulating B. subtilis cells (Fig. 4B, lanes 2–6). The ratio of σE to pro-σE in B. subtilis cells was higher than the ratio of σE-H6 to pro-σE(M17I)-H6 in E. coli, suggesting that processing is more efficient during sporulation. However, on a per cell basis, E. coli produced much more σE-H6 than B. subtilis produced σE.
To characterize the E. coli system with respect to protein localization, cell lysates produced using a French press were subjected to low speed centrifugation to remove cell debris, followed by high speed centrifugation to separate cytosolic (supernatant) from cell envelope (pellet) fractions. SpoIIGA (19, 20) and pro-σE (22, 24, 29) are membrane-associated during B. subtilis sporulation and therefore were expected to be in the E. coli cell envelope fraction, whereas σE was expected to be in the cytosolic fraction (22, 24, 29). The expectation was less clear for SpoIIR, as this protein is believed to be secreted from the B. subtilis forespore (6–8), so it may be secreted to the periplasm of E. coli, where it might interact with extracellular parts of SpoIIGA (21). As expected, SpoIIGA-GFP-F2 and pro-σE(M17I)-H6 were in the E. coli cell envelope fraction (Fig. 4C, lane 4), whereasσE-H6 was in the cytosolic fraction (Fig. 4C, lane 3). Interestingly, SpoIIR-F2 was in the cell envelope fraction (Fig. 4C, lane 4), consistent with the notion that it is secreted to the periplasm and interacts with a membrane protein(s).
Similarity of SpoIIGA to Aspartic Proteases—The E. coli system described above will facilitate mutagenesis studies aimed at defining features of SpoIIGA, SpoIIR, and pro-σE that are critical for accurate and efficient processing. Here, we focus on testing previous suggestions that the C-terminal intracellular domain of SpoIIGA forms an aspartic protease upon dimerization (16) or forms a serine protease (17).
BLAST analysis (45) did not reveal similarity of the predicted SpoIIGA intracellular domain (from Ser-147 to the C terminus of the protein at Ser-309) to known proteases. However, a short region of the domain exhibited similarity to aspartic proteases (supplemental Fig. 2) using the HHpred algorithm (46). Near the C-terminal end of most of the aligned sequences is the sequence DTG or DSG, in which Asp is known to be the catalytic aspartate residue for some of the proteases and therefore is predicted to be the catalytic aspartate residue for the others, including Asp-183 of SpoIIGA.
Further analysis of the C-terminal domain sequence of SpoIIGA with remote homology and fold recognition tools through the BioInfoBank MetaServer (47) identified structures of prophytepsin, an aspartic protease from barley (48), of a secreted aspartic protease from Candida albicans (49, 50), and of HIV-1 protease, also an aspartic protease. These aspartic proteases were identified primarily through a match of predicted secondary structures for the SpoIIGA sequence with structures from the Protein Data Bank in combination with a comparison of the primary sequences. The corresponding structures could be used as a template to predict models for the C-terminal structure of SpoIIGA. Closer inspection based on the agreement of the secondary structure elements suggested that the HIV-1 protease structure would be the most suitable template. The alignment of HIV-1 protease with SpoIIGA in Fig. 5 shows that the sheet regions and the short C-terminal helix of HIV-1 protease are matched well to the predicted secondary structure of SpoIIGA. However, SpoIIGA also appears to have a three-helix bundle at residues 200–230 inserted into the structure that is absent in HIV-1 protease. Based on this alignment, a model of the C-terminal domain of SpoIIGA was built as a dimer in complex with a short segment of pro-σE around the cleavage site (see “Experimental Procedures”).
FIGURE 5.
Alignment of the predicted secondary structure of the C-terminal domain of SpoIIGA with that for HIV-1 protease. Numbers at the top refer to residues in SpoIIGA. Predicted regions of α-helix (H) and β-sheet (E) are shown above the SpoIIGA sequence and below the HIV-1 protease (PR) sequence. Identical residues in the two proteins are indicated with an asterisk.
The final model is shown in Fig. 6. It resembles in part the familiar HIV-1 protease structure with the catalytic aspartate (Asp-183 in the SpoIIGA model) at the center of the dimer. The additional three-helix bundle is predicted to be packed on either side of the flaps that in HIV-1 protease control substrate access to the active site. Another difference is the longer loop at residues 281–292 of SpoIIGA that protrudes from the dimer near the substrate and is predicted to play a role in substrate binding and recognition. Also, the predicted structure appears more closed compared with HIV-1 protease; however, opening of the dimer to allow substrate entry by rotating the flap with the entire attached subdomain as seen in recent simulation studies of HIV-1 protease (51) remains possible.
FIGURE 6.
Model of the SpoIIGA C-terminal domain dimer with a short segment of pro-σE in the active site. The two chains of the SpoIIGA dimer are colored predominantly blue and light blue, with arrows and helices indicating β-sheet and α-helix secondary structures, respectively. Each chain has an extra three-helix bundle (residues 200–230; colored green and light green) not found in HIV-1 protease and a longer loop (residues 281–292; colored orange on both chains) than in HIV-1 protease. The putative catalytic aspartate residues (Asp-183; colored red on both chains and showing the van der Waals surfaces) are below a segment of the pro-σE substrate (residues 22–33; colored gray and shown as a ribbon at the ends, and with atoms shown and colored according to atom type for residues 25–30). Above the substrate are the β-hairpin flaps of each chain. The model is shown in the orientation typically shown for HIV-1 protease. This is upside down relative to Fig. 1, and the gray bar representing the MCM of B. subtilis or the IM of E. coli in Fig. 1 is shown below the model to emphasize this point and to indicate the parts of the C-terminal domain of SpoIIGA that are predicted to be near the membrane surface.
Mutational Analysis of SpoIIGA—The homology model (Fig. 6) and the HHpred result (supplemental Fig. 2) predict that Asp-183 is the catalytic aspartate residue of SpoIIGA. Moreover, Asp is perfectly conserved at the corresponding position in SpoIIGA orthologs and lies within the most highly conserved sequence, (GAS)-X(ILV)D(ST)GNXLXXP, in their C-terminal domains (supplemental Fig. 3). A D183A substitution in SpoIIGA-GFP-F2 abolished processing of pro-σE(M17I)-H6 when the two proteins were coexpressed with SpoIIR-F2 in E. coli (Fig. 7, lane 2, bottom panel). The D183A mutant protein accumulated normally (Fig. 7, lane 2, top panel). A conservative substitution, D183E, also abolished processing despite accumulation of the mutant SpoIIGA-GFP-F2 protein (Fig. 7, lane 3). A D183N substitution produced similar results (Fig. 7, lane 35). We conclude that Asp-183 is critical for SpoIIGA function, consistent with our predictions that SpoIIGA is an aspartic protease and that Asp-183 is the catalytic aspartate residue.
FIGURE 7.
Effects of mutations in spoIIGA on pro-σE processing in E. coli. E. coli cells bearing pID50 to produce SpoIIR-F2 and pro-σE(M17I)-H6, and pID48 to produce SpoIIGA-GFP-F2 (WT) or a derivative of pID48 to produce SpoIIGA-GFP-F2 with the indicated substitution(s) or truncation, were induced with IPTG for 2 h. Samples (5 μl) were subjected to Western blot analysis with antibodies against FLAG (upper) or σE (lower). Vertical gaps between panels separate different experiments. In some panels, intervening lanes were deleted from the image. For each mutation in spoIIGA, a representative result from at least two independent experiments is shown.
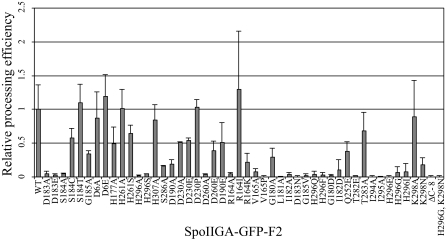
Substitutions in the highly conserved sequence around Asp-183 interfered with SpoIIGA function. Neither these substitutions nor others described below had much effect on the ability of the mutant SpoIIGA-GFP-F2 proteins to accumulate (Fig. 7, top panels), suggesting that the mutant proteins retain their overall structure. Some SpoIIGA orthologs have Ala at the position corresponding to Gly-180 in B. subtilis SpoIIGA (supplemental Fig. 3), as do some aspartic proteases (supplemental Fig. 2). G180A reduced but did not abolish processing (Fig. 7, lane 32, bottom panel). To quantify the effect, the ratio of the σE-H6 signal to the pro-σE(M17I)-H6 signal was measured in two or more separate experiments, and the average was normalized to that for wild-type SpoIIGA-GFP-F2 (Fig. 8). The “processing efficiency” of the G180A mutant SpoIIGA-GFP-F2 protein was about 30% of the wild type. In the model (Fig. 6), Gly-180 is located in the middle of the central β-sheet participating in hydrophobic core formation (with Val-167, Ala-169, Val-178, Ile-182, Ala-293, and Ile-295). The G180A substitution would maintain the hydrophobic core, but distortions because of the larger alanine side chain may affect the geometry of the nearby active site. On the other hand, the model predicted that changing Gly-180 to a residue with a polar or charged side chain would be more disruptive of the hydrophobic core and therefore greatly impair enzyme activity. In agreement with this prediction, G180D abolished pro-σE(M17I)-H6 processing (Fig. 7, lane 42). Similarly, at the position corresponding to Ile-182, which is part of the same hydrophobic cluster in the model, SpoIIGA orthologs have Val, Ile, or Leu, as do aspartic proteases with three exceptions that have Phe (supplemental Figs. 2 and 3). Consequently, nonconservative substitutions for Ile-182 were predicted to impair processing. Both I182A (Fig. 7, lane 34) and I182D (Fig. 7, lane 43) abolished processing, consistent with the predictions. At the position corresponding to Leu-181, aspartic proteases and SpoIIGA orthologs have an amino acid residue with a bulky hydrophobic (Val, Ile, or Leu) or aromatic (Phe or Tyr) side chain (supplemental Figs. 2 and 3). In the model (Fig. 6), Leu-181 is predicted to interact with both tyrosines adjacent to the cleavage site in pro-σE and would likely be involved in substrate recognition. Consistent with this specific role of Leu-181, L181A abolished processing (Fig. 7, lane 33). The Ser-184 side chain is predicted to stabilize the dimer interface immediately below the active site, forming the “fireman's grip” (52, 53). Based on mutational analysis of the HIV-1 protease (54), which has Thr-26 corresponding to Ser-184 in SpoIIGA (supplemental Fig. 2), S184T and S184C were predicted to allow processing, perhaps with reduced efficiency, and S184A was predicted to abolish processing. In agreement, processing was undiminished for S184T, reduced 2-fold for S184C, and abolished for S184A (Fig. 7, lanes 4–6, and Fig. 8). At the position corresponding to Gly-185, a Gly is perfectly conserved in SpoIIGA orthologs (supplemental Fig. 3) and almost perfectly conserved in the HHpred alignment (supplemental Fig. 2). G185A reduced processing efficiency to about 40% of the wild type, and G185V abolished processing (Fig. 7, lanes 7 and 38, and Fig. 8). Gly-185 is positioned immediately adjacent to Asp-183 in the model (Fig. 6) with its C-α hydrogen that is replaced by a larger side chain in other residues pointing directly at the catalytic aspartate. Consequently, any side chain other than the hydrogen of Gly-185 is expected to affect the active site, and larger side chains are expected to lead to more serious disruptions than smaller side chains, consistent with the greater effect we observed on processing for G185V than G185A. The HHpred alignment ends at the position corresponding to Gly-185 (supplemental Fig. 2), so we did not continue to make substitutions at every residue in the C-terminal direction, although it seems likely that Asn-186, Leu-188, and Pro-191 play important roles, given the conservation of these residues at the corresponding positions of SpoIIGA orthologs (supplemental Fig. 3). From the model, Asn-186 and Leu-188 are predicted to be involved in substrate recognition, whereas Pro-191 might be critical in maintaining stability of the central β-sheet. Our mutational analysis of amino acid residues adjacent to Asp-183 in the SpoIIGA sequence, together with their conservation in SpoIIGA orthologs and aspartic proteases, demonstrate their importance for SpoIIGA function and strongly support the model that SpoIIGA is an aspartic protease.
FIGURE 8.
Quantification of the effects of mutations in spoIIGA on pro-σE processing. Relative processing efficiency was determined as described under “Experimental Procedures” from two to five independent cultures. Bars indicate averages, and error bars indicate one S.D. Results for mutant SpoIIGA proteins are presented in the same order as in Fig. 7.
Several other amino acid residues crucial for SpoIIGA function were identified by making substitutions based on the homology model and sequence alignments. In the model (Fig. 6), Ile-294 and Ile-295 form part of a β-sheet secondary structure predicted to help position the catalytic Asp-183 in the active site of the enzyme. SpoIIGA orthologs have Ile or a similar residue (Val, Leu, or Met) at the corresponding positions (supplemental Fig. 3). We hypothesized that a residue with a bulky hydrophobic side chain is important at these positions. Indeed, I294A and I295A abolished processing (Fig. 7, lanes 47 and 48). In the HIV-1 protease dimer, a loop of each subunit acts as a mobile flap that controls substrate access to the active site (55, 56). In our homology model of the C-terminal domain of SpoIIGA (Fig. 6), Gln-252 is predicted to be in the center of the flap, with Gln-252 from both chains interacting directly with each other. We hypothesized that substitutions at this position would interfere with flap function. In particular, changing Gln-252 to a charged residue would be expected to hinder flap closure because of electrostatic repulsion of like charges. In agreement, Q252E reduced processing efficiency to about 40% of the wild type (Fig. 7, lane 44, and Fig. 8). In the model (Fig. 6), Thr-282 is semi-buried at the beginning of the longer loop (residues 281–292) as compared with the HIV-1 protease structure. Thr-282 interacts with the flaps from the side, but this residue may also be involved in proper positioning of the longer loop to interact with the substrate. T282E abolished processing (Fig. 7, lane 45), consistent with a disruption of the local structure because of the introduction of a charge into a semi-buried environment. On the other hand, T283A, which would be expected to be less disruptive based on the model (Fig. 6), had less of an effect on processing (Fig. 7, lane 46, and Fig. 8). Several retroviral proteases exhibit an RP sequence shifted by two amino acids from an RV sequence in SpoIIGA in the HHpred alignment (supplemental Fig. 2). In the secondary structure-based alignment (Fig. 5) that was used to construct the model (Fig. 6), the RP sequence is aligned with the RV sequence. Although the RV sequence is not highly conserved among SpoIIGA orthologs (supplemental Fig. 3), we found that R164A, V165A, and V165P abolished processing, and a conservative substitution, R164K, reduced processing efficiency to about 25% of the wild type (Fig. 7, lanes 27 and 29–31, and Fig. 8). A nonconservative substitution, R164I, did not impair processing (Fig. 7, lane 28). This substitution was made because several SpoIIGA orthologs have Ile at the position corresponding to Arg-164 in B. subtilis. According to the model (Fig. 6), Arg-164 and Val-165 would contribute to the overall stability of the structure, but it is not clear why SpoIIGA function would be sensitive to an R164A substitution and insensitive to an R164I substitution.
The C-terminal tip of SpoIIGA is also important for processing. In the HIV-1 protease dimer, the C-terminal tails of the two subunits interact with each other and with the N-terminal tails (57). This feature is conserved in the homology model of the SpoIIGA C-terminal domain (Fig. 6). Also conserved is the length of the C-terminal domain, especially for SpoIIGA orthologs of closely related species (Bacilli, Geobacilli, and Oceanobacillus in supplemental Fig. 3). These observations suggested that SpoIIGA might be sensitive to a small truncation at its C-terminal end. We found that deletion of the last eight amino acid residues (ΔC-8) abolished processing (Fig. 7, lane 54). Interestingly, the SpoIIGA orthologs of Clostridia and several other species lack 6–9 residues at their C-terminal end as compared with B. subtilis SpoIIGA (supplemental Fig. 3).
Whereas our mutational and computational analyses strongly support the model that SpoIIGA is an aspartic protease, neither approach supports the suggestion that it is a serine protease (17). Although serine proteases were not identified with any of the bioinformatics servers, one may entertain the possibility of modeling the C-terminal domain of SpoIIGA based on known serine protease structures. Known serine protease structures generally consist of folds with more than 200 or 300 residues, inconsistent with the relatively short C-terminal domain of SpoIIGA. Furthermore, the secondary structure pattern generally does not match the predicted secondary structure for SpoIIGA. In particular, the chymotrypsin-like family consists of two six-stranded β-barrels with an active site composed of serine from one barrel and histidine from the other one (58). The C-terminal region of SpoIIGA is predicted to also consist predominantly of β-strands, but the sequence is not long enough to form two β-barrels. The subtilisin family consists of an α/β fold with alternating β-strands and α-helices predominantly (59), which is inconsistent with the predicted secondary structure of SpoIIGA (Fig. 5). The α/β hydrolase fold family of serine proteases differs from subtilisin but also consists of alternating α-helices and β-strands (60), inconsistent with the predicted SpoIIGA secondary structure. Other known serine protease structures are also inconsistent with the SpoIIGA sequence length and predicted secondary structure. However, it is conceivable that SpoIIGA represents a new class of serine proteases.
Most serine proteases form a catalytic triad composed of Ser, Asp, and His residues (61). Conserved Ser and Asp residues, and all His residues, in the SpoIIGA C-terminal domain were subjected to mutational analysis. No Ser residue is perfectly conserved among SpoIIGA orthologs, but Ser is highly conserved at the position corresponding to Ser-286 in B. subtilis SpoIIGA (supplemental Fig. 3). However, S286A did not abolish processing, although it did reduce processing efficiency to about 20% of the wild type (Fig. 7, lane 18, and Fig. 8). We conclude that Ser-286 is not the catalytic serine of a serine protease, although it is fairly important for SpoIIGA function. In our homology model based on HIV-1 aspartic protease (Fig. 6), Ser-286 is located at the tip of the longer loop (residues 281–292 of SpoIIGA), in direct contact with the substrate. It is expected to be involved in substrate binding and/or recognition. Asp or Glu is highly conserved among SpoIIGA orthologs at the positions corresponding to Asp-190 and Asp-260 in B. subtilis SpoIIGA, and Asp is perfectly conserved among closely related species at the position corresponding to Asp-230 in B. subtilis SpoIIGA (supplemental Fig. 3). However, substitutions at these positions did not abolish processing, with the exception of D260A (Fig. 7, lanes 19–24 and 26, and Fig. 8). Conservative Asp to Glu substitutions at positions 190, 230, and 260 reduced processing efficiency about 2-fold, whereas D183E abolished processing (Fig. 7, lane 3). Clearly, Asp-183 is more sensitive to substitution than other conserved Asp residues in the SpoIIGA C-terminal domain. In our model (Fig. 6), Asp-190 forms a salt bridge with Arg-245 that might be important for structural stability in the substrate binding cavity, whereas Asp-260 interacts closely with both Lys-275 and Lys-258, again possibly stabilizing the overall structure. Of the four His residues in the SpoIIGA C-terminal domain, only the one at the position corresponding to His-296 in B. subtilis is conserved among orthologs, mainly in closely related species (supplemental Fig. 3). Nevertheless, substitutions were made at all four positions (His-177, His-261, His-296, and His-307), and only those for His-296 abolished processing (Fig. 7, lanes 10–12, 15–17, 39, 40, and 49–51, and Fig. 8). It seems unlikely that His-296 is directly involved in catalysis because Ser, Gly, Pro, or Asn is present at the corresponding position in nine SpoIIGA orthologs (supplemental Fig. 3). Based on our model (Fig. 6), we hypothesized that His-296 might indirectly influence catalysis by affecting the position of Lys-298 in the vicinity of the active site. However, K298A did not impair processing (Fig. 7, lane 52). On the other hand, K298N reduced processing efficiency to about 20% of the wild type (Fig. 7, lane 53, and Fig. 8). We speculated that H296G might compensate for the effect of the K298N substitution, but processing was not restored in the double mutant (Fig. 7, lane 55). Although we do not understand the role of His-296, we conclude that neither computational nor mutational approaches support the suggestion that SpoIIGA is a serine protease, whereas both approaches provide evidence that SpoIIGA is an aspartic protease.
Substitutions for Asp-6 did not impair processing in the E. coli system. The N-terminal six amino acid residues of SpoIIGA are predicted to be outside the B. subtilis cell (21) or in the periplasm when expressed in E. coli. A D6A substitution reduced sporulation of B. subtilis when the mutant protein was expressed weakly from a heterologous promoter at an ectopic site in the chromosome of a spoIIGA null mutant, whereas the wild-type SpoIIGA protein or D6E mutant protein complemented the spoIIGA mutant fully when expressed similarly (21). When coexpressed with pro-σE and SpoIIR more strongly from a different heterologous promoter on a multicopy plasmid during growth of B. subtilis, the D6A mutant protein allowed processing of pro-σE but not as much processing as wild-type SpoIIGA (21). In our E. coli system, neither D6A nor D6E impaired processing (Fig. 7, lanes 8 and 9), perhaps because proteins are highly expressed.
Self-association of SpoIIGA and Interaction of SpoIIGA with SpoIIR—If SpoIIGA is an aspartic protease with Asp-183 as its catalytic aspartate residue, the active form of the enzyme is expected to be a dimer to form the catalytic dyad. An attractive hypothesis is that SpoIIR activates SpoIIGA by promoting dimer formation (62). To test whether SpoIIGA self-associates, E. coli was engineered to coexpress two forms of SpoIIGA with different tags, SpoIIGA-GFP-H6 (allowing metal-affinity purification) and SpoIIGA-GFP-F2 (allowing assessment of copurification with SpoIIGA-GFP-H6). E. coli expressing only SpoIIGA-GFP-F2 served as a negative control. The two induced strains were lysed mechanically, and the lysates were treated with 1% Sarkosyl detergent to solubilize membranes. After high speed centrifugation, the soluble SpoIIGA-GFP-H6 was purified by Ni2+-affinity chromatography and analyzed for copurification of SpoIIGA-GFP-F2 by Western blot. Samples taken before Ni2+-affinity purification show that the two E. coli strains produced a similar amount of SpoIIGA-GFP-F2 (Fig. 9A, lanes 1 and 2). After Ni2+-affinity purification, SpoIIGA-GFP-F2 was detected only if SpoIIGA-GFP-H6 had been coexpressed (Fig. 9A, lanes 3 and 4). We attempted to coexpress SpoIIR-F2 (from pID88) with the two forms of SpoIIGA (from pID87) to test if SpoIIR promotes SpoIIGA self-association; however, SpoIIR-F2 did not accumulate to a detectable level (data not shown). The two copies of spoIIGA on pID87 appeared to inhibit SpoIIR-F2 expression from pID88 even more than one copy of spoIIGA on pID48 inhibited expression of SpoIIR-F2 and pro-σE(M17I)-H6 from pID50 (Fig. 4A). Therefore, we were unable to test the effect of SpoIIR on SpoIIGA self-association, but our data show that SpoIIGA can self-associate in E. coli in the absence of SpoIIR.
FIGURE 9.
Self-association of SpoIIGA and interaction of SpoIIGA with SpoIIR. A, SpoIIGA self-association. Detergent-solubilized proteins from E. coli bearing pID48 to produce SpoIIGA-GFP-F2 (lanes 1 and 3) or pID87 to produce SpoIIGA-GFP-F2 and SpoIIGA-GFP-H6 (lanes 2 and 4) were subjected to Western blot analysis using antibodies against FLAG to detect SpoIIGA-GFP-F2 before (lanes 1 and 2) or after (lanes 3 and 4) Ni2+-affinity purification. B, SpoIIGA interaction with SpoIIR. Detergent-solubilized proteins from E. coli cells bearing pID88 to produce SpoIIR-F2 (lanes 1 and 3) or pID88 and pID2 to produce SpoIIR-F2 and SpoIIGA-GFP-H6 (lanes 2 and 4) were subjected to Western blot analysis using antibodies against FLAG to detect SpoIIR-F2 before (lanes 1 and 2) or after (lanes 3 and 4) Ni2+-affinity purification. In both panels, the purification concentrated the samples 5-fold, and equal volumes were loaded in all lanes, so lanes 3 and 4 contain proteins from 5-fold more cells than lanes 1 and 2.
To test whether SpoIIGA interacts with SpoIIR in E. coli, the experiment described above was repeated except with cells engineered to coexpress SpoIIGA-GFP-H6 (from pID2) and SpoIIR-F2 (from pID88). E. coli expressing only SpoIIR-F2 served as a negative control. As expected from the results shown in Fig. 4A, expression of SpoIIGA-GFP-H6 inhibited expression of SpoIIR-F2, but SpoIIR-F2 accumulated to an easily detectable level (Fig. 9B, lanes 1 and 2). After membrane solubilization and Ni2+-affinity purification, SpoIIR-F2 was detected only if SpoIIGA-GFP-H6 had been coexpressed (Fig. 9B, lanes 3 and 4). As an additional negative control, we found that SpoIIR-F2 was not detected after membrane solubilization and Ni2+-affinity purification of E. coli coexpressing histidine-tagged SpoIVFB and SpoIIR-F2 (data not shown). SpoIVFB is the B. subtilis protease that cleaves pro-σK (63, 64), and SpoIVFB was shown previously to associate with membranes when expressed in E. coli (33). We conclude that SpoIIR interacts with SpoIIGA but not with SpoIVFB.
DISCUSSION
Our results provide evidence that SpoIIGA is an aspartic protease that is activated by SpoIIR to cleave pro-σE. Coexpression of SpoIIGA and SpoIIR with pro-σE in E. coli is sufficient for accurate processing of pro-σE, producing σE with the correct N terminus. No other B. subtilis proteins are required for pro-σE processing. Neither SpoIIGA nor SpoIIR alone is able to cleave pro-σE. The C-terminal domain of SpoIIGA is similar to aspartic proteases, but SpoIIR does not show significant similarity to any protein of known function. Modeling of the C-terminal domain of SpoIIGA based on HIV-1 protease yielded a structure with explanatory and predictive power for effects of mutations in spoIIGA on pro-σE processing in the E. coli system. The data strongly support the model that the C-terminal domain of SpoIIGA forms a dimer with Asp-183 as the catalytic aspartate and with several other key features of HIV-1 protease. Consistent with the notion that SpoIIGA dimerizes, association of differentially tagged forms of SpoIIGA was detected in E. coli. In addition, SpoIIGA was found to interact with SpoIIR. Therefore, we propose that SpoIIGA forms an inactive dimer and that SpoIIR interaction with the N-terminal domain of SpoIIGA on one side of the membrane leads to a conformational change in the C-terminal domain of SpoIIGA on the other side of the membrane, allowing formation of the active aspartic protease dimer capable of cleaving pro-σE.
In addition to accurate processing of pro-σE, the E. coli system exhibited several other noteworthy characteristics. First, processing was rapid. The σE-H6 cleavage product was present at the earliest time tested, 10 min after addition of IPTG to induce production of T7 RNAP, which transcribes the genes encoding SpoIIGA-GFP-F2, SpoIIR-F2, and pro-σE(M17I)-H6 (Fig. 4A). Second, processing was abundant. Dilution of the E. coli sample shown in Fig. 4B, followed by Western blot analysis, suggests that the engineered E. coli system produces 5-fold more σE than sporulating B. subtilis on a per cell basis (data not shown). Moreover, the σE-H6 produced in E. coli can be purified easily using metal-affinity chromatography, and the cleavage site can be inferred from its N-terminal amino acid sequence as determined by Edman degradation. Hence, the E. coli system will be useful for investigations of component alterations that might change the cleavage site. Third, processing appears to occur at the membrane. SpoIIGA-GFP-F2, SpoIIR-F2, and pro-σE(M17I)-H6 were found in the cell envelope fraction, and σE-H6 was released to the cytosolic fraction (Fig. 4C). These observations are consistent with the model depicted in Fig. 1, in which SpoIIR is secreted to the periplasm of E. coli and interacts with the IM-embedded N-terminal domain of SpoIIGA, stimulating cleavage of IM-associated pro-σE and releasing σE into the E. coli cytoplasm. The E. coli system will facilitate further testing of this model using a combination of mutational analysis, protein-protein interaction studies, and more sophisticated cell fractionation methods. Here we utilized the E. coli system to perform mutational analysis of spoIIGA and to investigate the ability of SpoIIGA to self-associate and to interact with SpoIIR.
Modeling and mutational analyses provide strong evidence that the C-terminal domain of SpoIIGA forms a dimer with Asp-183 as the catalytic aspartate and with several other key features of HIV-1 protease. Substitution of Ala, Glu, or Asn for Asp-183 abolished pro-σE processing (Fig. 7), consistent with the proposed critical role of Asp-183 as the catalytic aspartate residue. The three residues preceding Asp-183 in SpoIIGA are sensitive to substitution with Ala. In HIV-1 protease, conservative substitutions at the corresponding positions (A22G, L23V, and L24V) impair proteolytic activity (65). The catalytic aspartate residue, Asp-25, of HIV-1 protease is followed by Thr-26, whose side-chain hydroxyl stabilizes the dimer interface near the active site by forming a hydrogen bond network referred to as the fireman's grip (52, 53). A conservative T26S substitution reduces HIV-1 protease activity in vitro but exhibits undiminished polyprotein processing in vivo (54, 66). Consistent with the in vivo result, Thr and Ser are interchangeable at the corresponding position of SpoIIGA, because S184T exhibits undiminished pro-σE processing (Figs. 7 and 8). Moreover, both HIV-1 protease (54, 65) and SpoIIGA (Figs. 7 and 8) exhibit reduced processing when this residue is changed to Cys, and no processing when it is changed to Ala. In addition to residues near the active site in the primary sequence of HIV-1 protease, another key feature of its structure and function is a mobile flap from each subunit of the dimer that appears to play important roles in substrate binding, product release, and the evolution of drug resistance (55, 56, 67). In our model of the C-terminal domain of SpoIIGA (Fig. 6), the flap loop is predicted to begin at Ile-242 and end at Val-257. Interestingly, within this region the sequence (IV)P(YF) is perfectly conserved among SpoIIGA orthologs (supplemental Fig. 3), suggesting it might play an important role in flap function. Although this prediction remains to be tested, we did test another prediction of the model about the flap. Q252E was predicted to hinder flap closure, and this change did impair pro-σE processing (Figs. 7 and 8). Another prediction of the model was that the C-terminal tip of SpoIIGA would be important for function. The C- and N-terminal β-strands of each subunit of HIV-1 protease form a four-stranded antiparallel β-sheet that is critical for dimer formation (57, 68). In agreement with the prediction that the C-terminal tip of SpoIIGA participates in forming a similar β-sheet structure, deletion of the eight C-terminal residues abolished pro-σE processing (Fig. 7). Taken together, our mutational analysis of residues near the putative active site, within the putative flap, and at the C-terminal tip strongly support the model that the C-terminal domain of SpoIIGA resembles HIV-1 protease.
Although we think that the C-terminal domain of SpoIIGA resembles HIV-1 protease, there are some notable differences between our model (Fig. 6) and the HIV-1 protease structure. Most conspicuous is an extra three-helix bundle in each subunit of the dimer. These are predicted to be packed on either side of the flaps, so it is tempting to speculate that they affect flap function. The only substitutions we tested in the three-helix bundle were at its C-terminal end, Asp-230 to Ala, Glu, or Pro, and none of these severely impaired processing (Figs. 7 and 8). Another difference between our model (Fig. 6) and the HIV-1 protease structure is a longer loop at residues 281–292 of SpoIIGA. This loop is predicted to interact with the substrate. A T282E substitution abolished processing, and an S286A substitution reduced processing to 20% the wild-type level, whereas a T283A substitution that was predicted to be less disruptive had little effect on processing (Figs. 7 and 8). These results support the idea that the loop is important for SpoIIGA function. A major difference between SpoIIGA and HIV-1 protease is the membrane-embedded N-terminal domain of SpoIIGA. Deletion or addition of a few residues at the N-terminal end of HIV-1 protease interferes with dimer formation (55). It was attractive to think that SpoIIGA might be monomeric and that SpoIIR might activate SpoIIGA by promoting dimer formation (62). However, we found that differentially tagged forms of SpoIIGA self-associate in E. coli (Fig. 9A), suggesting that SpoIIGA forms inactive dimers or higher order oligomers in the absence of SpoIIR. We also found that SpoIIGA interacts with SpoIIR (Fig. 9B). Conceivably, SpoIIR could promote conversion of inactive SpoIIGA oligomers to active dimers. A simpler model would be that SpoIIR converts inactive SpoIIGA dimers to active dimers.
How might SpoIIR activate inactive SpoIIGA dimers? During sporulation of B. subtilis, SpoIIR is believed to be secreted from the FS into the space between the membranes of the polar septum (Fig. 1). This would put SpoIIR in a position to interact with the N-terminal tip and/or extracellular loops of the membrane-embedded domain of SpoIIGA. Deletion of the largest predicted extracellular loop did not prevent SpoIIGA function during sporulation; however, a D6A substitution in the predicted extracellular N-terminal tip of SpoIIGA decreased pro-σE processing, leading to the suggestion that this tip might interact with SpoIIR (21). In our E. coli system, SpoIIR is presumably secreted to the periplasm, allowing it to interact with extracellular parts of the N-terminal domain of SpoIIGA (Fig. 1). However, the D6A substitution in SpoIIGA did not significantly decrease pro-σE processing in E. coli (Figs. 7 and 8). Because the effect of the D6A substitution in B. subtilis was partially overcome by elevating expression of the mutant SpoIIGA protein (21), it seems likely that overexpression of the mutant protein in E. coli accounts for its activity. In any case, an interaction between the SpoIIGA Asp-6 residue and SpoIIR is not essential for pro-σE processing. On the other hand, our findings that SpoIIR and SpoIIGA are necessary and sufficient for pro-σE processing in E. coli are consistent with a model in which SpoIIR interacts with extracellular parts of the N-terminal domain of SpoIIGA on one side of the membrane, causing a conformational change in the C-terminal domain of SpoIIGA on the other side of the membrane that activates the protease. In particular, the β-sheet predicted to be formed by the N- and C-terminal β-strands of the C-terminal domain of each subunit (Fig. 6) might be affected by SpoIIR binding. Alternatively or in addition, SpoIIR might affect the longer loop of SpoIIGA (residues 281–292) because both ends of this loop are attached to parts of the protein predicted to be near the surface of the membrane (Fig. 6).
Membrane-associated aspartic proteases have been described previously, but our model for the structure and activation of SpoIIGA is unique. The yapsins are a family of five glycosylphosphatidylinositol-linked aspartic proteases required for cell wall integrity in Saccharomyces cerevisiae (69). BACE-1 and BACE-2 have a single transmembrane segment near their C terminus and an N-terminal aspartic protease domain that in the case of BACE-1 cleaves the amyloid precursor protein, which after further cleavage by presenilin forms the neurotoxic amyloid β-peptide implicated in Alzheimer disease (reviewed in Ref. 70). Presenilin and signal peptide peptidases are intramembrane-cleaving aspartic proteases, meaning that they are believed to cleave their substrates within a membrane (reviewed in Ref. 71). All of these aspartic proteases have both catalytic aspartate residues in a single polypeptide chain. In contrast, catalytic aspartate residues are contributed by each chain of the proposed SpoIIGA dimer. Also, SpoIIGA is proposed to have five transmembrane segments in its N-terminal domain (16), which function as a receptor for the SpoIIR signal (6–8, 21). Hence, SpoIIGA is proposed to be a novel type of signal transducing protease.
The proposed signal transduction pathway leading to pro-σE processing (Fig. 1) is also quite different from that leading to pro-σK processing later during B. subtilis sporulation. Cleavage of pro-σK requires the putative intramembrane-cleaving metalloprotease SpoIVFB (63, 64). Coexpression of SpoIVFB and pro-σK in E. coli results in accurate and abundant processing to σK (33). Activity of SpoIVFB is negatively regulated by SpoIVFA and BofA (33, 72–76). During B. subtilis sporulation, SpoIVFB becomes active after the FS is engulfed by the MC (reviewed in Ref. 1). The completion of engulfment triggers activation of σG RNAP in the FS, which transcribes the spoIVB gene, whose product is believed to cross the innermost membrane surrounding the FS. SpoIVB is a serine protease that cleaves SpoIVFA, which is located in the outermost membrane surrounding the FS in a complex that includes BofA and SpoIVFB. A second serine protease, CtpB, is also believed to be secreted primarily from the FS (77) into the intermembrane space as a backup mechanism to ensure cleavage of SpoIVFA (78) and to cleave BofA (79), although there appears to be another mechanism(s) to ensure cleavage of BofA, and it is uncertain whether BofA must be removed from the complex to activate SpoIVFB. In any case, σK is released by SpoIVFB from the outermost membrane surrounding the FS into the MC. As for pro-σE, processing of pro-σK occurs in response to a signal from the FS, but the mechanisms of signaling and pro-σ cleavage appear to be very different.
We attempted to inhibit processing of pro-σE in our E. coli system, but none of the protease inhibitors we tested impaired processing. Each potential inhibitor was added to the E. coli cells at the same time IPTG was added to induce production of recombinant SpoIIR, SpoIIGA, and pro-σE (supplemental Table S3). Although we tested inhibitors of other types of proteases, we focused on inhibitors of aspartic proteases; however, the aspartic protease inhibitors are at or above a size (600 daltons) expected to diffuse through pores in the E. coli OM, and permeability of these compounds to the OM (and in some cases the IM as well) may limit their access to the cytoplasm. For this reason, it would be premature to conclude that SpoIIGA exhibits unique resistance to protease inhibitors. The development of an in vitro assay for SpoIIGA protease activity would facilitate inhibitor testing. Removing the N-terminal membrane-embedded domain of SpoIIGA or replacing it with glutathione S-transferase did not permit pro-σE processing in E. coli. It remains to be tested whether an active “tethered dimer” of the C-terminal domain of SpoIIGA can be created, analogous to tethered dimers of HIV-1 protease (80, 81) and other retroviral proteases (82). Ultimately, however, proof of our model demands reconstitution of SpoIIGA into an artificial membrane and stimulation of pro-σE processing by SpoIIR in vitro, a daunting challenge.
Supplementary Material
Acknowledgments
We are grateful for the gifts of protease inhibitors from Dr. Ben M. Dunn (University of Florida School of Medicine, Gainesville) and Dr. Michael S. Wolfe (Brigham and Women's Hospital and Harvard Medical School, Boston, MA) and for the gift of monoclonal antibody against σE from Dr. William G. Haldenwang (University of Texas Health Science Center, San Antonio).
This work was supported, in whole or in part, by National Institutes of Health Grant GM43585 (to L. K.). This work was also supported by National Science Foundation Grant 0447799, a grant from the Alfred P. Sloan Foundation (to M. F.), and by the Michigan Agricultural Experiment Station. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3, Figs. 1–3, and additional references.
Footnotes
The abbreviations used are: MC, mother cell; HIV-1, human immunodeficiency virus type 1; FS, forespore; RNAP, RNA polymerase; MCM, mother cell membrane; FSM, forespore membrane; IM, inner membrane; IPTG, isopropyl β-d-thiogalactopyranoside; OM, outer membrane; PBS, phosphate-buffered saline.
References
- 1.Kroos, L. (2007) Annu. Rev. Genet. 41 13-39 [DOI] [PubMed] [Google Scholar]
- 2.Barak, I., and Wilkinson, A. J. (2005) Mol. Microbiol. 57 611-620 [DOI] [PubMed] [Google Scholar]
- 3.Yudkin, M. D., and Clarkson, J. (2005) Mol. Microbiol. 56 578-589 [DOI] [PubMed] [Google Scholar]
- 4.Steil, L., Serrano, M., Henriques, A. O., and Volker, U. (2005) Microbiology 151 399-420 [DOI] [PubMed] [Google Scholar]
- 5.Wang, S. T., Setlow, B., Conlon, E. M., Lyon, J. L., Imamura, D., Sato, T., Setlow, P., Losick, R., and Eichenberger, P. (2006) J. Mol. Biol. 358 16-37 [DOI] [PubMed] [Google Scholar]
- 6.Londono-Vallejo, J. A., and Stragier, P. (1995) Genes Dev. 9 503-508 [DOI] [PubMed] [Google Scholar]
- 7.Karow, M. L., Glaser, P., and Piggot, P. J. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 2012-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmeister, A. E. M., Londono-Vallejo, A., Harry, E., Stragier, P., and Losick, R. (1995) Cell 83 219-226 [DOI] [PubMed] [Google Scholar]
- 9.Feucht, A., Evans, L., and Errington, J. (2003) Microbiology 149 3023-3034 [DOI] [PubMed] [Google Scholar]
- 10.Eichenberger, P., Fujita, M., Jensen, S. T., Conlon, E. M., Rudner, D. Z., Wang, S. T., Ferguson, C., Haga, K., Sato, T., Liu, J. S., and Losick, R. (2004) Plos Biol. 2 1664-1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichenberger, P., Jensen, S. T., Conlon, E. M., van Ooij, C., Silvaggi, J., Gonzalez-Pastor, J. E., Fujita, M., Ben-Yehuda, S., Stragier, P., Liu, J. S., and Losick, R. (2003) J. Mol. Biol. 327 945-972 [DOI] [PubMed] [Google Scholar]
- 12.LaBell, T. L., Trempy, J. E., and Haldenwang, W. G. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 1784-1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyao, A., Theeragool, G., Takeuchi, M., and Kobayashi, Y. (1993) J. Bacteriol. 175 4081-4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenney, T. J., and Moran, C. P. (1987) J. Bacteriol. 169 3329-3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonas, R. M., Weaver, E. A., Kenney, T. J., Moran, C. P., Jr., and Haldenwang, W. G. (1988) J. Bacteriol. 170 507-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stragier, P., Bonamy, C., and Karmazyn-Campelli, C. (1988) Cell 52 697-704 [DOI] [PubMed] [Google Scholar]
- 17.Masuda, E. S., Anaguchi, H., Sato, T., Takeuchi, M., and Kobayashi, Y. (1990) Nucleic Acids Res. 18 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shazand, K., Frandsen, N., and Stragier, P. (1995) EMBO J. 14 1439-1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters, H. K., and Haldenwang, W. G. (1991) J. Bacteriol. 173 7821-7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fawcett, P., Melnikov, A., and Youngman, P. (1998) Mol. Microbiol. 28 931-943 [DOI] [PubMed] [Google Scholar]
- 21.Londono-Vallejo, J. A. (1997) Microbiol. 143 2753-2761 [DOI] [PubMed] [Google Scholar]
- 22.Ju, J., Luo, T., and Haldenwang, W. (1997) J. Bacteriol. 179 4888-4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ju, J., and Haldenwang, W. G. (2003) J. Bacteriol. 185 5897-5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmeister, A. (1998) J. Bacteriol. 180 2426-2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlson, H. C., Lu, S., Kroos, L., and Haldenwang, W. G. (1996) J. Bacteriol. 178 546-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters, H. K., Carlson, H. C., and Haldenwang, W. G. (1992) J. Bacteriol. 174 4629-4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters, H. K., and Haldenwang, W. G. (1994) J. Bacteriol. 176 7763-7766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gholamhoseinian, A., and Piggot, P. J. (1989) J. Bacteriol. 171 5747-5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita, M., and Losick, R. (2002) Mol. Microbiol. 43 27-38 [DOI] [PubMed] [Google Scholar]
- 30.Pogliano, K., Hofmeister, A., and Losick, R. (1997) J. Bacteriol. 179 3331-3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ju, J., Luo, T., and Haldenwang, W. (1998) J. Bacteriol. 180 1673-1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride, S. M., Rubio, A., Wang, L., and Haldenwang, W. G. (2005) Mol. Microbiol. 57 434-451 [DOI] [PubMed] [Google Scholar]
- 33.Zhou, R., and Kroos, L. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6385-6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroos, L., Yu, Y. T., Mills, D., and Ferguson-Miller, S. (2002) J. Bacteriol. 184 5393-5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youngman, P., Perkins, J. B., and Losick, R. (1984) Plasmid 12 1-9 [DOI] [PubMed] [Google Scholar]
- 36.Harwood, C. R., and Cutting, S. M. (eds) (1990) Molecular Biological Methods for Bacillus, John Wiley & Sons Ltd., Chichester, UK
- 37.Pillai, B., Kannan, K. K., and Hosur, M. V. (2001) Proteins 43 57-64 [DOI] [PubMed] [Google Scholar]
- 38.Sali, A., and Blundell, T. L. (1993) J. Mol. Biol. 234 779-815 [DOI] [PubMed] [Google Scholar]
- 39.Kolinski, A., and Skolnick, J. (1994) Proteins 18 338-352 [DOI] [PubMed] [Google Scholar]
- 40.Feig, M., Karanicolas, J., and Brooks, C. L., III (2004) J. Mol. Graphics 22 377-395 [DOI] [PubMed] [Google Scholar]
- 41.Laco, G. S., Schalk-Hihi, C., Lubkowski, J., Morris, G., Zdanov, A., Olson, A., Elder, J. H., Wlodawer, A., and Gustchina, A. (1997) Biochemistry 36 10696-10708 [DOI] [PubMed] [Google Scholar]
- 42.Brooks, B., Bruccoleri, R., Olafson, B., States, D., Swaminathan, S., and Karplus, M. (1983) J. Comput. Chem. 4 187-217 [Google Scholar]
- 43.MacKerell, A., Jr., Bashford, D., Bellott, M., Dunbrack, J., Evanseck, M., Field, M., Fischer, S., Gao, J., Guo, H., Ha, S., Joseph-McCarthy, D., Kuchnir, L., Kuczera, K., Lau, F., Mattos, C., Michnick, S., Ngo, T., Nguyen, D., Prodhom, B., Reiher, W., Roux, B., Schlenkrich, M., Smith, J., Stote, R., Straub, J., Watanabe, M., Wiorkiewicz-Kuczera, J., Yin, D., and Karplus, M. (1998) J. Phys. Chem. B 102 3586-3616 [DOI] [PubMed] [Google Scholar]
- 44.Lee, M. S., Feig, M., Salsbury, F. R., Jr., and Brooks, C. L., III (2003) J. Comput. Chem. 24 1348-1356 [DOI] [PubMed] [Google Scholar]
- 45.Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990) J. Mol. Biol. 215 403-410 [DOI] [PubMed] [Google Scholar]
- 46.Soding, J., Biegert, A., and Lupas, A. N. (2005) Nucleic Acids Res. 33 W244-W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ginalski, K., Elofsson, A., Fischer, D., and Rychlewski, L. (2003) Bioinformatics (Oxf.) 19 1015-1018 [DOI] [PubMed] [Google Scholar]
- 48.Kervinen, J., Tobin, G. J., Costa, J., Waugh, D. S., Wlodawer, A., and Zdanov, A. (1999) EMBO J. 18 3947-3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cutfield, S. M., Dodson, E. J., Anderson, B. F., Moody, P. C., Marshall, C. J., Sullivan, P. A., and Cutfield, J. F. (1995) Structure (Lond.) 3 1261-1271 [DOI] [PubMed] [Google Scholar]
- 50.Abad-Zapatero, C., Goldman, R., Muchmore, S. W., Hutchins, C., Stewart, K., Navaza, J., Payne, C. D., and Ray, T. L. (1996) Protein Sci. 5 640-652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hornak, V., Okur, A., Rizzo, R. C., and Simmerling, C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 915-920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearl, L., and Blundell, T. (1984) FEBS Lett. 174 96-101 [DOI] [PubMed] [Google Scholar]
- 53.Pearl, L. H., and Taylor, W. R. (1987) Nature 329 351-354 [DOI] [PubMed] [Google Scholar]
- 54.Strisovsky, K., Tessmer, U., Langner, J., Konvalinka, J., and Krausslich, H. G. (2000) Protein Sci. 9 1631-1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishima, R., Freedberg, D. I., Wang, Y. X., Louis, J. M., and Torchia, D. A. (1999) Structure (Lond.) 7 1047-1055 [DOI] [PubMed] [Google Scholar]
- 56.Wlodawer, A., and Vondrasek, J. (1998) Annu. Rev. Biophys. Biomol. Struct. 27 249-284 [DOI] [PubMed] [Google Scholar]
- 57.Weber, I. T. (1990) J. Biol. Chem. 265 10492-10496 [PubMed] [Google Scholar]
- 58.Birktoft, J. J., and Blow, D. M. (1972) J. Mol. Biol. 68 187-240 [DOI] [PubMed] [Google Scholar]
- 59.Alden, R. A., Birktoft, J. J., Kraut, J., Robertus, J. D., and Wright, C. S. (1971) Biochem. Biophys. Res. Commun. 45 337-344 [DOI] [PubMed] [Google Scholar]
- 60.Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I., Schrag, J., Sussman, J. L., Verschueren, K. H. G., and Golman, A. (1992) Protein Eng. 5 197-211 [DOI] [PubMed] [Google Scholar]
- 61.Rawlings, N. D., and Barrett, A. J. (1994) Methods Enzymol. 244 19-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kroos, L., Zhang, B., Ichikawa, H., and Yu, Y.-T. N. (1999) Mol. Microbiol. 31 1285-1294 [DOI] [PubMed] [Google Scholar]
- 63.Yu, Y.-T. N., and Kroos, L. (2000) J. Bacteriol. 182 3305-3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rudner, D., Fawcett, P., and Losick, R. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14765-14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loeb, D. D., Hutchison, C. A., III, Edgell, M. H., Farmerie, W. G., and Swanstrom, R. (1989) J. Virol. 63 111-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konvalinka, J., Litterst, M. A., Welker, R., Kottler, H., Rippmann, F., Heuser, A. M., and Krausslich, H. G. (1995) J. Virol. 69 7180-7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heaslet, H., Rosenfeld, R., Giffin, M., Lin, Y. C., Tam, K., Torbett, B. E., Elder, J. H., McRee, D. E., and Stout, C. D. (2007) Acta Crystallogr. Sect. D. Biol. Crystallogr. 63 866-875 [DOI] [PubMed] [Google Scholar]
- 68.Ishima, R., Torchia, D. A., Lynch, S. M., Gronenborn, A. M., and Louis, J. M. (2003) J. Biol. Chem. 278 43311-43319 [DOI] [PubMed] [Google Scholar]
- 69.Krysan, D. J., Ting, E. L., Abeijon, C., Kroos, L., and Fuller, R. S. (2005) Eukaryot. Cell 4 1364-1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haass, C. (2004) EMBO J. 23 483-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolfe, M. S., and Kopan, R. (2004) Science 305 1119-1123 [DOI] [PubMed] [Google Scholar]
- 72.Cutting, S., Oke, V., Driks, A., Losick, R., Lu, S., and Kroos, L. (1990) Cell 62 239-250 [DOI] [PubMed] [Google Scholar]
- 73.Cutting, S., Roels, S., and Losick, R. (1991) J. Mol. Biol. 221 1237-1256 [DOI] [PubMed] [Google Scholar]
- 74.Ricca, E., Cutting, S., and Losick, R. (1992) J. Bacteriol. 174 3177-3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Resnekov, O., and Losick, R. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3162-3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rudner, D. Z., and Losick, R. (2002) Genes Dev. 16 1007-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Campo, N., and Rudner, D. Z. (2007) J. Bacteriol. 189 6021-6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campo, N., and Rudner, D. Z. (2006) Mol. Cell 23 25-35 [DOI] [PubMed] [Google Scholar]
- 79.Zhou, R., and Kroos, L. (2005) Mol. Microbiol. 58 835-846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng, Y. S., Yin, F. H., Foundling, S., Blomstrom, D., and Kettner, C. A. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 9660-9664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DiIanni, C. L., Davis, L. J., Holloway, M. K., Herber, W. K., Darke, P. L., Kohl, N. E., and Dixon, R. A. (1990) J. Biol. Chem. 265 17348-17354 [PubMed] [Google Scholar]
- 82.Bizub, D., Weber, I. T., Cameron, C. E., Leis, J. P., and Skalka, A. M. (1991) J. Biol. Chem. 266 4951-4958 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.