Abstract
The combined application of ion-trap mass spectrometry and peptide-specific antibodies for the isolation and structural analysis of collagen cross-linking domains is illustrated with examples of results from various types of collagen with the emphasis on bone and cartilage. We highlight the potential of such methods to advance knowledge on the importance of post-translational modifications (e.g., degrees of lysine hydroxylation and glycosylation) and preferred intermolecular binding partners for telopeptide and helical cross-linking domains in regulating cross-link type and placement.
Keywords: Collagen, Cross-links, Hydroxylysine, Pyridinoline, Lysyl oxidase, Mass spectrometry
1. Introduction
The basic mechanism and principal pathways of collagen cross-linking were defined in the late 1960s, 1970s, and early 1980s [1–9]. Techniques included labeling reducible cross-links with tritiated borohydride, isolating them for structural analysis after proteolysis or acid hydrolysis as tritiated peptides or amino acids and similarly pursuing 3-hydroxypyridinium cross-linking residues by their inherent fluorescence [9,10]. Different tissue types showed distinctive patterns of cross-linking chemistry, though all were essentially based on the reactions of peptide-bound aldehydes created from specific lysine and hydroxylysine side chains by the action of lysyl oxidase(s) during the assembly of collagen subunits into fibrils. All the fibril-forming collagens (types I, II, III, V, and XI) and the fibril-associated type IX collagen rely on this mechanism of cross-linking to provide tissue structural integrity and material function [11]. Using relatively standard methods of peptide isolation by HPLC and other separation techniques followed by sequence analysis, many of the cross-linking interaction sites in the major tissue collagens have been worked out [9–11]. There are still many questions and gaps in knowledge, however, including whether for any tissue the full complement of cross-linking maturation products has been identified and the manner and extent to which evolved tissue differences in cross-linking confer functional advantages. For example, bone collagen has a unique and characteristic cross-linking phenotype [10–12]. Is this relevant to its ability to mineralize as we suspect [13], and, if so, what is the molecular benefit bestowed by the distinctive cross-linking?
In order to explore these and related questions in collagen biology, we have refined protein mass spectrometric methods to give a new approach. This promises greatly increased sensitivity, selectivity, and speed for profiling collagen cross-linking domains from small samples of polymeric collagen. We focus here on collagens of the skeletal tissues, bone, and cartilage, but the methods exemplified are applicable to all forms of cross-linked collagen. In addition, antibodies raised against peptide sequences that adjoin cross-linking sites have proven to be very useful tools for probing the extent to which heterotypic cross-linking can occur between different chains and molecular types of collagen in different tissues [14–16]. This added level of tissue and fibril complexity is emerging as a common evolutionary theme in matrix biology. Finally, we provide examples of applying related methods to identify the structure of cross-linked collagen peptides recovered from urine and potentially other body fluids. Such peptides have seen extensive use as biomarkers of the normal turnover and pathological breakdown of bone, cartilage, and other tissues.
These evolving techniques are summarized as a series of examples of results from bone and cartilage collagens.
2. Methods
2.1. Electrospray mass spectrometry
Ion-trap instruments seem particularly well suited to giving informative fragmentation patterns with the type of cross-linked peptides derived from collagen. For the results presented here we used an LCQ Deca XP ion-trap mass spectrometer with in-line liquid chromatography (LC) (ThermoFinnigan) [15,17]. A C8 capillary column 300 μm i.d. × 150 mm long (Grace Vydac 208MXS5.315) flowing at 4.5 μl/min feeds directly into the mass spectrometer. We use a C8 column rather than the standard C18 because we have found that cross-linked structures and some of the larger collagenous peptides elute more readily from this less hydrophobic column. The mobile phase consists of buffer A: 0.1% formic acid in MilliQ water with 2% buffer B, and buffer B: 0.1% formic acid in 3:1 acetonitrile:N-propanol (v/v). Samples are loaded in 0.1% formic acid containing 5% buffer B and run with a complex gradient of 2–52% buffer B over 40 min. The sample stream is introduced into the mass spectrometer with an atmospheric pressure electrospray ionization source (ESI). The spray voltage is set at 3 kV and the inlet capillary temperature is 160 °C. We routinely set the Deca XP to run in an automatic triple-play mode, which cycles through a full scan, zoom scan and MS/MS every few milliseconds.
Samples can be generated for mass spectrometry by standard protein purification methods such as chromatography, or samples can come directly from SDS–PAGE. A band is cut from the gel lane, washed, and digested with an enzyme (trypsin, endo Asp-N, etc.) to extract peptides from the gel slice [18,19]. We find that a crucial step in sample preparation is adequate peptide solubilization prior to loading on the mass spectrometer. Cross-linked peptides require 5% organic solvent in the LC loading buffer for efficient recovery.
2.2. Antibody production
Antisera were raised in rabbits against synthetic peptides (DGSKGPTISA and GGDKGPVVAA) that were the human homologues of the N-telopeptide sequences found to contain the aldehyde-forming cross-linking lysines in bovine α1(XI) and α2(XI) chains, respectively [20]. The peptides were conjugated to keyhole limpet hemocyanin with glutaraldehyde, which tends to result in the production of antibodies that recognize an epitope that features the free carboxyterminal sequence of amino acids. Thus, the mouse monoclonal antibody, 2B4, recognizes the C-terminus of EKGPDP where K is involved in cross-linking [20], and which can be used to immunopurify cross-linked collagen type II C-telopeptides from urine (see later). A mouse monoclonal antibody (1H11) that recognizes cross-linked N-telopeptides (NTx) from human bone collagen bearing the α2(I) sequence, JYDGKGVG [21], was used similarly to purify such peptides from urine.
3. Tissue-specific patterns of cross-linking
Collagen fibrils in all tissues are heteropolymers, in that the bulk collagen (type I or II) is copolymerized on a template of either type V collagen for type I collagen-based connective tissues [22] or type XI collagen for type II collagen-based cartilages and related tissues [23]. Fig. 1 illustrates this concept for cartilage type II collagen where collagen XI is polymerized in the interior (not necessarily as a single central core as depicted, but, for example, as proposed in the recent model of Holmes and Kadler [24]) and collagen IX is covalently bonded to surface type II molecules and to other type IX molecules [15]. Tissues such as cartilage, bone, high-load-bearing tendon, ligaments, fibrocartilage, etc., make collagen that is extensively cross-linked by trifunctional pyridinolines or pyrroles, the formation of which requires lateral molecular packing in the 3D structure that includes an underlying element of nearest neighbor molecular relationships portrayed in the lower two schemes.
Fig. 1.
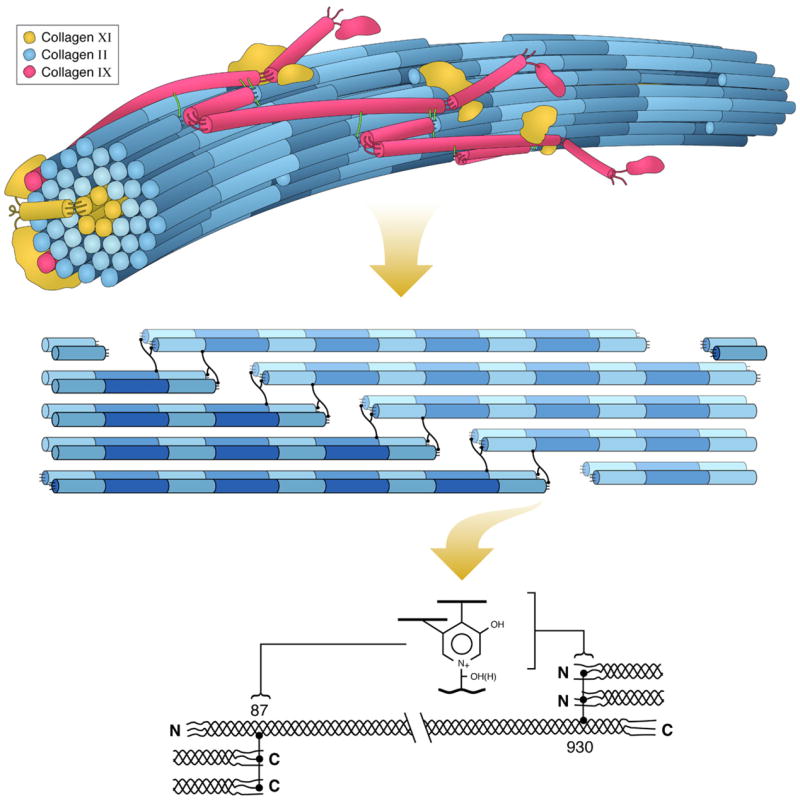
Hierarchical depiction of a heterotypic collagen fibril, emphasizing the internal axial relationships required for mature cross-link formation. Upper: Three-dimensional concept of the type II/IX/XI heterotypic fibril of developing cartilage matrix. Middle: Detail illustrating required nearest neighbor axial relationships for trifunctional intermolecular cross-links to form in collagens of cartilage, bone, and other high-tensile strength tissue matrices. The exact 3D spatial pattern of cross-linking bonds is still unclear for any tissue. Lower: Detail of the axial stagger of individual collagen molecules required for pyridinoline cross-linking.
The essential chemistry of cross-link formation for bone, cartilage, and many other tough connective tissue collagens is summarized in Fig. 2. Pyrroles and pyridinolines in about equal amounts are characteristic maturation products of bone collagen, whereas cartilages go on to form predominantly hydroxylysyl pyridinoline. The ultimate chemistry is determined by the degree of hydroxylation of lysine residues at the two telopeptide and two helical cross-linking sites found in the major fibrillar collagen molecules (types I, II, and III), where the cross-links are located as shown in Fig. 1.
Fig. 2.
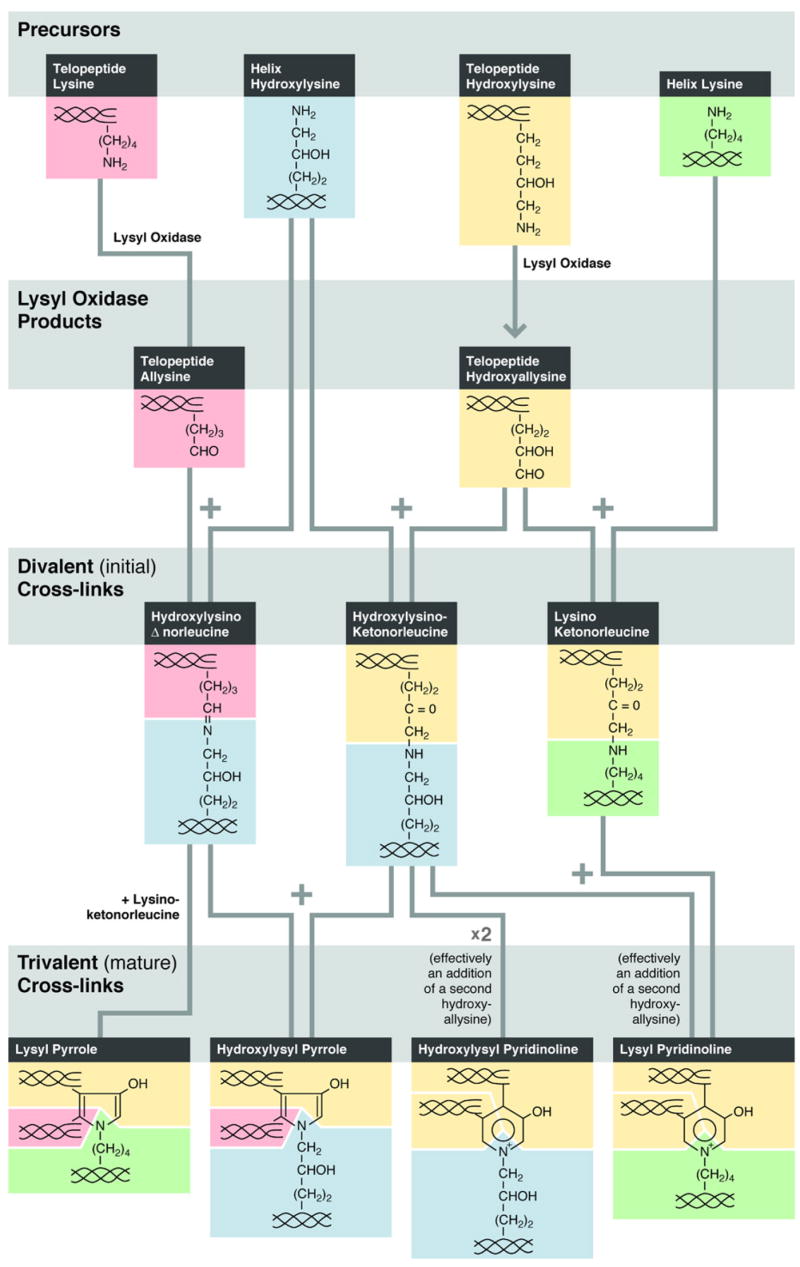
Chemical pathway of cross-linking interactions initiated by lysyl oxidase that predominates in skeletal tissue collagens. Bone collagen features both pyrrole and pyridinoline cross-links whereas mature cartilage contains only pyridinolines. Lysines of triple-helix and telopeptide origin are tracked by color to the mature cross-linking structures.
3.1. In-gel trypsin digested collagen
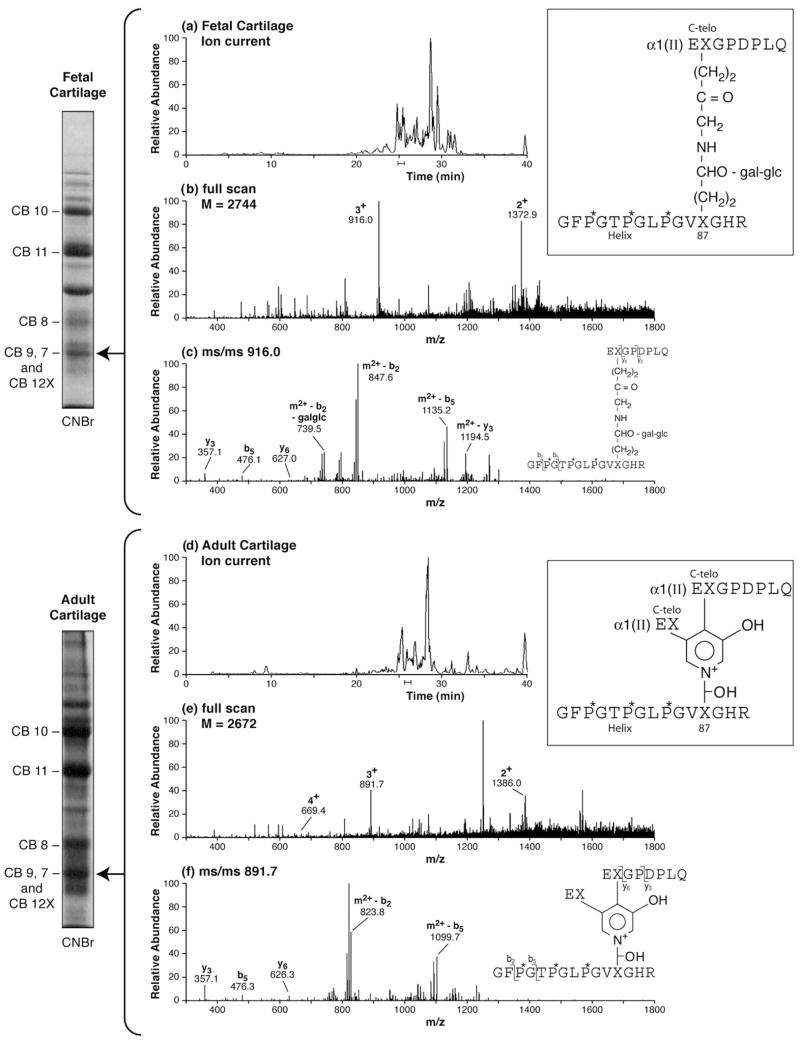
Ion-trap mass spectrometry can detect cross-linked peptides in in-gel trypsin digests of collagen chains and peptide fragments. In the two examples shown in Fig. 3, the main cross-linked peptides from the C-telopeptide-to-helix site are recovered and identified from CNBr-digests of pepsin-solubilized type II collagen from fetal epiphyseal and adult articular human cartilages. A band containing the appropriate CB12X cross-linked fragment was digested in-gel with trypsin [18,19] and run on LCMS. In each example, panel a is the ion-current of the LC eluent, panel b is the mass spectrum for the eluent region indicated and panel c is the MS/MS fragmentation pattern showing the origins of the major cleavage products as C-terminal (y) and N-terminal (b) ions. From fetal cartilage the cross-links are mostly the divalent keto-amine in which glucosyl galactosyl hydroxylysine was the reactant at triple-helix residue K87. From adult articular cartilage the main cross-link at this locus was now hydroxylysyl pyridinoline in the peptide identified but there are no hexoses on the hydroxylysine at K87. The probable explanation is a decreasing extent of hydroxylysine glycosylation with increasing tissue maturity, but it could also be a preference for non-glycosylated hydroxylysine residues in the maturation reaction from divalent cross-links to pyridinolines.
Fig. 3.
Examples of cross-linked peptides recovered on in-gel trypsin digestion of type II collagen (CNBr-digested) from fetal and adult human cartilage. The collagens were extracted from tissue by pepsin digestion and purified by 0.9 M NaCl precipitation before CNBr digestion [17]. (a) Divalent cross-links are prominent in fetal cartilage and in this example the cross-link in the identified peptide is glucosyl galactosyl hydroxylysino-ketonorleucine derived from glcgalHyl at K87 in the triple-helix. (b) The trifunctional cross-link hydroxylysyl pyridinoline predominates in cross-linked peptides from adult cartilage. The structure of the peptide shown, with one telopeptide arm cleaved immediately after the cross-linking hydroxylysine (X), probably reflects cleavage by pepsin at this peptide bond. Note that from the MS results, it is not possible to determine the relative positions of these two telopeptide arms on the pyridinol ring. The lowest panel in each example (c and f) shows the principal fragment ions on MS/MS from the 3+ parent ion and their origins as b-ion (aminoterminal) and y-ion (carboxyterminal) cleavage products.
3.2. Cathepsin K digested bone collagen
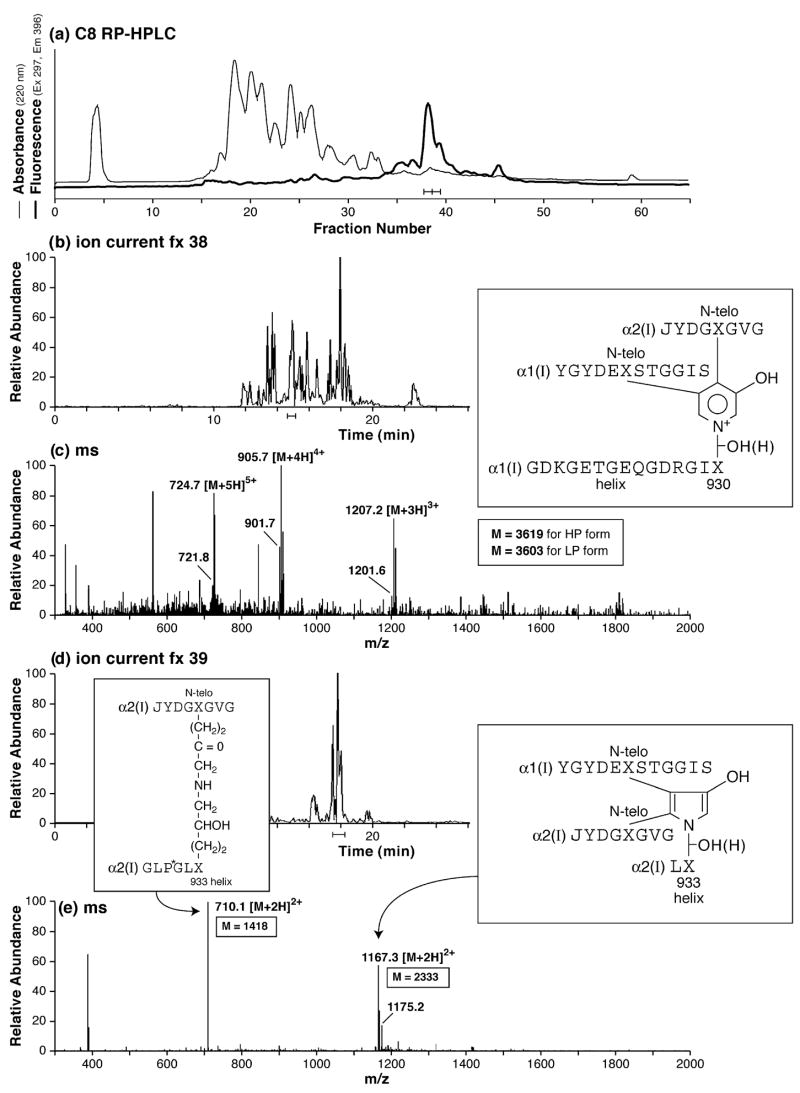
Similarly, mass spectrometry can be used to identify cross-linked peptides in protease digests of bone collagen. Fig. 4 shows the identification of major peptide products from head-to-tail cross-linking sites after complete digestion of decalcified adult human bone collagen using recombinant cathepsin K. Digests were prepared as described by Atley et al. [25]. Cross-linked peptides were resolved from the bulk collagen peptides by reverse-phase HPLC as a retarded series of minor UV-absorbing peaks distinguished by the fluorescence of pyridinoline components. The characteristic ratio of lysyl pyridinoline to hydroxylysyl pyridinoline for bone collagen is evident in the ratio of molecular ion pairs differing by 16 mass units, reflecting an oxygen atom and the degree of partial hydroxylation of residue K930 in the α1(I) collagen chains from which the structure shown was derived. Major forms of divalent (keto-amine) cross-linked peptide and pyrrole-based peptide were also identified in this late-eluting pool. The cross-linking structure shown is based on a 3-hydroxypyrrole, which we now believe is the core structure of the pyrrole cross-links in bone collagen, rather than a pyrrole lacking a hydroxyl on the ring as depicted earlier [26] and so revising the mechanism of formation.
Fig. 4.
RP-HPLC resolution and mass spectrometric structural identification of cross-linked peptides from human bone collagen digested by cathepsin K in vitro. (a) RP-HPLC and detection by fluorescence of trivalent pyridinoline-containing peptides from the cathepsin K digest. (b and c) LC/MS resolution and detection of a major pyridinoline-containing peptide from fraction 38. The HP and LP forms of the same peptide are resolved in the mass spectrum as ions that differ by 16 mass units (= oxygen atom), e.g., 905.7 and 901.7 for the 4+ charge state. (d and e) LC/MS resolution and detection of a major pyrrole-containing cross-linked peptide from fraction 39 of the HPLC profile. The predominant 2+ charge state of the pyrrole structure is a useful distinguishing feature from the predominant 3+ charge of the equivalent peptide in which the cross-link is a pyridinoline residue (latter not shown).
3.3. Collagenase-digested cartilage collagen
If sample size is not an issue, column chromatography after proteolysis to small peptides is a preferred initial enrichment step over in-gel digestion after SDS–PAGE. Fig. 5 summarizes results for adult human articular cartilage collagen digested with crude bacterial collagenase. The enriched cross-linked peptides are concentrated in retarded fractions on reverse-phase HPLC and can be identified from their MS/MS fragmentation behavior (not shown). Typical losses are the y ions of the full-length C-terminal telopeptide arms. Two molecular species are shown, one from each of the two cross-linking sites. Various molecular species reflecting a range of ragged ends plus or minus one or more residues presumably arise from the variability of collagenase action as can be seen for the peptide identified from fraction 24.
Fig. 5.
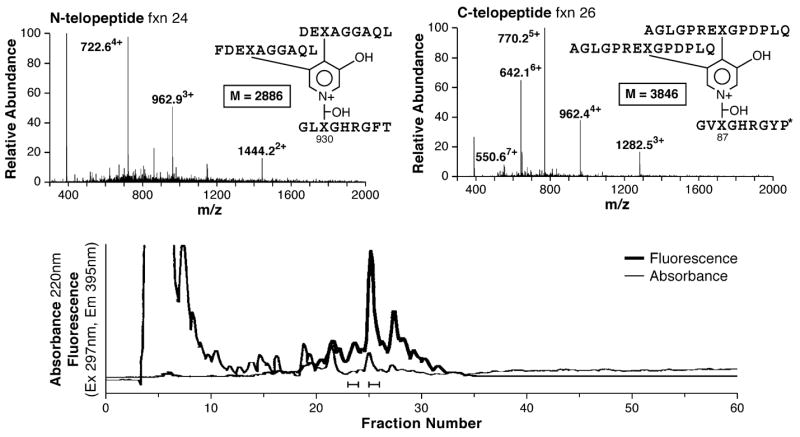
Examples of cross-linked peptides recovered from a bacterial collagenase digest of human cartilage type II collagen. Both N-telopeptide and C-telopeptide forms, from each of the two intermolecular cross-linking sites (Fig. 1) can be identified.
4. Collagen type XI
We earlier reported the identification of the main cross-linking interaction sites in type XI collagen using tritiated borohydride to label the reducible keto-amines [27]. Collagen XI, when solubilized by pepsin and purified by salt precipitation, contains only divalent keto-amine cross-links, with essentially no pyridinolines [10,27]. Fig. 6 illustrates in the upper two panels how tritiated telopeptides can be released by periodate from such purified radiolabeled collagen XI α-chains and identified by sequence analysis. Taking this a step further we made rabbit polyclonal antisera against synthetic peptides matching the human α1(XI) and α2(XI) N-telopeptide domains flanking the lysine residues identified as cross-linking sites and therefore predicted to be lysyl oxidase substrates. Use of these two polyclonal antisera on Western blots confirmed the highly selective nature of the cross-linking affinities exhibited by these domains. In the lower left panel, the α1(XI) N-telopeptide antiserum selectively bound to the α3(XI) α-chain on a Western blot and the α2(XI) N-telopeptide antiserum to the α1(XI) α-chain. These antisera can be used to determine the molecular binding partners of the N-telopeptides of type XI collagen chains present in more complex cartilages [28] or, for example, engineered tissue constructs induced in vivo or grown in vitro.
Fig. 6.
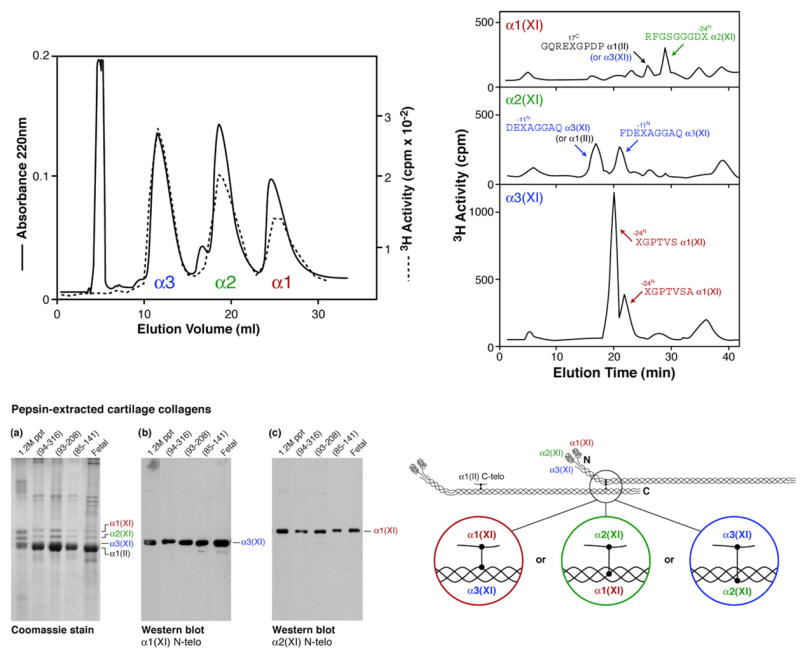
Summary of methods used to locate the sites of intermolecular cross-linking and the interacting sequences in cartilage type XI collagen polymers. The lower right and upper panels summarize published results on bovine type XI collagen [27]. Lower left panel shows Western blot results on human cartilage using two polyclonal antisera, one against the human α1(XI) cross-linking N-telopeptide sequence, the other against the human α2(XI) N-telopeptide.
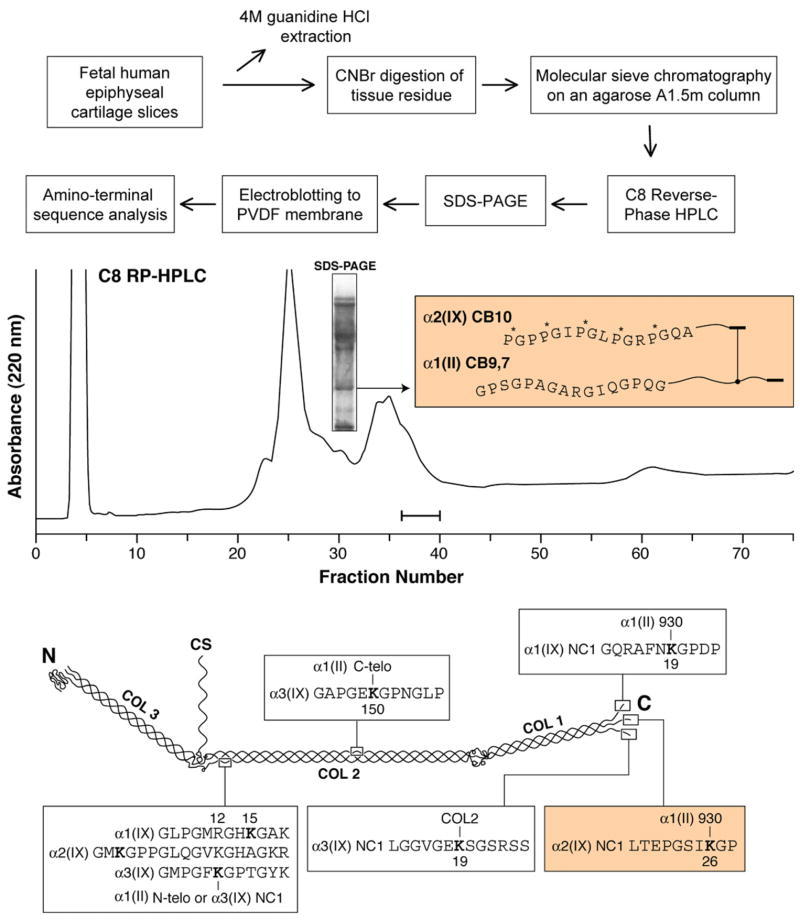
5. Collagen type IX
Type IX collagen is a heavily cross-linked fibril-modifying molecule in the FACIT (Fibril-Associated Collagen with Interrupted Triple-helix) family of homologous gene products that has evolved through vertebrate species to function exclusively in association with and covalently cross-linked to type II collagen fibrils. Our knowledge of the cross-linking properties of type IX collagen, including the location of its intermolecular cross-linking sites is built on conventional peptide fractionation methods and sequence analysis [14,15]. Fig. 7 illustrates how we used such an approach to investigate whether a remaining candidate locus for lysyl oxidase-mediated cross-linking (the α2(IX) NC1 domain) anticipated from past results on α1(IX) and α2(IX), was indeed cross-linked. The results of N-terminal sequence analysis of a purified CNBr-derived fragment that gave two running sequences (middle panel) indicates that the α2(IX) NC1 domain was cross-linked to type II collagen. The molecular size of the peptide on SDS–PAGE fits the size of the cross-linked peptide based on methionine locations in α1(II) and α2(IX) chains. All three NC-1 domains (α1(IX), α2(IX), and α3(IX)) therefore participate in cross-linking to collagen II (at K930).
Fig. 7.
Collagen IX. Flow diagram (upper) showing the methods, results (middle), and molecular interpretation (lower) indicating that the α2(IX)NC1 domain of collagen IX (and, combined with previous results, all three NC1 domains) participates in covalent cross-linking to the surface of type II collagen fibrils.
6. Cross-linked collagen peptides in urine
Pyridinoline cross-linking residues are excreted in urine as products of tissue collagen degradation and, since pyridinolines cannot be metabolized, they can provide a measure of the amount of tissue collagen degraded [29,30]. Because bone turns over rapidly, it is the principal source. Roughly two-thirds of urinary pyridinolines are in the form of small peptides (<2000 MW). Immunoassays that target such peptides in urine and serum have been used extensively in clinical studies for monitoring bone resorption [31,32]. Our original studies that identified such bone-derived peptides also revealed that type II collagen peptides contributed to the urinary pyridinoline pool [20,33].
It is now possible to use tandem mass spectrometry to profile the spectrum of peptide degradation products derived from such collagen domains, for example, after enrichment by immunoaffinity chromatography. Fig. 8 shows mass spectrometric results from a growing child’s urine, demonstrating NTx peptides from type I collagen and CTx-II peptides from type II collagen. Such methods have proved useful for examining non-invasively the post-translational status of lysine/hydroxylysine residues at cross-linking sites in types I and II collagens of patients with suspected Ehlers–Danlos subtype-VIA and potentially other, related disorders caused by collagen defects [34].
Fig. 8.

Cross-linked peptides isolated by immuno-affinity chromatography from adolescent human urine. (a) NTx-peptides, originating from bone resorption, affinity enriched on mAb 1H11. (b) CTx-II peptides, originating from mineralized growth plate resorption, affinity enriched on mAb 2B4.
7. Concluding remarks
We demonstrate the potential of mass spectrometric analysis to reveal details of the post-translational cross-linking chemistry of collagen, not possible by traditional methods and with great sensitivity in terms of sample size. Though at first daunting to interpret, mass spectra from complex cross-linked peptides can reveal characteristic patterns of fragmentation and differences in charge envelope (e.g., between pyrrole- and pyridinoline-based structures for essentially the same linked peptides) that can aid in the structural identification. The ability to differentiate glycosylated and non-glycosylated cross-links is revealing interesting variations between tissue types, with increasing maturational age and in heritable diseases that affect collagen structure. From the examples provided here, it should be evident that such approaches can provide new insights on unresolved structural questions and in re-evaluating proposed mechanisms and molecular sites of cross-link formation based on earlier, indirect methods. By avoiding harsh hydrolysis conditions and working with peptides produced under mild proteolytic conditions, we can now probe native cross-link environments and local post-translational chemistry more reliably. This offers the potential to reveal any post-translational variations at cross-linking loci likely to be involved in directing the ensuing interaction chemistry, preferred chain involvements and hence varying the properties of the polymer.
Acknowledgments
Grant support was provided by NIH NIAMS AR36794 and AR37318 and the Burgess Chair Endowment of the University of Washington. We thank Tom Eykemans for the figures and Kae Ellingsen for manuscript preparation.
References
- 1.Rojkind M, Blumenfeld OO, Gallop P. J Biol Chem. 1966;241:1530–1536. [PubMed] [Google Scholar]
- 2.Bornstein P, Kang AH, Piez KA. Proc Natl Acad Sci USA. 1966;55:417–424. doi: 10.1073/pnas.55.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey AJ, Peach CM. Biochem Biophys Res Commun. 1968;33:812–819. doi: 10.1016/0006-291x(68)90233-7. [DOI] [PubMed] [Google Scholar]
- 4.Tanzer ML. J Biol Chem. 1968;243:4045–4054. [PubMed] [Google Scholar]
- 5.Mechanic G, Tanzer ML. Biochem Biophys Res Commun. 1970;41:1597–1604. doi: 10.1016/0006-291x(70)90571-1. [DOI] [PubMed] [Google Scholar]
- 6.Tanzer ML, Housely T, Berube L, Fairweather R, Franzblau C, Gallop PM. J Biol Chem. 1973;248:393–402. [PubMed] [Google Scholar]
- 7.Robins SP, Shimokomaki M, Bailey AJ. Biochem J. 1973;131:771–780. doi: 10.1042/bj1310771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimoto D, Akiba K, Nakamura N. Biochem Biophys Res Commun. 1977;76:1124–1129. doi: 10.1016/0006-291x(77)90972-x. [DOI] [PubMed] [Google Scholar]
- 9.Eyre DR, Paz MA, Gallop PM. Annu Rev Biochem. 1984;53:717–748. doi: 10.1146/annurev.bi.53.070184.003441. [DOI] [PubMed] [Google Scholar]
- 10.Eyre D. Meth Enzymol. 1987;144:115–139. doi: 10.1016/0076-6879(87)44176-1. [DOI] [PubMed] [Google Scholar]
- 11.Eyre DR, Wu JJ. In: Topics in Current Chemistry. Brinckmann J, Notbohm H, Müller PK, editors. Springer Publishers; 2005. pp. 207–229. [Google Scholar]
- 12.Eyre DR, Dickson IR, Van Ness K. Biochem J. 1988;252:495–500. doi: 10.1042/bj2520495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamauchi M, Katz EP, Otsubo K, Teraoka K, Mechanic GL. Connect Tissue Res. 1989;21:159–167. doi: 10.3109/03008208909050006. [DOI] [PubMed] [Google Scholar]
- 14.Wu JJ, Woods PE, Eyre DR. J Biol Chem. 1992;267:23007–23014. [PubMed] [Google Scholar]
- 15.Eyre DR, Pietka T, Weis MA, Wu JJ. J Biol Chem. 2004;279:2568–2574. doi: 10.1074/jbc.M311653200. [DOI] [PubMed] [Google Scholar]
- 16.Eyre DR, Weis MA, Wu JJ. Eur Cell Mater. 2006;12:57–63. doi: 10.22203/ecm.v012a07. [DOI] [PubMed] [Google Scholar]
- 17.Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, Monticone M, Catagnola P, Rauch F, Glorieux FH, Vranka J, Bächinger HP, Pace JM, Schwarze U, Byers PH, Weis MA, Fernandes RJ, Eyre DR, Yao Z, Boyce BF, Lee B. Cell. 2006;127:291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 18.Hanna SL, Sherman NE, Kinter MT, Goldberg JB. Microbiology. 2000;146:2495–2508. doi: 10.1099/00221287-146-10-2495. [DOI] [PubMed] [Google Scholar]
- 19.Kinter M, Sherman NE. Protein Sequencing and Identification Using Mass Spectrometry. John Wiley & Sons, Inc; New York: 2000. pp. 152–157. [Google Scholar]
- 20.Eyre DR, Atley LA, Vosberg-Smith K, Shaffer K, Ochs V, Clemens D. In: Chemistry and Biology of Mineralized Tissues. Goldberg M, Boskey A, Robinson C, editors. AAOS Publishers; 2000. pp. 347–350. [Google Scholar]
- 21.Hanson DA, Weis MA, Bollen AM, Maslan SL, Singer FR, Eyre DR. J Bone Miner Res. 1992;7:1251–1258. doi: 10.1002/jbmr.5650071119. [DOI] [PubMed] [Google Scholar]
- 22.Wenstrup RJ, Florer JB, Brunskill EW, Bell SM, Chervoneva I, Birk DE. J Biol Chem. 2004;279:53331–53337. doi: 10.1074/jbc.M409622200. [DOI] [PubMed] [Google Scholar]
- 23.Eyre D. Arthritis Res. 2002;4:30–35. doi: 10.1186/ar380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes DF, Kadler KE. Proc Natl Acad Sci USA. 2006;103:17249–17254. doi: 10.1073/pnas.0608417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atley LM, Mort JS, Lalumiere M, Eyre DR. Bone. 2000;26:241–247. doi: 10.1016/s8756-3282(99)00270-7. [DOI] [PubMed] [Google Scholar]
- 26.Hanson DA, Eyre DR. J Biol Chem. 1996;271:26508–26516. doi: 10.1074/jbc.271.43.26508. [DOI] [PubMed] [Google Scholar]
- 27.Wu JJ, Eyre DR. J Biol Chem. 1995;270:18865–18870. doi: 10.1074/jbc.270.32.18865. [DOI] [PubMed] [Google Scholar]
- 28.Fernandes RJ, Weis MA, Scott MA, Seegmiller RE, Eyre DR. Matrix Biol. 2007;26:597–603. doi: 10.1016/j.matbio.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robins SP, Stewart P, Astbury C, Bird HA. Ann Rheum Dis. 1986;45:969–973. doi: 10.1136/ard.45.12.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beardsworth LJ, Eyre DR, Dickson IR. J Bone Miner Res. 1990;5:671–676. doi: 10.1002/jbmr.5650050702. [DOI] [PubMed] [Google Scholar]
- 31.Eyre DR. Spine. 1997;22(24 Suppl):17S–24S. doi: 10.1097/00007632-199712151-00004. [DOI] [PubMed] [Google Scholar]
- 32.Seibel MJ. Arq Bras Endocrinol Metabol. 2006;50:603–620. doi: 10.1590/s0004-27302006000400006. [DOI] [PubMed] [Google Scholar]
- 33.Eyre DR. 5,140,103. Peptide fragments containing HP and LP cross-links. US Patent. 1992
- 34.Eyre D, Shao P, Weis MA, Steinmann B. Mol Genet Metab. 2002;76:211–216. doi: 10.1016/s1096-7192(02)00036-7. [DOI] [PubMed] [Google Scholar]