Abstract
Improved strategies for synthesis make it possible to expand the range of glycopeptides available for detailed conformational studies. The glycopeptide 1 was synthesized using a new solid phase synthesis of carbohydrates and a convergent coupling to peptide followed by deprotection. Its conformational properties were subjected to NMR analysis and compared with a control peptide 2 prepared by conventional solid phase methods. Whereas peptide 2 fails to manifest any appreciable secondary structure, the glycopeptide 1 does show considerable conformational bias suggestive of an equilibrium between an ordered and a random state. The implications of this ordering effect for the larger issue of protein folding are considered.
Although glycosylation of many natural products is a widespread phenomenon, the consequences of this modification on the molecular properties of proteins are not well understood. An intriguing possibility is that glycosylation of the protein effects the conformation of the nascent chain and influences its rate of folding (1–3). In spite of this and many other outstanding issues pertaining to glycosylated species, studies have been relatively few due to the limited availability of suitable material and model compounds. Some relief is now being realized with improvements in methodology for synthesis of oligosaccharides and glycopeptides (4–11). For instance, access to the glycopeptide 1 studied here (vide infra) is a consequence of these advances (12). Its pentapeptide segment was designed to incorporate the Asn–Xxx–Thr consensus sequence for N-glycosylation, and a trisaccharide was linked through the side chain amide of the asparagine residue to the peptide.
A detailed picture of the organization of a pendant oligosaccharide on a fully folded protein in solution has been obtained from the recent study of the glycosylated CD2 protein (13). However, in trying to determine the extent to which glycosylation influences early stages of the protein folding process, glycopeptides provide more appropriate models since the conformation of the system is not dominated by the overall fold of the ultimately organized protein structure. Some recent studies on glycopeptides have provided insights into potential conformational consequences of glycosylation for both O-linked and N-linked species (14–19).
In the case of O-linked glycopeptides (14, 15, 20), NMR studies have shown that the peptide backbone responds to glycosylation, as evidenced by changes in sequential amide–amide nuclear Overhauser effect (NOE) interactions. The response is further modulated by whether the sugar component is a mono- or disaccharide. Direct evidence for local interaction between sugar and peptide backbone has been indicated by the existence of an NOE crosspeak between the amide proton of the N-acetylglucosamine attached to the O-linked residue and the backbone amide of the threonine to which the sugar was attached (14). In the case of N-linked species in aqueous solution, fluorescence energy transfer and NMR studies support the notion of a change in the distribution of conformations in response to glycosylation (16–19). For N-linked glycopeptides in aqueous solution, NOE data that might indicate proximity between the saccharide and peptide backbone sites have been lacking. In a recent study of a glycosylated tripeptide in organic solvent, NOE interactions indicating an ordered structure have been observed (21).
A problem that had been undercutting research pertaining to this important question was that molecules of the size that can in practice be readily prepared often do not show a clear preference for a single conformation in aqueous solution. Such lack of structural definition complicates NMR studies in particular, and conformational analysis in general. Given the sorts of advances discussed above, we were now in a posture to gain access to more promising constructs. Thus, the glycopeptide 1 used in this study was synthesized using our convergent solid-phase synthesis of a trisaccharide (made via the glycal assembly method) followed by convergent coupling to a pentapeptide and global deprotection. The reference peptide 2 was synthesized using standard solid phase peptide synthesis (22).
RESULTS
Our goal was to conduct studies in aqueous solution that might emulate, as closely as possible, a biologically relevant environment. Although NOE crosspeaks in the amide region were not seen in H2O down to 0°C, they were observed at −12°C using a mixture of 10% C3D6O in 90% H2O as solvent, allowing the increased stabilization of low energy conformers. The addition of up to 15% of acetone had no significant impact on the resonance positions in the spectrum, thus indicating that the level of cosolvent used does not perturb the conformation. To understand how glycosylation influenced the conformational properties of the peptide portion, the unglycosylated peptide 2 was examined under these conditions to provide a frame of reference (see Fig. 1).
Figure 1.

The β-N-linked glycopeptide 1 and the pentapeptide 2 investigated. The arrows indicate some key NOE interactions observed to amidic protons on the glycopeptide.
For the glycopeptide at −12°C, a continuous string of NOEs was observed among the NH protons of the backbone as well as out along the asparagine side chain to its NH and to the N-acetyl group of the N-linked sugar residue. This finding is illustrated via a section of the two-dimensional NOE spectroscopy data from glycopeptide in Fig. 2A. By contrast, the NOE interactions of the peptide are fewer and less intense (Fig. 2B). Along the backbone of the free peptide, a crosspeak of medium intensity connects the NH of the C-terminal Thr residue to the NH of the preceding Leu-4. Also, a weak crosspeak between the NH of this leucine and the α NH of the preceding asparagine residue can be observed. Correcting for the intrinsic variation in intensities of NOEs between the two molecules using an internal standard such as the alanine α to methyl proton crosspeak, the NH–NH NOE intensities of the peptide are still smaller than those of the glycopeptide. In addition the NOE between the backbone amides of Leu-2 and Asn-3 is entirely missing for the peptide. To check for the presence of NOEs in the peptide that might have been missed due to the effects of molecular size (23), a rotating-frame Overhauser effect spectroscopy experiment was conducted and no additional NOEs were observed.
Figure 2.
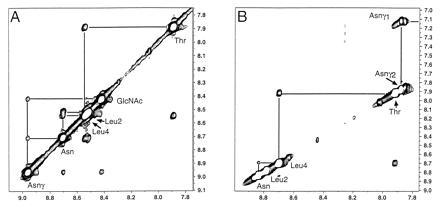
The amide proton portions of 600 MHz 2D NOE spectroscopy experiments with 350 ms mixing time at a temperature of −12°C in 90% H2O/10% C3D6O, using watergate water suppression (24) for the glycopeptide 1 (A) at ≈1 mM and pH 5.6, and the unmodified peptide 2 (B) at ≈4 mM and pH 5.6.
The patterns of amidic to aliphatic NOEs for the reference peptide component of both molecules are rather similar, both in terms of interactions to other protons in the same residue and to α and side chain protons of the previous residue. For the glycopeptide there are NOEs evident between the Asn-γ amide and the amide of the N-acetyl group, with nonexchangeable protons on the sugar. NOE interactions in the glycopeptide are observed between the methyl of the N-acetyl group and the backbone NH of the Leu-2 residue, with evidence for interaction with the Leu-4 NH as well.
This finding is important as it provides a means of identifying the orientation of the N-acetylglucosamine residue relative to the peptide. NOEs between a component of the oligosaccharide and a backbone proton have not been noted in the other N-linked glycopeptide models whose studies in aqueous solution have been reported (17–19). The shifts of the β protons and 3JCαCβ couplings for the asparagine residue are rather similar both in the glycopeptide and free peptide, indicating that the χ1 torsion angle is largely in one of two staggered conformations, χ1 = −60° or 180°, with one favored over the other. Because the β protons of Asn are not discriminated, it is possible that one of these is preferred in the peptide and the other in the glycopeptide. Analysis of peaks arising from the more distal second and third sugar residues was not made because of complications from spectral overlap.
Measurement of the NH to Hα couplings were in the range of 6.5–7.5 Hz with the exception of the threonine amide, which had a coupling of 8.7 Hz, indicating an extended or random structure (25). The effect of temperature on chemical shift of the amides ranged from −7.9 to −9.2 parts per billion/°C. These measurements tend to suggest the absence of strong specific hydrogen bonding interactions for the peptide amide protons (26).
The most likely structural conclusion for the peptide portion of the glycopeptide is a conformational equilibrium between at least one ordered or partially ordered structure, and a random state. To explore whether NOEs would allow the putative ordered state of the glycopeptide to be consistent with a single preferred conformer, 15 simulated annealing calculations were carried out with the x-plor program (27), using 52 NOEs as constraints. There were 15 interresidue NOEs for the peptide portion, and seven between the peptide and the N-linked sugar. These calculations started from an extended structure for the peptide backbone with the side chain amide linkage and the N-acetyl on the glucosamine in the trans orientation. The latter arrangement is consistent with coupling constants to the side chain and N-acetyl amides of 9.8 and 9.5 Hz, respectively. An extended structure seemed to be the most appropriate as well since there were neither the intermediate range NOEs nor coupling constants in the peptide backbone that would suggest a turn or more compact structure.
Of the resulting structures, two were discarded for NOE violations and unfavorable energies, of the remaining 13, 11 fell into a family of rather similar conformations (pairwise rms 1.35 ± 1.11 Å). The family of similar conformations is shown in Fig. 3. These results indicate that a single conformational class is compatible with the pattern of NOEs. In analyzing influences exerted by the oligosaccharide, one might be concerned with the fact that the termini of the peptide were not capped. However, earlier work on conformational preferences of peptides suggest that this is not a dominating effect (26). The conclusions drawn here and that from earlier work (14–19) indicates that glycosylation at least limits the conformational range available to the neighboring peptide. Even if a single well defined secondary structure in this size peptide domain is not induced, the conformational restriction has the effect of reducing the range of possibilities explored by this domain. This restriction could likely be extrapolated to the behavior of a glycosylated segment in a protein being transcribed.
Figure 3.

Rendering of the 11 similar structures derived from the simulated annealing calculations. The two distal sugars are omitted from the illustration since there were no experimentally determined NOE constraints for them.
Of course, the effects shown here may not be solely attributable to specific carbohydrate contacts. In principle other noninteractive, noncarbohydrate moieties mounted at the γ carboxyl of the asparganine residue might well have induced their own conformational restrictions on the peptide. However, our work in concert with earlier findings (14–21) tends to suggest the existence of such specific contacts as biasing element in glycopeptides. It has been argued theoretically that a modest change in biasing the energy landscape can have a significant effect on reducing the number of folding paths a protein may choose with concomitant increase in the rate of productive folding (3). Thus, even if glycosylation only prejudiced the conformation at an early stage of folding, it could have the impact of substantially speeding up the process of arriving at the correct folded structure. The results described are certainly supportive of such a scenario. Further advances in synthetic methodology will allow for assessment of the full range of the effects of glycosylation on secondary structure.
Acknowledgments
This research was supported by National Institutes of Health Grant AI16943. We also acknowledge a Miriam and Benedict Wolf Cancer Research Fund Grant to R.A.K. and a National Institutes of Health Postdoctoral Training Grant T32 CA62948 to X.B.
Footnotes
Abrreviation: NOE, nuclear Overhauser effect.
References
- 1.Levinthal C. In: Mossbauer Spectroscopy in Biological Systems. Debrunner P, Tsibris J C M, Munch E, editors. Urbana: Univ. of Illinois Press; 1969. pp. 22–24. [Google Scholar]
- 2.Wetlaufer D B, Ristow S. Annu Rev Biochem. 1973;42:135–158. doi: 10.1146/annurev.bi.42.070173.001031. [DOI] [PubMed] [Google Scholar]
- 3.Zwanzig R, Szabo A, Bagchi B. Proc Natl Acad Sci USA. 1992;89:20–22. doi: 10.1073/pnas.89.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunz H. Pure Appl Chem. 1993;65:1223–1232. [Google Scholar]
- 5.Meldal M, Bock K. Glycoconjugates J. 1994;11:59–63. doi: 10.1007/BF00731144. [DOI] [PubMed] [Google Scholar]
- 6.Meldal M. In: Neoglycoconjugates: Preparation and Applications. Lee Y C, Lee R T, editors. London: Academic; 1994. pp. 145–198. [Google Scholar]
- 7.Danishefsky S J, Roberge J Y. In: Glycopeptides and Related Compounds: Chemical Synthesis, Analysis and Applications. Large D G, Warren C D, editors. New York: Dekker; 1996. in press. [Google Scholar]
- 8.Paulsen H. Angew Chem Int Ed Engl. 1990;29:823–938. [Google Scholar]
- 9.Kunz H. Angew Chem Int Ed Engl. 1987;26:294–308. [Google Scholar]
- 10.Cohen-Anisfeld S T, Lansbury P T., Jr J Am Chem Soc. 1993;115:10531–10537. [Google Scholar]
- 11.Anisfeld S T, Lansbury P T., Jr J Org Chem. 1990;55:5560–5562. [Google Scholar]
- 12.Roberge J Y, Beebe X, Danishefsky S J. Science. 1995;269:202–204. doi: 10.1126/science.7618080. [DOI] [PubMed] [Google Scholar]
- 13.Wyss D F, Choi J S, Li J, Knoppers M H, Willis K J, Arulanasdam A R N, Smolyar A, Reinherz E L, Wager G. Science. 1995;269:1273–1278. doi: 10.1126/science.7544493. [DOI] [PubMed] [Google Scholar]
- 14.Andreotti A H, Kahne D. J Am Chem Soc. 1993;115:3352–3353. [Google Scholar]
- 15.Liang R, Andreotti A H, Kahne D. J Am Chem Soc. 1995;115:10395–10396. [Google Scholar]
- 16.Imperiali B, Rickert K W. Proc Natl Acad Sci USA. 1995;92:97–101. doi: 10.1073/pnas.92.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis J T, Hirsani S, Bartlett C, Reid B R. J Biol Chem. 1994;269:3331–3338. [PubMed] [Google Scholar]
- 18.Wormald M R, Wooten E W, Bazzo R, Edge C J, Feinstein A, Rademacher T W, Dwek R A. Eur J Biochem. 1991;198:131–139. doi: 10.1111/j.1432-1033.1991.tb15995.x. [DOI] [PubMed] [Google Scholar]
- 19.Rickert K W, Imperiali B. Chem Biol. 1995;2:751–759. doi: 10.1016/1074-5521(95)90103-5. [DOI] [PubMed] [Google Scholar]
- 20.Huang X, Smith M C, Berzofsky J A, Barchi J. FEBS Lett. 1996;393:280–286. doi: 10.1016/0014-5793(96)00912-x. [DOI] [PubMed] [Google Scholar]
- 21.Lee K-C, Falcone M L, Davis J T. J Org Chem. 1996;61:4198–4199. doi: 10.1021/jo960590c. [DOI] [PubMed] [Google Scholar]
- 22.Grant G A, editor. Synthetic Peptides: A User’s Guide. New York: Freeman; 1992. [Google Scholar]
- 23.Neuhaus D, Williamson M P. The Nuclear Overhauser Effect in Structural and Conformational Analysis. New York: VCH; 1989. [Google Scholar]
- 24.Piotto M, Saudek V, Sklenar V. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 25.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 26.Dyson H J, Rance M, Houghton R A, Lerner R, Wright P E. J Mol Biol. 1988;201:161–200. doi: 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]
- 27.Brunger, A. T. (1992) x-plor: A System for X-ray Crystallography and NMR (Yale Univ. Press, New Haven, CT), version 3.1.


