Abstract
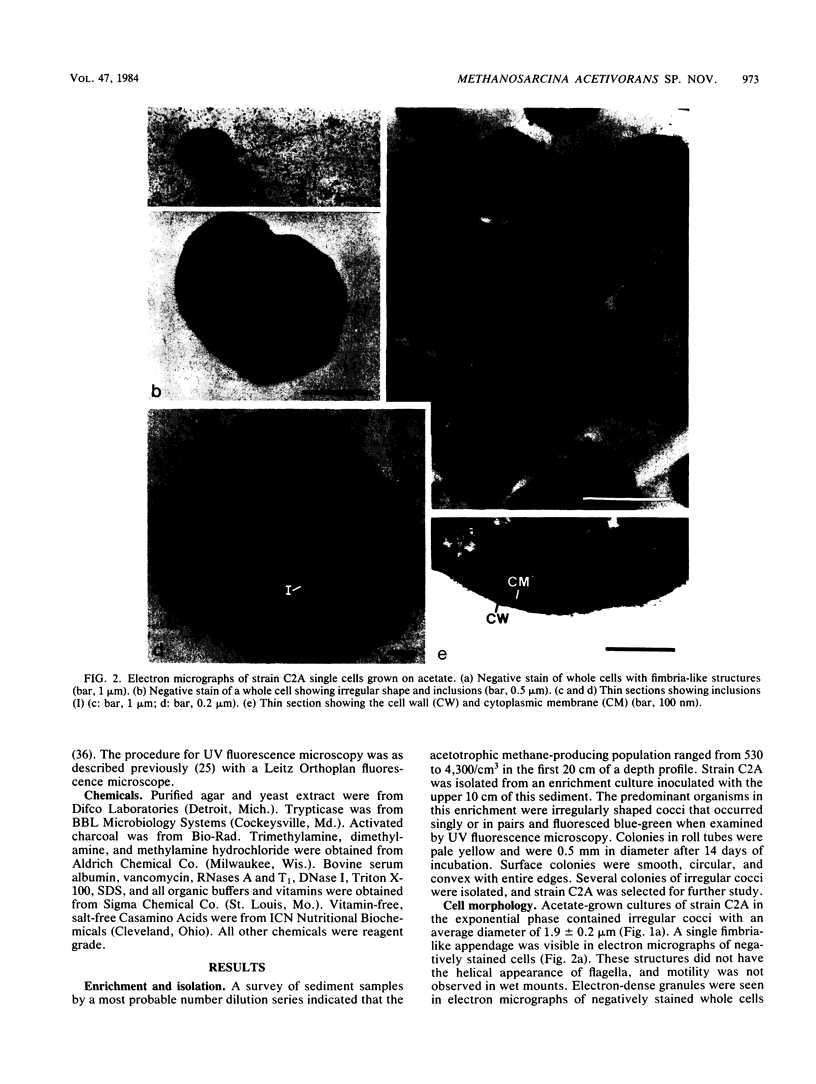
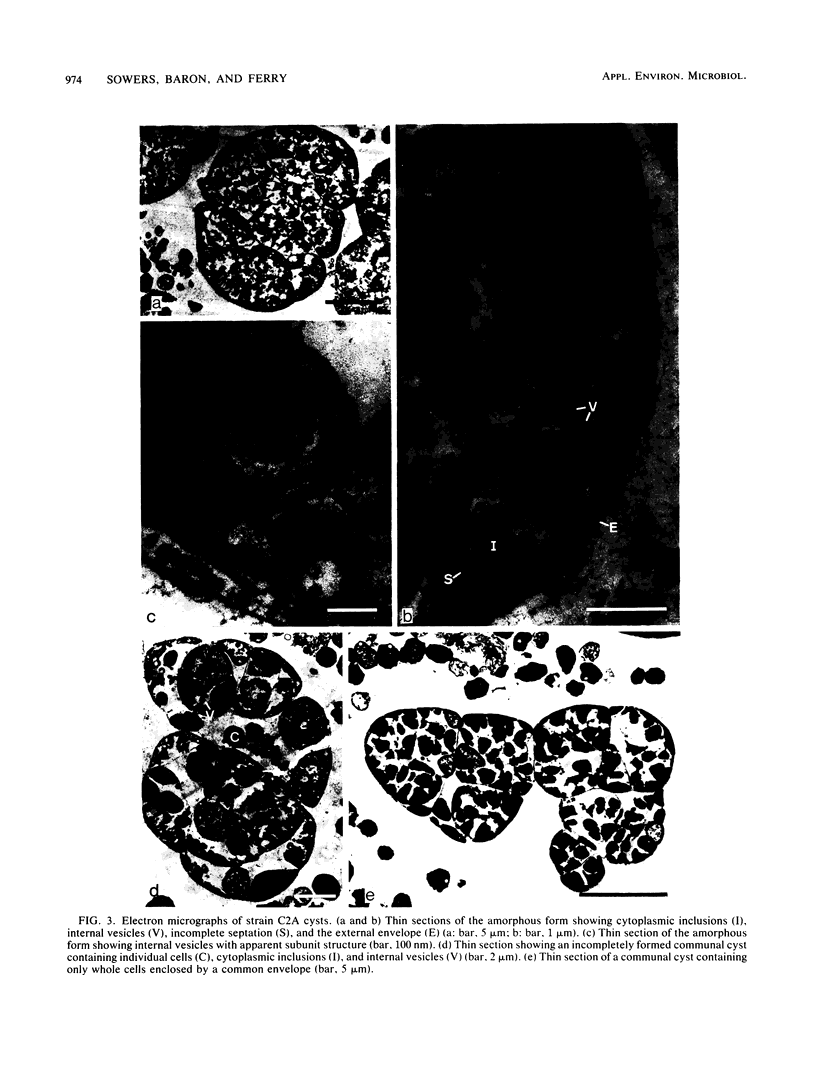
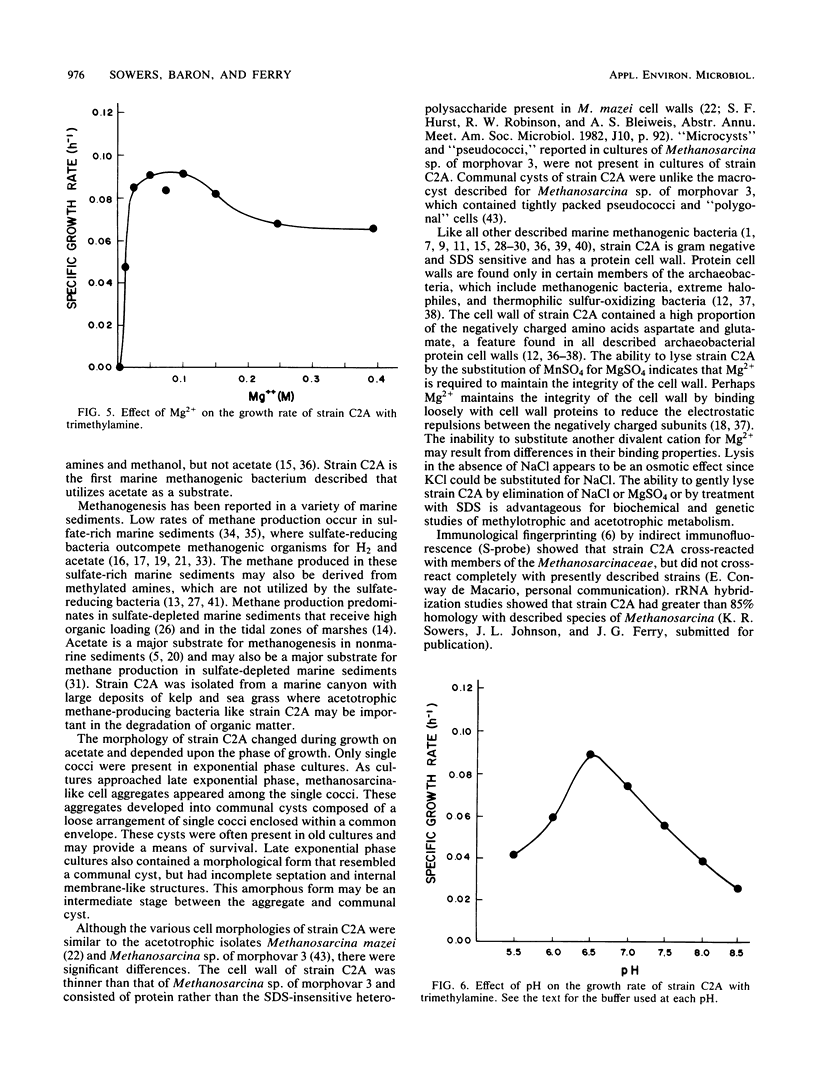
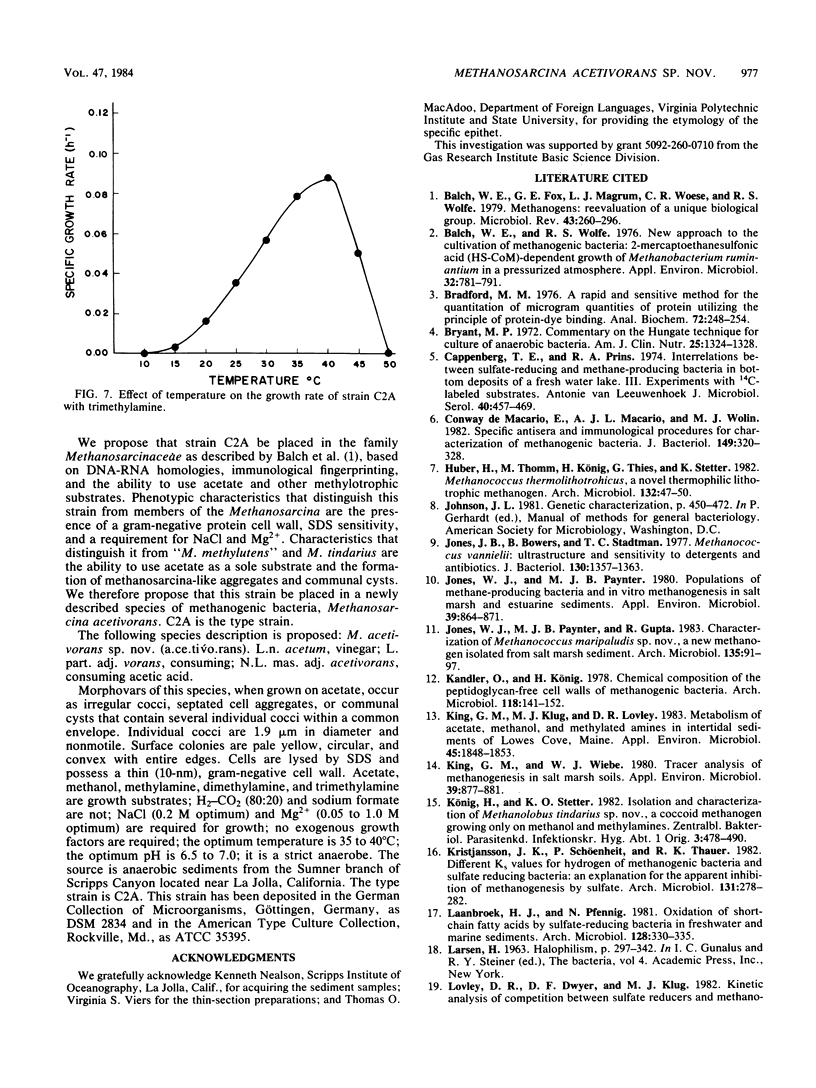
A new acetotrophic marine methane-producing bacterium that was isolated from the methane-evolving sediments of a marine canyon is described. Exponential phase cultures grown with sodium acetate contained irregularly shaped cocci that aggregated in the early stationary phase and finally differentiated into communal cysts that released individual cocci when ruptured or transferred to fresh medium. The irregularly shaped cocci (1.9 ± 0.2 mm in diameter) were gram negative and occurred singly or in pairs. Cells were nonmotile, but possessed a single fimbria-like structure. Micrographs of thin sections showed a monolayered cell wall approximately 10 nm thick that consisted of protein subunits. The cells in aggregates were separated by visible septation. The communal cysts contained several single cocci encased in a common envelope. An amorphous form of the communal cyst that had incomplete septation and internal membrane-like vesicles was also present in late exponential phase cultures. Sodium acetate, methanol, methylamine, dimethylamine, and trimethylamine were substrates for growth and methanogenesis; H2-CO2 (80:20) and sodium formate were not. The optimal growth temperature was 35 to 40°C. The optimal pH range was 6.5 to 7.0. Both NaCl and Mg2+ were required for growth, with maximum growth rates at 0.2 M NaCl and 0.05 M MgSO4. The DNA base composition was 41 ± 1% guanine plus cytosine. Methanosarcina acetivorans is the proposed species. C2A is the type strain (DSM 2834, ATCC 35395).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Wolfe R. S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl Environ Microbiol. 1976 Dec;32(6):781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bryant M. P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. 1972 Dec;25(12):1324–1328. doi: 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- Cappenberg T. E., Prins R. A. Interrelations between sulfate-reducing and methane-producing bacteria in bottom deposits of a fresh-water lake. 3. Experiments with 14C-labeled substrates. Antonie Van Leeuwenhoek. 1974;40(3):457–469. doi: 10.1007/BF00399358. [DOI] [PubMed] [Google Scholar]
- Conway de Macario E., Macario A. J., Wolin M. J. Specific antisera and immunological procedures for characterization of methanogenic bacteria. J Bacteriol. 1982 Jan;149(1):320–328. doi: 10.1128/jb.149.1.320-328.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. B., Bowers B., Stadtman T. C. Methanococcus vannielii: ultrastructure and sensitivity to detergents and antibiotics. J Bacteriol. 1977 Jun;130(3):1357–1363. doi: 10.1128/jb.130.3.1357-1363.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. J., Paynter M. J. Populations of methane-producing bacteria and in vitro methanogenesis in salt marsh and estuarine sediments. Appl Environ Microbiol. 1980 Apr;39(4):864–871. doi: 10.1128/aem.39.4.864-871.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O., König H. Chemical composition of the peptidoglycan-free cell walls of methanogenic bacteria. Arch Microbiol. 1978 Aug 1;118(2):141–152. doi: 10.1007/BF00415722. [DOI] [PubMed] [Google Scholar]
- King G. M., Klug M. J., Lovley D. R. Metabolism of acetate, methanol, and methylated amines in intertidal sediments of lowes cove, maine. Appl Environ Microbiol. 1983 Jun;45(6):1848–1853. doi: 10.1128/aem.45.6.1848-1853.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. M., Wiebe W. J. Tracer analysis of methanogenesis in salt marsh soils. Appl Environ Microbiol. 1980 Apr;39(4):877–881. doi: 10.1128/aem.39.4.877-881.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laanbroek H. J., Pfennig N. Oxidation of short-chain fatty acids by sulfate-reducing bacteria in freshwater and in marine sediments. Arch Microbiol. 1981 Jan;128(3):330–335. doi: 10.1007/BF00422540. [DOI] [PubMed] [Google Scholar]
- Lovley D. R., Klug M. J. Intermediary metabolism of organic matter in the sediments of a eutrophic lake. Appl Environ Microbiol. 1982 Mar;43(3):552–560. doi: 10.1128/aem.43.3.552-560.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Klug M. J. Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations. Appl Environ Microbiol. 1983 Jan;45(1):187–192. doi: 10.1128/aem.45.1.187-192.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C. S., Berner R. A. Methane production in the interstitial waters of sulfate-depleted marine sediments. Science. 1974 Sep 27;185(4157):1167–1169. doi: 10.1126/science.185.4157.1167. [DOI] [PubMed] [Google Scholar]
- Mink R. W., Dugan P. R. Tentative identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 Mar;33(3):713–717. doi: 10.1128/aem.33.3.713-717.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountfort D. O., Asher R. A. Role of sulfate reduction versus methanogenesis in terminal carbon flow in polluted intertidal sediment of waimea inlet, nelson, new zealand. Appl Environ Microbiol. 1981 Aug;42(2):252–258. doi: 10.1128/aem.42.2.252-258.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland R. S., Polcin S. Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl Environ Microbiol. 1982 Dec;44(6):1270–1276. doi: 10.1128/aem.44.6.1270-1276.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivard C. J., Henson J. M., Thomas M. V., Smith P. H. Isolation and Characterization of Methanomicrobium paynteri sp. nov., a Mesophilic Methanogen Isolated from Marine Sediments. Appl Environ Microbiol. 1983 Aug;46(2):484–490. doi: 10.1128/aem.46.2.484-490.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone F. J., Martens C. S. Methane production from acetate and associated methane fluxes from anoxic coastal sediments. Science. 1981 Feb 13;211(4483):707–709. doi: 10.1126/science.211.4483.707. [DOI] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Metabolism of formate in Methanobacterium formicicum. J Bacteriol. 1980 Jun;142(3):800–807. doi: 10.1128/jb.142.3.800-807.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior E., Lindström E. B., Banat I. M., Nedwell D. B. Sulfate reduction and methanogenesis in the sediment of a saltmarsh on the East coast of the United kingdom. Appl Environ Microbiol. 1982 May;43(5):987–996. doi: 10.1128/aem.43.5.987-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Ferry J. G. Isolation and Characterization of a Methylotrophic Marine Methanogen, Methanococcoides methylutens gen. nov., sp. nov. Appl Environ Microbiol. 1983 Feb;45(2):684–690. doi: 10.1128/aem.45.2.684-690.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensland H., Larsen H. A study of the cell envelope of the halobacteria. J Gen Microbiol. 1969 Mar;55(3):325–336. doi: 10.1099/00221287-55-3-325. [DOI] [PubMed] [Google Scholar]
- Sørensen J., Christensen D., Jørgensen B. B. Volatile Fatty acids and hydrogen as substrates for sulfate-reducing bacteria in anaerobic marine sediment. Appl Environ Microbiol. 1981 Jul;42(1):5–11. doi: 10.1128/aem.42.1.5-11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOLIN E. A., WOLIN M. J., WOLFE R. S. FORMATION OF METHANE BY BACTERIAL EXTRACTS. J Biol Chem. 1963 Aug;238:2882–2886. [PubMed] [Google Scholar]
- Weiss R. L. Subunit cell wall of Sulfolobus acidocaldarius. J Bacteriol. 1974 Apr;118(1):275–284. doi: 10.1128/jb.118.1.275-284.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Ankwanda E., Wolfe R. S. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982 Mar;149(3):852–863. doi: 10.1128/jb.149.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfrey M. R., Ward D. M. Substrates for sulfate reduction and methane production in intertidal sediments. Appl Environ Microbiol. 1983 Jan;45(1):193–199. doi: 10.1128/aem.45.1.193-199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhilina T. N., Zavarzin G. A. Obrazovanie tsist metanosartsinoi. Mikrobiologiia. 1979 May-Jun;48(3):451–456. [PubMed] [Google Scholar]