Abstract
We have used two monovalent phage display libraries containing variants of the Zif268 DNA-binding domain to obtain families of zinc fingers that bind to alterations in the last 4 bp of the DNA sequence of the Zif268 consensus operator, GCG TGGGCG. Affinity selection was performed by altering the Zif268 operator three base pairs at a time, and simultaneously selecting for sets of 16 related DNA sequences. In this way, only four experiments were required to select for all possible 64 combinations of DNA triplet sequences. The results show that (i) for high-affinity DNA binding in the range observed for the Zif268 wild-type complex (Kd = 0.5–5 nM), finger 1 specifically requires the arginine at the carboxy terminus of its recognition helix that forms a bidentate hydrogen-bond with the guanine base (G) in the crystal structure of Zif268 complexed to its DNA operator sequence GCG TGG GCG; (ii) when the guanine base (G) is replaced by A, C, or T, a lower-affinity family (Kd ⩾ 50 nM) can be detected that shows an overall tendency to bind G-rich DNA; (iii) the residues at position 2 on the finger 2 recognition helix do not appear to interact strongly with the complementary 5′ base in the finger 1 binding site; and (iv) unexpected substitutions at the amino terminus of finger 1 can occasionally result in specificity for the 3′ base in the finger 1 binding site. A DNA recognition directory was constructed for high-affinity zinc fingers that recognize all three bases in a DNA triplet for seven sequences of the type GNN. Similar approaches may be applied to other zinc fingers to broaden the scope of the directory.
The zinc finger motif is the first of the protein–DNA structures for which detailed structural information is available (1, 2) to be extensively studied in terms of its DNA base recognition properties (3–9). The motif is widespread in eukaryote cells where it can be identified according to a conserved zinc-chelating sequence of the type -Cys-(Xaa)2-4-Cys-(Xaa)3-Phe-(Xaa)5-Leu-(Xaa)2-His-(Xaa)3-5-His (10). Each finger is ≈30 amino acid residues long and is folded into a compact module that comprises an α helix containing the invariant histidine residues coordinated through zinc to the cysteines of a single β-turn (11–13). Inside cells, zinc fingers are often contained as repetitive arrays in transcription factors that direct sequence-specific binding to various DNA operators (14). In vitro, certain zinc fingers can also be shown to bind RNA (15, 16) or DNA–RNA hybrids (17).
The crystal structures of the DNA complexes of both Zif268 [a three-finger domain from a murine transcription factor (18)] and Tramtrack [a two-finger domain from Drosophila (19)] reveal that the recognition helices of individual fingers are inserted into the major groove of DNA at three base pair intervals, and that DNA recognition is mediated through base contacts with the side-chains of amino acids located at four positions on the recognition helix (amino acids at positions −1, 2, 3, and 6 in Fig. 1a) (1, 2). The modular arrangement of the recognition helices has prompted speculation that DNA recognition operates according to a code involving only a limited number of amino acid substitutions at the four positions on the recognition helix (1, 4).
Figure 1.
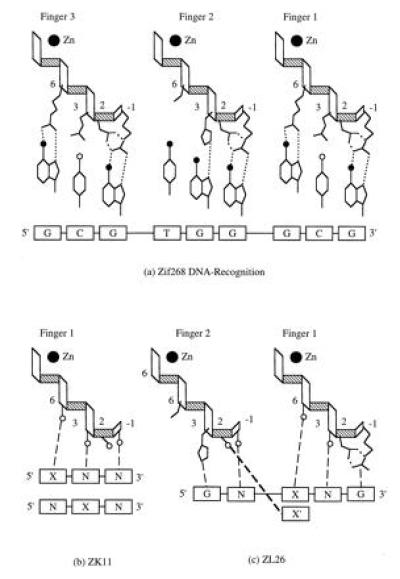
(a) Zif268 DNA recognition (1). Each of the three zinc finger recognition helices is shown in relation to its 3 base pair binding site. Hydrogen bonds (dotted lines) are formed when side-chains at positions −1, 2, 3, and 6 contact bases in the DNA major groove. Each helix is oriented such that position −1 on the helix amino terminus interacts at the 3′ end of the binding site. Fingers 1 and 3 contain the side-chains Arg-1, Asp-2, Glu-3, and Arg-6. Finger 2 contains Arg-1, Asp-2, His-3, and Thr-6. The DNA bases are differentiated according to filled circles (carbonyl groups) and open circles (amide groups exposed in the DNA major groove). (b) The ZK11 phage display library. The library contains amino acid substitutions at positions −1, 2, 3, and 6 (shown as open circles) on finger 1 and are shown in relation to two sets of DNA base replacements, XNN and NXN. X is fixed as A, C, G, or T, and N stands for a mixture of four bases. Dashed lines represent the expected interactions between side-chains and DNA bases according to the Zif268 crystal structure (1). (c) The ZL26 library. The library substitutions are located at positions −1 and 2 on finger 2 and at positions 3 and 6 on finger 1 (shown as four open circles). The thick dashed line represents the potential interaction (1, 2) between the side-chain at position 2 (finger 2) and the complementary 5′ base in the finger 1 binding site.
Experiments involving Zif268 phage display libraries have shown that it is possible to alter the binding specificities of individual fingers for their respective DNA triplets, and the specificity of each finger can often be inferred from the amino acids present at the four positions of a given zinc finger helix (6–9). Not all of the possible 64 triplets are specifically recognized by individual fingers under affinity selection, either due to the content of particular phage display libraries or as a general property of the protein–DNA complex under investigation. Furthermore, the extent to which DNA recognition is modified according to the position of the finger in a multifingered complex is not known. To address these questions, we have used sets of 16 related DNA triplets (9) that together encode all 64 base combinations to screen two Zif268 libraries for families of fingers that recognize DNA with different binding affinities. The results should aid in the design of zinc finger domains that recognize biologically important DNA sequences both in vivo and in vitro.
MATERIALS AND METHODS
Design of Zif268 Phage Display Libraries.
The construction of a phage display library (ZK11) to select for Zif268 finger 1 variants with altered nucleotide triplet specificity has been described (7). A second phage display library (ZL26) was constructed by Kunkel mutagenesis (20). A frameshifted derivative of the Zif268 phage display vector, pZF910 (7), which contains a SalI restriction endonuclease site at the frameshift, was used for single-stranded template preparation. The library was constructed using two mutagenic oligonucleotides: (i) 5′TGC GAT CGT CGA TTT TCT CGC TCG GAT NNS CTT ACC NNS CAT ATC CGC ATC CAC3′, where N is any of the four possible bases and S is either G or C, which restores the reading frame for zinc finger expression and also introduces random substitutions at positions 3 and 6 on finger 1; (ii) 5′ATG CGT AAC TTC AGT NNS AGT NNS CAC CTT ACC ACC CAC3′, which introduces random substitutions at positions −1 and 2 on the finger 2 recognition helix (see Fig. 1a for amino acid positions). These substitutions were introduced primarily to select for residues at position 2 on finger 2 that potentially interact with the complementary base in the 5′ position of the finger 1 binding site, as suggested by Pavletich and Pabo (1) and Fairall et al. (2) (Fig. 1c). The residues at positions −1 and 2 are unchanged in finger 1 to form the specific interaction with guanine (G) at the 3′ end of the Zif268 operator sequence, GCG TGG GCG.
The mutagenesis reaction mixture was transformed into Escherichia coli XL-1 Blue cells (Stratagene) and titered as 8 × 107 independent transformants. Plasmid DNA from the library was restricted with SalI to detect the amount of background molecules (pZF910). The mutagenesis efficiency was estimated to be at least 30%. The total number of clones expressing zinc finger variants (2.4 × 107) was, therefore, over 20-fold higher than the theoretical library size (324), and 4-fold higher than the number required to ensure 99% probability that all clones are represented.
Affinity Matrix Construction and Library Selections.
The construction of an affinity matrix has been described (7). Briefly, biotinylated DNA containing the Zif268 binding site or a variant thereof is bound to streptavidin-coated microtiter wells, and the wells were subsequently blocked with acetylated BSA. Selections were performed in a buffer containing 25 mM LiCl2/10 mM Tris·HCl (pH 7.0). Phagemid particles [≈5 × 1011 colony forming units (cfu)] propagated from a zinc finger phage display library (ZK11 or ZL26) were added to the wells and incubated for 1 hr at 4°C. The wells were rinsed with 15 3-sec exposures to a buffer containing 10 mM NaCl/10 mM Tris·HCl (pH 8.0). Bound particles were eluted by incubating the well with 0.5 M NaCl/10 mM Tris·HCl (pH 8.0) for 30 min at room temperature. Phagemid binding was monitored by titering an aliquot (10 μl) from each well on microtiter plates and the remainder (90 μl) was propagated by infecting E. coli XL-1 Blue cells. Background levels were calculated by binding phagemids in wells containing either duplex DNA in which the Zif268 binding site (GCG TGG GCG) was replaced with TAT GTT TAT, or in wells containing no DNA. We did not see any significant difference between these two negative controls.
ZK11 library selections were performed in affinity wells containing the entire Zif268 binding site in which the finger 1 site (GCG in GCG TGG GCG) was replaced by DNA triplets in which one of the bases at either the 5′ or middle location in the finger 1 binding site was fixed: ANN, CNN, GNN, TNN, NAN, NCN, NGN, NTN, where A, C, G, T, and N are adenine, cytosine, guanine, thymine, and all four bases, respectively. Each target cell therefore contained a set of 16 altered Zif268 binding sites. Library selections were performed in the presence of 10 nM competitor DNA containing altered Zif268 binding sites with fixed bases other than the base for which selection was performed (e.g., if selection was for a GNN, competitor DNA contained ANN, CNN, and TNN). ZL26 selections were for triplets of the type N-AN(G), N-CN(G), N-GN(G), N-TN(G), where the fixed base occupies the 5′ position of the finger 1 binding site and the triplets are contained within both finger 1 and 2 binding sites.
Zinc Finger Specificity and Affinity Determinations.
To determine the base specificities of individual clones, phagemids were incubated in affinity wells containing fixed bases at one of the three locations in the finger 1 binding site, then rinsed and eluted as described above. Phagemid titers were calculated by plating infected E. coli XL-1 Blue cells on Luria–Bertani agar plates containing 100 μg/ml ampicillin. After incubation overnight at 37°C, the binding signatures (9) were read by identifying which of the fixed bases for each of the three base locations in the binding site gave the highest binding affinity. A semiquantitative estimation of the binding affinities was also performed by binding these phagemids in wells containing sites in which all three base positions were fixed. Binding affinities were calibrated by comparing the binding titers of wild-type Zif268 phagemids against previously reported Kd values for various substitutions of the finger 1 binding site (7). As shown in Table 1, the relative binding titers correlate closely with the relative Kd values from band-shift assays. For clones with high-affinity binding, phagemid titers were in the range of 5 × 107 to 5 × 108 cfu/ml, indicating that these fingers bind DNA with Kd values in the range 0.5–5 nM. Two or more base pair substitutions of the Zif268 binding site lower phagemid titers to background levels, indicating that DNA binding affinity was reduced substantially, with Kd greater than 50 nM.
Table 1.
Calibration of zinc finger binding affinities
| DNA triplet | Phagemid titer, cfu/ml | Kd, nM | Relative Kd (vs GCG) | Relative titer |
|---|---|---|---|---|
| GAG | 7 × 107 | 2.8 | 5.6 | 7.1 |
| GCG | 5 × 108 | 0.5 | 1 | 1 |
| GGG | 6 × 107 | 5.6 | 11.2 | 8.3 |
| GTG | 7 × 107 | 3.4 | 6.8 | 7.1 |
| GCA | 2 × 108 | 2.4 | 4.8 | 2.5 |
| GCC | 6 × 107 | 3.0 | 7.4 | 8.3 |
| GCT | 6 × 107 | 3.7 | 7.5 | 8.3 |
| GTT | 5 × 106 | ND | — | ≥100 |
| ATA | 5 × 106 | ND | — | ≥100 |
Binding affinities for zinc finger variants were calibrated by comparing the binding titers of Zif268 phagemids against previously reported Kd values for the free peptide (7). Two or more base substitutions of the Zif268 finger 1 triplet (GCG) result in background levels of affinity binding (5 × 106 cfu/ml). ND, not determined.
RESULTS AND DISCUSSION
Zif268 is a zinc finger DNA binding domain that consists of three independent finger motifs adapted to bind a G-rich sequence of DNA, GCG TGG GCG. X-ray crystallography of both the Zif268 and Tramtrack DNA complexes shows that the fingers can be aligned over 3-bp intervals by a periodic rotation and translation of each finger with respect to the main DNA axis (1, 2). The recognition helix of each finger interacts with bases along one strand of the major groove, with the side-chains at positions −1, 2, 3, and 6 on the recognition helix making analogous contacts with the bases over 3-bp intervals (Fig. 1a).
Residues at position −1 usually contact the 3′ base of each DNA triplet. Position 3 contains residues that recognize the middle base of the triplet, and if position 6 contains arginine, it makes a highly specific bidentate hydrogen bond with 5′ guanine. Position 2 seems to play an auxiliary role in DNA recognition. In Zif268, aspartate at position 2 (Asp-2) is on all three recognition helices, where it appears to buttress the interaction between Arg-1 and a 3′ guanine. In Tramtrack, Ser-2 on the first finger contacts the 3′ base directly due to a local distortion in the DNA (2) (Fig. 2), and in both structures Asp-2 may also interact with cytosine when it is complementary to the 5′ guanine in the binding site of a preceding finger (see below).
Figure 2.
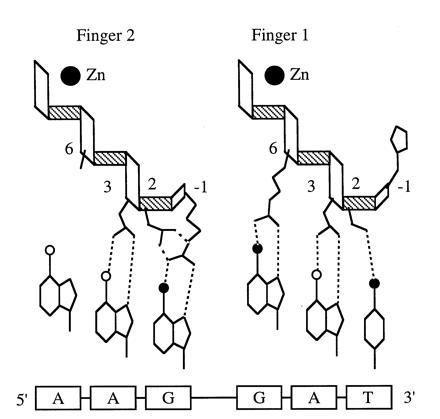
Tramtrack DNA recognition (2). Interactions between the side-chains on each of the recognition helices and the DNA bases are presented according to the scheme in Fig. 1. The finger 1 helix contains His-1, Ser-2, Asn-3, and Arg-6, and the finger 2 helix contains Arg-1, Asp-2, Asn-3, and Ala-6.
To characterize the extent that finger motifs from phage display libraries can be adapted to bind different DNA triplets, we used affinity selection to screen for families of clones that vary in their ability to bind sets of 16 related DNA triplets simultaneously. Each set contains a fixed base at one position in the DNA triplet (for example, GNN, where the fixed base is guanine at the 5′ position of the triplet and N is any base) so that only four sets of selection experiments need be performed to cover the entire range of 64 base combinations (in this case, ANN, CNN, GNN, and TNN). Also, by varying the identity of the fixed base (X) and its position in the DNA base triplet (XNN or NXN, etc.), we can test for “recognition rules” between side chains at a particular position on the recognition helix and the fixed base in the DNA triplet.
The Role of Arg-6 in DNA Recognition.
In Zif268, arginine at position 6 (Arg-6) on the finger 1 recognition helix makes a bidentate hydrogen-bond with guanine (G) in the Zif268 binding site GCG TGG GCG. An analogous interaction also occurs with Arg-6 on the finger 3 recognition helix with G in GCG TGG GCG (Fig. 1a). Affinity sorting selects for a family of clones that contain arginine at position 6 on the finger 1 recognition helix when the finger 1 binding site (GCG in GCG TGG GCG) is replaced by the related set of DNA triplets, GNN (upper part of Table 2). Selection for Arg-6 is accompanied by a significant increase in the level of DNA binding in target wells that contain the altered Zif268 binding sites (about 50-fold over background binding). However, if the 5′ G is altered to A, C, or T (i.e., selection is for DNA triplets containing ANN, CNN, and TNN), the phage display library cannot be enriched for clones that bind DNA above background levels. Although affinity selection could still result in a consensus selection for bases other than 5′G under these conditions (see below), the upper part of Table 2 shows that at position 6 on the recognition helix, which should respond to the identity of the 5′ base, there is also no selection-driven consensus for these replacements of the finger 1 binding site. The results suggest that the Arg-6/5′ G interaction found in the crystal structure is a unique, and energetically important component of DNA recognition in the finger 1 binding site of Zif268 that may limit the extent to which Zif268 can be adapted to recognize altered DNA sequences.
Table 2.
Selection for base replacements of the finger 1 binding site of Zif268
| DNA triplet | Sort | Amino acid at
helix positions
|
Predicted net charge at pH 7.0 | |||
|---|---|---|---|---|---|---|
| −1 | 2 | 3 | 6 | |||
| ANN | 5 | Q | G | S | G | 0 |
| A | V | G | R | 0 | ||
| E | S | E | R | −1 | ||
| G | G | Q | G | 0 | ||
| E | L | N | S | −1 | ||
| V | Q | E | N | −1 | ||
| E | V | G | E | −2 | ||
| K | G | E | G | 0 | ||
| CNN | 5 | P | A | G | K | +1 |
| V | W | R | Q | +1 | ||
| G | E | K | G | 0 | ||
| D | K | G | Q | 0 | ||
| G | G | R | E | 0 | ||
| S | H | R | A | +1 | ||
| G | D | E | R | −1 | ||
| L | D | R | D | −1 | ||
| E | R | G | G | 0 | ||
| GNN | 5 | T | Q | S | R | +1 |
| S | S | E | R | 0 | ||
| E | Q | R | R | +1 | ||
| E | S | S | R | 0 | ||
| TNN | 5 | T | R | G | Q | +1 |
| T | G | G | K | +1 | ||
| S | A | V | T | 0 | ||
| N | S | G | Q | 0 | ||
| R | S | D | K | 0 | ||
| NAN | 3 | S | G | V | R | +1 |
| T | Q | G | R | +1 | ||
| K | A | T | R | +2 | ||
| R | A | A | R | +2 | ||
| 5 | S | G | G | R (3) | +1 | |
| E | R | S | R | +1 | ||
| 6 | S | A | N | R | +1 | |
| E | A | N | R | 0 | ||
| S | G | G | R (4) | +1 | ||
| S | T | G | R | +1 | ||
| S | S | G | R | +1 | ||
| S | A | G | R | +1 | ||
| E | G | S | R | 0 | ||
| E | G | A | R | 0 | ||
| NCN | 3 | D | W | G | R | +1 |
| S | T | G | R | +1 | ||
| D | W | S | R | 0 | ||
| G | W | E | R | 0 | ||
| 6 | R | S | E | R (4) | +1 | |
| Q | G | E | R | 0 | ||
| R | V | D | R | +1 | ||
| A | R | D | R | +1 | ||
| E | G | A | R | 0 | ||
| E | G | S | R | 0 | ||
| E | A | G | R | 0 | ||
| A | A | Q | R | +1 | ||
| NGN | 3 | S | G | G | R (2) | +1 |
| T | T | G | R | +1 | ||
| E | K | S | R | +1 | ||
| 6 | R | E | H | R | +1 | |
| K | E | H | R | +1 | ||
| S | G | G | R (2) | +1 | ||
| T | S | G | R | +1 | ||
| T | G | G | R | +1 | ||
| T | F | G | R | +1 | ||
| E | S | K | R (3) | +1 | ||
| NTN | 3 | E | T | S | R | 0 |
| E | R | S | R | +1 | ||
| T | S | A | R | +1 | ||
| E | G | S | R | 0 | ||
| E | T | S | R | 0 | ||
| T | G | S | R | +1 | ||
| 6 | T | N | A | R | +1 | |
| T | W | M | R | +1 | ||
| E | G | A | R (3) | 0 | ||
| T | G | M | R | +1 | ||
| S | S | S | R | +1 | ||
| T | H | A | R | +1 | ||
| S | G | G | R (3) | +1 | ||
Finger Coding Relationships.
We also tested position 3 on the finger 1 recognition helix for any DNA recognition properties that might emerge when affinity selection is for GCG TGG NXN. In fingers 1 and 3 of Zif268, a specific complex is formed with the cytosines in its binding site GCG TGG GCG, even though Glu-3 on each recognition helix does not interact directly with DNA. His-3 in finger 2, however, makes a direct contact with G in GCG TGG GCG, and in Tramtrack, Asn-3 on each finger interacts with A in AAG GAT (Fig. 2). We have also previously demonstrated that Ala3 can confer specificity to T in GCG TGG GTG (7). The ZK11 library that contains substitutions at positions −1, 2, 3, and 6 can be enriched for clones that bind DNA for any fixed base X in GCG TGG NXN. The lower part of Table 2 shows that they are all members of the Arg6 family that binds to GCG TGG GNN. Unexpectedly, under these conditions selection at position 6 operates freely on mixed bases at the 5′ position and yet results in the same family of high-affinity binding clones with specificity for the 5′ G.
A closer analysis of the data (Table 3) reveals that there are only a limited number of substitutions at position 3 on the recognition helix that vary (with the exception of glycine, which appears in all four selections) according to the identity of the fixed base. Among these are Asn-3, Glu-3 (or Asp-3), His-3, and Ala-3 selected for A, C, G, and T, respectively. The results of a binding signature analysis (9) for clones containing these position 3 substitutions have been summarized in Table 4, where they are presented in the form of a directory that should aid in the design of zinc fingers to recognize biologically important sequences. The fingers can be used to recognize 7 of the 16 possible GNN finger 1 binding sites, and each of the four side-chains at position 3 can be used to specify the corresponding base identified by affinity selection [for example, Asn-3 in Glu-1 Ala-2 Asn-3 Arg-6 (E A N R) selected for NAN (lower part of Table 2) specifies A in the context of GAC). There is no a priori reason, however, that these four side-chains will specify the same bases when they occur in the context of different DNA triplets.
Table 3.
Selection pressures for the middle base of the finger 1 binding site of Zif268
| DNA triplet | Sort | Amino acid (position 3) | Pe | Pf | σ32 | (Pf − Pe)/σ32 |
|---|---|---|---|---|---|---|
| NAN | 6 | Asn | 0.031 | 0.18 | 0.030 | 5.0 |
| Gly | 0.062 | 0.63 | 0.043 | 13.2 | ||
| Ala | 0.062 | 0.09 | 0.043 | <1 | ||
| Ser | 0.094 | 0.09 | 0.052 | <1 | ||
| NCN | 6 | Glu | 0.062 | 0.45 | 0.043 | 9.0 |
| Asp | 0.031 | 0.18 | 0.030 | 5.0 | ||
| Gly | 0.062 | 0.09 | 0.043 | <1 | ||
| Ser | 0.094 | 0.09 | 0.052 | <1 | ||
| NGN | 6 | His | 0.031 | 0.20 | 0.030 | 5.6 |
| Lys | 0.031 | 0.33 | 0.030 | 9.9 | ||
| Gly | 0.062 | 0.50 | 0.043 | 10.1 | ||
| NTN | 6 | Ala | 0.062 | 0.45 | 0.043 | 9.0 |
| Met | 0.031 | 0.18 | 0.030 | 5.0 | ||
| Gly | 0.062 | 0.27 | 0.043 | 4.8 |
The frequencies of amino acids at position 3 on the finger 1 recognition helix are shown as their fractional representation (Pf) in clones isolated after six rounds of selection. Expected frequencies (Pe) were calculated from the number of corresponding NNS codons, assuming a completely random starting library. The standard deviation (σ) for each residue was calculated as σ = [Pe(1 − Pe)/n]1/2, with n = 32.
Table 4.
Zinc finger DNA recognition directory
| DNA triplet | Amino acid at
helix positions
|
|||
|---|---|---|---|---|
|
| ||||
| −1 | 2 | 3 | 6 | |
| GAC | E | A | N | R |
| GCA | Q | G | E | R |
| GCG | R | D | E | R (Zif268) |
| R | D | D | R* | |
| G | W | E | R | |
| GGG | R | E | H | R |
| GGA | K | E | H | R |
| GTG | E | R | A | R* |
| GTT | T | S | A | R |
Substitutions of the Zif268 finger 1 recognition helix required to alter specificity for an assortment of binding sites were derived from phage display and affinity selection data. The base specificities were derived from binding signatures (9) as described.
The specificity was previously deduced from band-shift assays (7).
Finger-Tip Interactions.
All the fingers entered into Table 4 can be used to specify all three bases in their respective binding sites according to their binding signature analysis. If we consider the substitutions at positions −1 and 2 in relation to the 3′ base, then a number of side-chains can be utilized to build the required specificity. For instance, either Lys-1 or Qln-1 can specify 3′ A in the context of GGA or GCA, respectively, even though only Qln-1 would be expected to specify this residue through the formation of a specific bidentate hydrogen bond (21). The binding signature of Gly-1 Trp-2 Glu-3 Arg-6 (GWER) (Fig. 3) also shows that Trp-2 specifically recognizes 3′G so that both Trp-2 and Arg-1 (in RDDR, or in Zif268 itself) specify 3′ G in the context of the same binding site, GCG. An intriguing possibility is that Trp-2 introduces a deformation at the 3′ end of the triplet by intercalating with DNA (22, 23).
Figure 3.
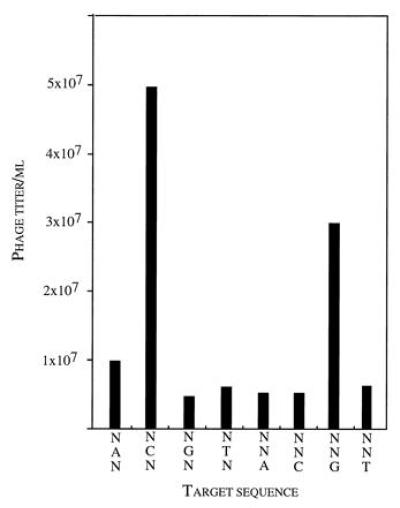
Partial binding signature of zinc finger 1 variant Gly-1 Trp-2 Glu-3 Arg-6 (GWER). Phagemids propagated from the clone were bound in affinity wells containing fixed bases at set positions in the finger 1 binding site (NAN, NCN, etc.). The resulting titers after infecting E. coli F+ cells show that GWER specifically recognizes (G)CG.
Limitations in Zif268 DNA Recognition.
It would be useful in zinc finger design to adapt these fingers to recognize DNA sequences containing ANN, CNN, or TNN. The crystal structures of the DNA complexes of Zif268 (1) and Tramtrack (2) suggest that Asp-2 can interact with cytosine when it is complementary to the 5′ guanine in the binding site of a preceding finger (Fig. 1c). To test if side-chains at position 2 in finger 2 of Zif268 are sensitive to the identity of the 5′ base (actually to the complementary base on the other DNA strand) in the finger 1 binding site, we designed a new phage display library (ZL26, Fig. 1c). Substitutions were introduced at positions −1 and 2 on the finger 2 recognition helix and at positions 3 and 6 on the finger 1 recognition helix, and the Zif268 binding site was replaced by GCG TGN XNG [where X is the fixed base in the set of DNA triplets N XN(G)]. Hence, position −1 on finger 2 is free to interact with any base N in GCG TGN XNG, position 3 on finger 1 is free to interact with N in GCG TGN XNG and the residues at positions 2 (finger 2) and 6 (finger 1) should respond to the identity of the 5′ base in the finger 1 DNA triplet (XNG).
Binding enrichment from the ZK26 library does not occur when X is fixed as either A, C, or T in GCG TGN-XNG. Sequencing clones isolated after affinity selection shows, however, that a family of weaker-binding clones can be detected that contain arginine at position −1 (Arg-1) on the finger 2 recognition helix (Table 5). These fingers bind DNA with an apparent Kd in a range higher than 50 nM and lack the Arg-6 interaction with 5′ G that determines high-affinity binding. The residues at position 2 (on finger 2), however, did not respond to the identity of the 5′ base in the finger 1 binding site in a manner that suggests base-specific recognition. In selection for N-AN(G), these residues reverted to aspartic acid, presumably indicating that the Arg-1/Asp-2 salt bridge observed in the x-ray structure of Zif268 (1) is restored, even though this is incompatible with the formation of a hydrogen bond with thymine as the complementary 5′ base. Otherwise, there was a strong preference for serine. Serine is conserved at this position in over 50% of zinc finger sequences (24).
Table 5.
Selection for a low affinity binding population
| DNA triplet | Sort | Amino acid at
helix positions
|
Predicted net charge at pH 7.0 | |||
|---|---|---|---|---|---|---|
| Finger 1
|
Finger 2
|
|||||
| 3 | 6 | −1 | 2 | |||
| N-AN(G) | 4 | G | A | R | D | 0 |
| W | S | G | N | 0 | ||
| W | I | R | D | 0 | ||
| E | R | R | D | 0 | ||
| L | R | R | D | 0 | ||
| N-CN(G) | 4 | H | D | R | S | 0 |
| H | N | N | V | 0 | ||
| H | H | R | D | 0 | ||
| H | A | R | S | +1 | ||
| N-TN(G) | 4 | H | R | R | S | +2 |
| H | E | R | S | 0 | ||
| H | A | R | S | +1 | ||
| G | R | R | D | +1 | ||
| H | G | R | S | +1 | ||
Selection was for the 5′ base in the finger 1 binding site using a library containing substitutions on both finger 1 (positions 3 and 6) and finger 2 (positions −1 and 2). Selection for N-AN(G), N-CN(G), and N-TN(G) (where the fixed base occupies the 5′ position in the finger 1 binding site) results in a low affinity consensus class without binding enrichment. The upper part of the table shows selection for DNA triplet replacements of the finger 1 binding site using a library containing substitutions at positions −1, 2, 3, and 6 on the finger 1 recognition helix. The finger 1 binding site (GCG in GCG TGG GCG) was replaced by DNA sequences containing XNN, where X is a fixed base (A, C, G, or T) and N is any base. Binding enrichment only occurs for GNN, and results in complete consensus for Arg at position 6. The lower part of the table shows the selection for fixed middle bases. All clones in these high-affinity populations contain Arg at position 6 on the recognition helix, even though selection is for mixed bases at the 5′ position in the finger 1 binding site. Numbers in parentheses refer to multiple isolates of a particular clone.
Selection for Arg-1 from this library suggests that guanine present as a mixed base N in the GCG TGN-XNG sequences is selecting Arg-1 as observed for the Arg-1/3′ G interaction of finger 2 in the Zif268 crystal structure. A similar effect is also seen at His-3 in finger 1 when affinity selection is for either GCG TGN-CNG or GCG TGN-TNG (Table 3). His-3 on finger 2 in Zif268 interacts with G in GCG TGG GCG (Fig. 1a) and can also specify this base in the finger 1 binding site (see below). The results suggest that when affinity binding is not driven by the Arg-6/5′ G interaction, the propensity of the fingers is to bind G-rich sequences, GCG TGG XGG (25). Interestingly, selection for His-3 does not occur when A is fixed in GCG TGN-ANG, indicating that base recognition can also be limited by the context of the binding triplet.
Additional electrostatic constraints are also required for DNA binding (7) (Tables 2 and 5). In all clones isolated from affinity binding populations, the predicted charge balance at positions −1, 2, 3, and 6 on the recognition helix is maintained at either 0 or +1, whereas negative charge is only occasionally accommodated when clones are isolated from populations which retain background levels of binding affinity.
Conclusions.
The zinc fingers listed in Table 4 constitute a family that was recurrently selected from the phage display library for different sets of DNA base triplets (GNN, NAN, NCN, NGN, and NTN). Each of the fingers contains arginine at position 6 on its recognition helix. In the Zif268 crystal structure, Arg-6 makes a bidentate hydrogen-bond with guanine (G) in the Zif268 binding site GCG TGG GCG, and each of the fingers also recognize 5′ G in the DNA base triplets of their respective binding sites. Since these fingers were isolated by affinity selection, their corresponding DNA triplets are likely to represent those sequences that the finger can be adapted to bind with the highest binding affinity. Each of the fingers bind their DNA triplets with affinities similar to Zif268 itself, with Kd values in the range of 0.5–5 nM (see Materials and Methods), and are able to specify all three bases in the finger 1 binding site.
Table 4 shows that many of the “recognition rules,” inferred from affinity selection for finger 2 binding sites (9) or by site-directed mutagenesis of the middle finger of a zinc finger consensus sequence framework (26), can be used to predict the sequence of the DNA triplets recognized by finger 1 in Zif268. Zif268-like complexes usually require the Arg-6/5′ G interaction in finger 1 to form a high-affinity complex, whereas this side-chain can be replaced in the middle finger of Zif268 (Fig. 1a) and either 5′ G or 5′ T can be accommodated as the 5′ base in the DNA triplets recognized by the middle finger. A comparison of the DNA triplets recognized by altered zinc fingers shows, however, that if the fingers are required to recognize all three bases in the DNA triplet, they are usually limited to a subset of sequences containing GNN (Fig. 4).
Figure 4.
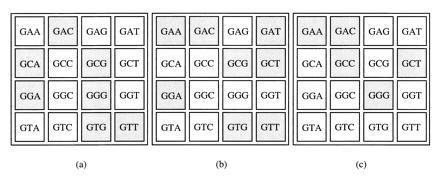
Zinc fingerprints. DNA triplets recognized by either Zif268 finger 1 (a), Zif268 finger 2 (b) (9), and a three-finger consensus sequence framework (c) (26) are represented as shaded boxes. The data include only those sequences for which zinc fingers were altered to recognize all three bases in the DNA base triplet.
Although the fingers can be adapted to bind a number of identical GNN triplets, DNA recognition may also be modified according to the position of the finger in each multifingered complex. The finger 1 amino terminus appears less constrained under selection than finger 2, which shows an almost unique preference for the Arg-1/3′ G interaction even when selection is for mixed 3′ bases. The analogous Arg-1/3′ G interactions between fingers may therefore differ in their contributions to binding energy, as shown by alanine-scanning of ARD1 zinc fingers (27). A related effect can also be seen in mutations of zinc fingers when they operate in vivo (28). These effects should be kept in mind when attempting to redesign an entire three-finger DNA complex utilizing the kind of directory described here.
Zif268 fingers also show a tendency to select G-rich sequences even when the guanines are present as mixed bases at various positions in the binding site. G-rich sequences are recognized by a number of related fingers, and may have a different conformation from canonical B-form DNA (29). In a G-rich sequence, base sliding between successive guanines distends the DNA major groove to make it deeper (as in A-form DNA) while maintaining a width closer to that of B-form DNA (30). The overall conformation of the binding site may thus be an important determinant for keeping the fingers in register when they bind DNA.
Limiting zinc finger recognition to sequences that contain 5′ G in each DNA triplet places some restrictions on the number of biologically important sequences Zif268-based fingers will recognize. However, other naturally occurring zinc fingers bind to predominantly A-rich sequences (31, 32) and these too may prove amenable to further adaptation by the methods described here. Furthermore, although the residues at positions −1, 2, 3, and 6 are directly implicated in DNA binding, it is also known that other residues in Zif268 can control DNA affinity. In particular, x-ray crystallography suggests that the linker sequences between fingers play only a passive role in DNA recognition, but alterations of this sequence in TFII also affect DNA binding (33). Alternatively, zinc fingers may be used to constitute a core DNA binding domain that can be appended with additional DNA binding motifs to extend the region of base specificity (34).
Acknowledgments
We thank James Wells (Genentech) for the monovalent phage display library developed in his laboratory by A.C.J, and Paul Park for technical assistance. This work was supported by the Office of Health and Environmental Research, U.S. Department of Energy (DE-AC03-76SF00098).
Footnotes
Abbreviation: cfu, colony-forming unit.
References
- 1.Pavletich N, Pabo C O. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 2.Fairall L, Schwabe J W R, Chapman L, Finch J T, Rhodes D. Nature (London) 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 3.Nardelli J, Gibson T, Charnay P. Nucleic Acids Res. 1992;20:4137–4144. doi: 10.1093/nar/20.16.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desjarlais J R, Berg J M. Proc Natl Acad Sci USA. 1992;89:7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desjarlais J R, Berg J M. Proc Natl Acad Sci USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebar E J, Pabo C O. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 7.Jamieson A C, Kim S-H, Wells J A. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 8.Choo Y, Klug A. Proc Natl Acad Sci USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo Y, Klug A. Proc Natl Acad Sci USA. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller J, McLachlan A D, Klug A. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee M S, Gippert G P, Soman K V, Case D A, Wright P E. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- 12.Klevit R E, Herriot J R, Horvath S J. Proteins. 1990;7:215–226. doi: 10.1002/prot.340070303. [DOI] [PubMed] [Google Scholar]
- 13.Omichinski J G, Clore G M, Apella E, Sakaguchi K, Gronenborn A M. Biochemistry. 1990;29:9324–9334. doi: 10.1021/bi00492a004. [DOI] [PubMed] [Google Scholar]
- 14.Berg J M. Annu Rev Biophys Biophys Chem. 1990;19:405–421. doi: 10.1146/annurev.bb.19.060190.002201. [DOI] [PubMed] [Google Scholar]
- 15.Theunissen O, Rudt F, Guddat U, Mentzel H, Pieler T. Cell. 1992;71:679–690. doi: 10.1016/0092-8674(92)90601-8. [DOI] [PubMed] [Google Scholar]
- 16.Clemens K R, Wolf V, McBryant S J, Zhang P, Liao X, Wright P E, Gottesfeld J M. Science. 1993;260:530–533. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Berg J M. Science. 1995;268:282–284. doi: 10.1126/science.7536342. [DOI] [PubMed] [Google Scholar]
- 18.Christy B A, Lau L F, Nathans D. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison S D, Travers A A. EMBO J. 1990;9:207–216. doi: 10.1002/j.1460-2075.1990.tb08097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel T A, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 21.Seeman N D, Rosenberg J M, Rich A. Proc Natl Acad Sci USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner M H, Clore G M, Fisher C L, Fisher R J, Trinh L, Shiloach J, Gronenborn A M. Cell. 1995;83:761–771. doi: 10.1016/0092-8674(95)90189-2. [DOI] [PubMed] [Google Scholar]
- 23.Werner M H, Gronenborn A M, Clore G M. Science. 1996;271:778–783. doi: 10.1126/science.271.5250.778. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs G H. EMBO J. 1992;11:4507–4517. doi: 10.1002/j.1460-2075.1992.tb05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg J M. Proc Natl Acad Sci USA. 1992;89:11109–11110. doi: 10.1073/pnas.89.23.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desjarlais J R, Berg J M. Proc Natl Acad Sci USA. 1994;91:11099–11103. doi: 10.1073/pnas.91.23.11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thukral S K, Morrison M L, Young E T. Proc Natl Acad Sci USA. 1991;88:9188–9192. doi: 10.1073/pnas.88.20.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blumberg H, Eisen A, Sledziewski A, Bader D, Young E T. Nature (London) 1987;328:443–445. doi: 10.1038/328443a0. [DOI] [PubMed] [Google Scholar]
- 29.Fairall L, Martin S, Rhodes D. EMBO J. 1989;8:1809–1817. doi: 10.1002/j.1460-2075.1989.tb03575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nekludova L, Pabo C O. Proc Natl Acad Sci USA. 1994;91:6948–6952. doi: 10.1073/pnas.91.15.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanojevic D, Hoey T, Levine M. Nature (London) 1989;341:331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- 32.Gogos J A, Jin J, Wan H, Kokkinidis M, Kafatos F C. Proc Natl Acad Sci USA. 1996;93:2159–2164. doi: 10.1073/pnas.93.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choo Y, Klug A. Nucleic Acids Res. 1993;21:3341–3346. doi: 10.1093/nar/21.15.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomerantz J L, Sharp P A, Pabo C O. Science. 1995;267:93–96. doi: 10.1126/science.7809612. [DOI] [PubMed] [Google Scholar]


