Abstract
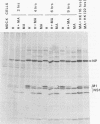
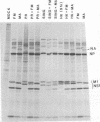
On mouse adaption of A/FM/1/47, a variant, A/FM/1/47-MA (FM-MA), that had acquired the properties of increased virulence and interference was produced. Coinfection of cells with FM-MA and prototype strains of influenza virus yielded > 100-fold more FM-MA virus than prototype virus, whereas coinfection with the same prototype strains and the parental A/FM/1/47 virus produced equivalent yields, indicating that FM-MA had acquired mutations that confer the property of interference during mouse adaption. FM-MA is a nondefective interfering virus that grows to a high titer in vivo and in vitro. It has previously been shown that segments 4, 7, and 8 and possibly segment 5 account for the increased virulence. In this study we show by genetic analysis of FM-MA x A/HK/1/68 reassortants that segment 2, coding for the polymerase-associated protein PB1, and possibly segment 8, encoding the NS1 and NS2 proteins, control the ability of FM-MA to interfere. Interference could not be overcome by increasing the titer of the coinfecting strain, but delaying FM-MA infection by 4 to 6 h did avoid interference. During interference of A/HK/1/68, protein synthesis was inhibited by less than 65% throughout coinfection. Given the kinetics of interference and the small perturbation in protein synthesis, interference appeared to occur at the level of late genome replication or virus assembly. Virulence and interference in FM-MA were not linked. An interfering avirulent FM-MA x A/HK/1/68 reassortant, E07, was capable of protecting mice against lethal pneumonia due to a virulent noninterfering reassortant, H04.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett A. D., Dimmock N. J. Defective interfering viruses and infections of animals. Curr Top Microbiol Immunol. 1986;128:55–84. doi: 10.1007/978-3-642-71272-2_2. [DOI] [PubMed] [Google Scholar]
- Braam J., Ulmanen I., Krug R. M. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell. 1983 Sep;34(2):609–618. doi: 10.1016/0092-8674(83)90393-8. [DOI] [PubMed] [Google Scholar]
- Brown E. G. Increased virulence of a mouse-adapted variant of influenza A/FM/1/47 virus is controlled by mutations in genome segments 4, 5, 7, and 8. J Virol. 1990 Sep;64(9):4523–4533. doi: 10.1128/jvi.64.9.4523-4533.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. M., Webster R. G. Protection of chickens from lethal influenza virus infection by influenza A/chicken/Pennsylvania/1/83 virus: characterization of the protective effect. Virology. 1991 Jul;183(1):427–432. doi: 10.1016/0042-6822(91)90160-d. [DOI] [PubMed] [Google Scholar]
- Frielle D. W., Huang D. D., Youngner J. S. Persistent infection with influenza A virus: evolution of virus mutants. Virology. 1984 Oct 15;138(1):103–117. doi: 10.1016/0042-6822(84)90151-x. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987 Sep 17;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Nayak D. P., Chambers T. M., Akkina R. K. Defective-interfering (DI) RNAs of influenza viruses: origin, structure, expression, and interference. Curr Top Microbiol Immunol. 1985;114:103–151. doi: 10.1007/978-3-642-70227-3_3. [DOI] [PubMed] [Google Scholar]
- Noronha-Blob L., Schulze I. T. Viral interference-mediated selection of a plaque-type variant of influenza virus. Virology. 1976 Jan;69(1):314–322. doi: 10.1016/0042-6822(76)90218-x. [DOI] [PubMed] [Google Scholar]
- Perrault J. Origin and replication of defective interfering particles. Curr Top Microbiol Immunol. 1981;93:151–207. doi: 10.1007/978-3-642-68123-3_7. [DOI] [PubMed] [Google Scholar]
- Richardson J. C., Akkina R. K. NS2 protein of influenza virus is found in purified virus and phosphorylated in infected cells. Arch Virol. 1991;116(1-4):69–80. doi: 10.1007/BF01319232. [DOI] [PubMed] [Google Scholar]
- Shapiro G. I., Gurney T., Jr, Krug R. M. Influenza virus gene expression: control mechanisms at early and late times of infection and nuclear-cytoplasmic transport of virus-specific RNAs. J Virol. 1987 Mar;61(3):764–773. doi: 10.1128/jvi.61.3.764-773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. H., Betts R. F., DeBorde D., Tierney E. L., Clements M. L., Herrington D., Sears S. D., Dolin R., Maassab H. F., Murphy B. R. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J Virol. 1988 Feb;62(2):488–495. doi: 10.1128/jvi.62.2.488-495.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. H., Clements M. L., Betts R. F., Dolin R., Buckler-White A. J., Tierney E. L., Murphy B. R. Evaluation of live avian-human reassortant influenza A H3N2 and H1N1 virus vaccines in seronegative adult volunteers. J Clin Microbiol. 1986 May;23(5):852–857. doi: 10.1128/jcm.23.5.852-857.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker-Dowling P., Lucas W., Youngner J. S. Cold-adapted vaccine strains of influenza A virus act as dominant negative mutants in mixed infections with wild-type influenza A virus. Virology. 1990 Apr;175(2):358–364. doi: 10.1016/0042-6822(90)90420-v. [DOI] [PubMed] [Google Scholar]
- Whitaker-Dowling P., Maassab H. F., Youngner J. S. Dominant-negative mutants as antiviral agents: simultaneous infection with the cold-adapted live-virus vaccine for influenza A protects ferrets from disease produced by wild-type influenza A. J Infect Dis. 1991 Dec;164(6):1200–1202. doi: 10.1093/infdis/164.6.1200. [DOI] [PubMed] [Google Scholar]
- Whitaker-Dowling P., Youngner J. S. Viral interference-dominance of mutant viruses over wild-type virus in mixed infections. Microbiol Rev. 1987 Jun;51(2):179–191. doi: 10.1128/mr.51.2.179-191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker-Dowling P., Zvolenski R., Youngner J. S. The genes associated with trans-dominance of the influenza A cold-adapted live virus vaccine. Virology. 1991 Jan;180(1):81–87. doi: 10.1016/0042-6822(91)90011-y. [DOI] [PubMed] [Google Scholar]