Abstract
Using truncated forms of recombinant yeast karyopherins α and β in in vitro binding assays, we mapped the regions of karyopherin α that bind to karyopherin β and the regions of karyopherin β that interact with karyopherin α and with Ran-GTP. Karyopherin α’s binding region for karyopherin β was localized to its N-terminal domain, which contains several clusters of basic residues, whereas karyopherin β’s binding region for karyopherin α was localized to an internal region containing two clusters of acidic residues. Karyopherin β’s binding region for Ran-GTP overlaps with that for karyopherin α and comprises at least one of the two acidic clusters required for karyopherin α binding in addition to further downstream determinants not required for karyopherin α binding. Overexpression in yeast of fragments containing either karyopherin β’s binding region for α and Ran-GTP or karyopherin α’s binding region for β resulted in sequestration of most of the cytosolic karyopherin α or karyopherin β, respectively, in complexes containing the truncated proteins. As these binding region-containing fragments lack other domains required for function of the corresponding protein, the overexpression of either fragment also inhibited in vivo nuclear import of a model reporter protein as well as cell growth.
Keywords: solution binding assay, binding determinants, green fluorescent protein
Import of a nuclear localization sequence (NLS)-containing protein into nuclei of digitonin-permeabilized cells requires the exogenous addition of four distinct transport factors: karyopherin α (also termed NLS receptor or importin α), karyopherin β (also termed p97 or importin β) (1–11), Ran, and p10 (12–15).
In the cytosol, karyopherins α and β exist as a heterodimeric complex (2, 5, 7–11). The α subunit of this complex binds the NLS substrate (1, 9, 11), whereas the β subunit mediates docking of the trimeric complex to the nuclear pore complex (NPC) (10, 16–18). Ran and p10 mediate translocation of both the substrate and karyopherin α into the nucleoplasm, while karyopherin β stays behind at the NPC (6, 18). Hence, transport into the nucleoplasm is accompanied by dissociation of the karyopherin heterodimer. Overlay blots using immobilized nuclear envelope proteins showed that karyopherin β binds specifically to a subgroup of nucleoporins that contain peptide repeats and are located on the cytoplasmic and nucleoplasmic fibers of the NPC (16–19).
In vitro binding studies using recombinant transport factors and peptide repeat-containing nucleoporins, all from Saccharomyces cerevisiae, have provided further insights into the mechanism of nuclear import (20). It was proposed that multiple rounds of docking and release of NLS substrate may serve to concentrate the NLS substrate near the NPC for subsequent diffusion across the central tube of the NPC, down a chemical concentration gradient (20).
The studies using yeast recombinant proteins also showed that Ran-GTP, but not Ran-GDP, dissociated the karyopherin heterodimer and associated with karyopherin β to form a dimeric complex (20, 21). Ran-GTP in this dimeric complex was found to be resistant to GTP hydrolysis by the Ran-specific GTPase-activating protein (21). Therefore, much of the cytosolic Ran may be kept in the GDP-bound form by the cytosolic Ran-specific GTPase-activating protein to prevent futile dissociation of the cytosolic karyopherin heterodimer by Ran-GTP (22).
However, Ran-GTP and GTP hydrolysis are required for import (12, 13). If Ran-GDP is the dominant form in the cytosol, how and where would Ran-GTP then be generated? A partial answer to these questions came from binding studies using recombinant yeast p10 (22). In overlay assays using SDS/PAGE-separated polypeptides of yeast nuclear envelopes, p10 bound to similar peptide repeat-containing nucleoporins as did karyopherin β. In liquid-phase binding assays, p10 bound to Ran-GDP, not Ran-GTP. Moreover, p10 also bound to karyopherin β. Hence, a pentameric complex consisting of a peptide repeat-containing nucleoporin, p10, Ran-GDP, and the karyopherin heterodimer could be assembled. Incubation of this complex with GTP yielded dissociation of karyopherin α, presumably by in situ conversion of Ran-GDP to Ran-GTP and subsequent association of Ran-GTP with karyopherin β, which remained bound to the nucleoporin. Thus, p10 docks Ran-GDP next to the nucleoporin-bound karyopherin heterodimer to allow in situ generation of Ran-GTP for dissociation of karyopherin α from docked karyopherin β (22). These data are consistent with the observed dissociation of the karyopherin heterodimer at the nuclear envelope and the transport of karyopherin α, but not of karyopherin β, into the nucleoplasm (6, 18).
Insights into how transport factors interact with the NLS substrate and nucleoporins have come from recent mapping studies using vertebrate recombinant proteins. It was shown that the N-terminal region of karyopherin α binds to karyopherin β (23–25) and that a C-terminal region of karyopherin α binds to the NLS substrate (24). Moreover, karyopherin α’s N-terminal binding site for karyopherin β also contains an NLS (24). Interestingly, karyopherin α’s N-terminal binding region for karyopherin β, when fused to a reporter protein, allowed import of the fusion protein into nuclei of digitonin-permeabilized cells in the absence of karyopherin α (23, 25).
Mapping studies with mammalian recombinant karyopherins also identified a discontinuous stretch of acidic residues as a major determinant of karyopherin β’s binding site for karyopherin α (26). Interestingly, this site overlaps with karyopherin β’s binding site for Ran-GTP. As Ran-GTP has a higher affinity for this site than karyopherin α, it displaces karyopherin α and forms a heterodimer with karyopherin β (26).
Here we report the mapping of yeast karyopherin α’s binding site for yeast karyopherin β and of karyopherin β’s binding site for karyopherin α and for Ran-GTP. As observed for the mammalian counterparts, the N-terminal region of yeast karyopherin α mediates binding to yeast karyopherin β. Moreover, karyopherin β’s binding site for karyopherin α and Ran-GTP overlap. Consistent with the in vitro mapping data, galactose-induced overexpression of these segments in yeast showed sequestration of the cytosolic karyopherin α and β, respectively, and inhibition of nuclear protein import and cell growth.
MATERIALS AND METHODS
Preparation of Recombinant Fragments of Karyopherins α and β.
Glutathione S-transferase (GST)-tagged yeast karyopherins α and β (7) and their GST-tagged fragments were generated by using pGEX-4T-3 (Pharmacia).
Plasmid pGEX-4T-3-α1-110 contains the PCR-amplified codons 1 to 110 as a BamHI/EcoRI fragment. pGEX-4T-3-α107-542, pGEX-4T-3-α1-61, and pGEX-4T-3-α62-542 contain the PCR-amplified codons 107 to 542, 1 to 61, and 62 to 542, respectively, as BamHI fragments.
pGEX-4T-3-β1-288 was generated by excision of an EcoRI fragment within pGEX-4T-3-β1-861 (7) that eliminates codons 289 to 861. pGEX-4T-3-β1-353 was generated by excision of a SphI/SmaI fragment within pGEX-4T-3-β1-861 and religation of the blunted ends that eliminates codons 354 to 861. pGEX-4T-3-β1-457 was generated by excision of a HindIII/SmaI fragment within pGEX-4T-3-β1-861 and religation of the blunted ends that eliminates codons 458 to 861. pGEX-4T-3-β348-861 contains the PCR-amplified codons 348 to 861 as a BamHI fragment. pGEX-4T-3-β288-457 contains the PCR-amplified codons 288 to 457 as a BamHI fragment. The expression in Escherichia coli BL21(DE3) and the purification of the soluble GST fusion proteins were performed as described (7, 20).
In Vitro Binding Studies.
The solution binding assay was performed as described (20). For the competition experiment with protein fragment α1-61, the αProtA/β complex was isolated from yeast cytosol and immobilized to IgG Sepharose (7). The fragment α1-61 was obtained by thrombin cleavage of the GST–α1-61 fusion protein according to refs. 7 and 20 and added in an ≈10-fold excess (0.5 μg protein) to the immobilized αProtA/β complex {2 μg protein bound to 20 μl resin in a final volume of 200 μl transport buffer [20 mM Hepes·KOH, pH 7.5/110 mM KOAc/2 mM Mg(OAc)2/2 mM EDTA/2 mM DTT]}. After 1 h incubation at room temperature, the released fraction was collected. The bound fraction was eluted with 0.5 M HOAc/NH4OAc (pH 3.4).
Overexpression of Protein A Fusion Proteins in Yeast.
For the galactose-induced expression of protein A-tagged proteins, the GAL1/2 μm/URA3 plasmid pYES2 (Invitrogen) was used.
To generate α1-61-ProtA, codons 2 to 61 were amplified by PCR using oligonucleotides (5′-AATTGGTACCAAAATGGATAATGGTACAGATTCT-3′ and 5′-AAAAGGATCCTGGGGGAATAAAGTTTCT-3′) and pRS315-Kap60-ProtA (7) as template. The KpnI/BamHI-digested PCR product was cloned into pYES resulting in pYES-α1-61. The BamHI fragment encoding five IgG binding domains of protein A was excised from pRS315-Kap60-ProtA and inserted into BamHI-digested pYES-α1-61, resulting in pYES-α1-61-ProtA. To generate α62-542-ProtA, codons 62 to 542 were amplified by PCR using oligonucleotides (5′-AATTGGTACCAAAATGACTGATGGCGCTGATTCT-3′ and 5′-AAAAGGATCCGTTAAAATTGAATTGTG-3′) and processed as above. To generate pYES2-ProtA, the gene encoding five IgG binding domains was amplified by PCR and processed as above.
To generate β288-457-ProtA, codons 288 to 457 were amplified by PCR using oligonucleotides (5′-AATTGGTACCAAAATGGAATTCTGGTCCACTATT-3′ and 5′-AAAAGGATCCTTGAACGACACCTGGAAG-3′) and pGEX-4T-3-KAP95 as template and processed as above.
To generate GST-β288-457-ProtA, the gene encoding the GST–β288-457 fusion protein was amplified by PCR using oligonucleotides (5′-AATTGGTACCAAAATGTCCCCTATACTAGGT-3′ and 5′-AAAAGGATCCTTGAACGACACCTGGAAG-3′) and pGEX-4T-3-β288-457 as template. The PCR product was digested with KpnI and SacI, yielding a 880-bp fragment encoding GST-β288-304. The KpnI/SacI fragment encoding β288-304 was excised from pYES2-β288-457-ProtA and replaced by the 880-bp fragment, resulting in pYES2-GST-β288-457-ProtA.
The resulting plasmids were transformed into yeast strain WCG (MATa/Matα his3-11, 15/his3-11, 15 leu2-3, 112/leu2-3, 112 ura3/ura3 can/can GAL/GAL) (27). Ura+ transformants were selected and analyzed for the expression of the protein A chimera (7). Cell cultures were grown at 30°C to mid-log phase in SC (synthetic complete)-ura, 2% glucose, and shifted for 4–6 h at 30°C to SC-ura, 2% galactose. Cells were disintegrated in a French pressure cell, the high speed supernatant was passed over IgG Sepharose, and the bound fraction was processed for SDS/PAGE as described (7). Blots were probed with rabbit antibodies raised against recombinant GST fusion proteins of yeast karyopherin α or β (a kind gift of M. Rexach, Rockefeller University). For the detection of anti-α or anti-β antibodies, protein A-HRP conjugates were used; for the detection of protein A chimera, anti-rabbit IgG-HRP conjugates were used as described (7).
Localization Study of H2B Histone Fused to the Green Fluorescent Protein (GFP).
Strain WCG was cotransformed with pYES-α1-61-ProtA, pYES-GST-β288-457-ProtA, or pYES-ProtA and a CEN-HIS3 plasmid containing the GAL1 promoter-driven gene fusion of histone H2B1 to the GFP (a generous gift of J. Loeb, Massachusetts Institute of Technology, Boston; ref. 28). Ura+His+ transformants were analyzed as above, grown to mid-log phase in SC-ura-his, 2% glucose, and shifted up for 10 h at 30°C to SC-ura-his, 2% galactose. The subcellular distribution of the H2B-GFP reporter protein was analyzed by direct fluorescence microscopy. Images were photographed on a Zeiss Axiophot microscope using the Image-1/AT image processing and analysis system.
RESULTS
An N-Terminal Region of Karyopherin α Contains the Binding Site for Karyopherin β.
The primary structure of yeast karyopherin α suggests three distinct regions (29, 30): a central hydrophobic domain containing eight “armadillo” repeats is flanked by hydrophilic N- and C-terminal regions. To map karyopherin α’s binding site for karyopherin β, we expressed various regions of karyopherin α as N-terminal GST-tagged proteins in E. coli (Fig. 1A). The GST-tagged proteins were purified, immobilized on glutathione agarose, and incubated with recombinant yeast karyopherin β. The bound material was analyzed by SDS/PAGE and Coomassie blue staining (Fig. 1B). Fusion proteins that contained the N-terminal region of karyopherin α bound karyopherin β (lanes 1 and 3), whereas proteins that contained only the armadillo repeats and the C-terminal region of karyopherin α did not bind (lanes 2 and 4). Moreover, it appeared that the major determinants for binding to karyopherin β were located within the N-terminal 61 residues of karyopherin α (Fig. 1B, lanes 3 and 4).
Figure 1.
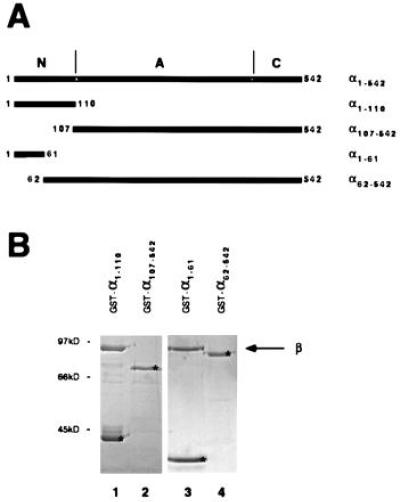
Karyopherin β is bound by α1–110 and α1-61, but not by α107-542 and α62-542. (A) Schematic drawing of karyopherin α and fragments that were isolated as recombinant GST fusion proteins from E. coli. The N-terminal (N), the armadillo repeat (A), and the C-terminal (C) regions are indicated. (B) Immobilized GST-α1–110 (lane 1), GST-α107-542 (lane 2), GST-α1-61 (lane 3), and GST-α62-542 (lane 4) were incubated with recombinant karyopherin β. Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. The immobilized GST-tagged fragments are marked by asterisks.
We have previously constructed a yeast strain in which the karyopherin α gene is replaced by a gene encoding staphylococcal protein A-tagged karyopherin α. A heterodimeric complex of protein A-tagged karyopherin α and karyopherin β can be isolated from the cytosol of these cells by IgG Sepharose chromatography (Fig. 2, lane 1; ref. 7). If the N-terminal 61 residues of karyopherin α contain the major determinants for binding karyopherin β, then a karyopherin α fragment comprising these residues might competitively displace ProtA-karyopherin α from the in vivo-formed heterodimer, leaving karyopherin β associated with the unbound karyopherin α fragment. Indeed, incubation of the IgG Sepharose-immobilized ProtA-karyopherin α/β heterodimer with a 10-fold excess of the karyopherin α fragment α1-61 resulted in partial release of karyopherin β into the unbound fraction (Fig. 2).
Figure 2.
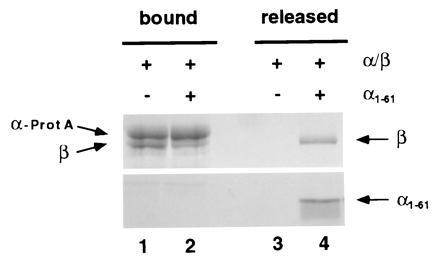
In vitro dissociation by α1-61 of in vivo-assembled karyopherin α/β heterodimer. In vivo-formed α-ProtA/β heterodimer was purified from yeast cytosol by IgG Sepharose chromatography (7). The immobilized heterodimer was incubated for 1 h at room temperature in the absence or in the presence of an approximately 10-fold excess of α1-61. The released material was collected and the bound material was eluted at pH 3 as described. The fractions were analyzed by SDS/PAGE and Coomassie blue staining.
Likewise, a protein A-tagged α1-61, when overexpressed in yeast from a galactose inducible promoter, might in vivo competitively displace karyopherin α and form a complex with karyopherin β. Indeed, 6 hr after galactose induction essentially all cytosolic karyopherin β was coisolated with protein A-tagged α1-61 (Fig. 3). We conclude that α1-61 is able to bind karyopherin β.
Figure 3.
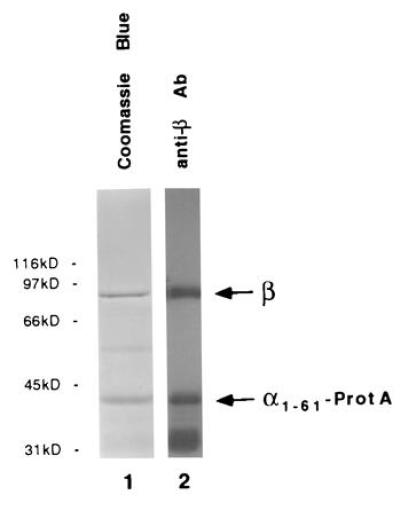
In vivo association of karyopherin β with in vivo-expressed α1-61. Following galactose-induction for 6 h, α1-61-ProtA was isolated from yeast cytosol by IgG Sepharose chromatography as described. The bound fraction was analyzed by SDS/PAGE and Coomassie blue staining (lane 1) or ECL Western blotting using anti-β antibodies and protein A-horseradish peroxidase conjugates (lane 2).
Acidic Clusters of Karyopherin β Are Binding Determinants for Karyopherin α.
As clusters of basic residues constitute karyopherin α’s principal binding determinants for karyopherin β, clusters of acidic residues may constitute karyopherin β’s binding determinants for karyopherin α. Tertiary structure predictions for karyopherin β suggest the existence of repetitive pairs of antiparallel amphipathic alpha helices, termed HEAT repeats (31) (Fig. 4B). Two clusters of acidic residues (termed a′ and a in Fig. 4B) are located in loops connecting the HEAT repeats, and these could be determinants for binding to karyopherin α. We expressed regions of karyopherin β that did or did not contain these regions as N-terminally-tagged GST fragments in E. coli (Fig. 4A). The GST-tagged proteins were purified, immobilized on glutathione agarose, and incubated with recombinant karyopherin α. Indeed, N-terminal fragments of karyopherin β that contained the two acidic regions bound to karyopherin α (Fig. 4C, lanes 3 and 4), whereas either an N-terminal fragment (Fig. 4C, lane 2) or a C-terminal fragment (Fig. 4C, lane 5) that did not contain these two acidic regions did not bind to karyopherin α. Moreover, an N-terminal fragment that contained the two acidic loops as well as two more downstream loops did not yield increased binding of karyopherin α (Fig. 4C, compare lanes 3 and 4) suggesting that the principal binding determinants for karyopherin α comprise the two acidic loops a′ and a (Fig. 4B).
Figure 4.
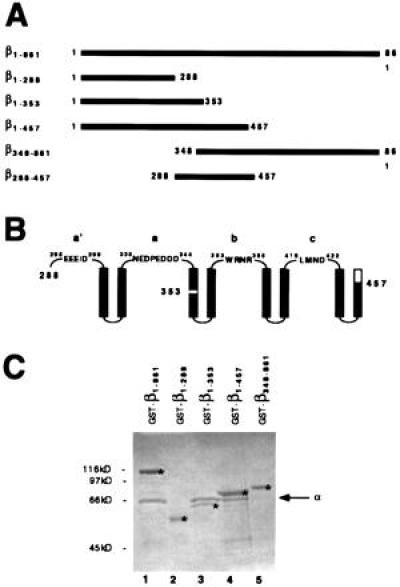
Karyopherin α is bound by β1-861, β1-353, and β1-457 but not by β1-288 and β348-861. (A) Schematic drawing of karyopherin β and β fragments that were isolated as recombinant GST fusion proteins from E. coli. (B) Schematic drawing of predicted amphipathic antiparallel alpha helices and connecting loops (labeled a′, a, b, and c); large numbers indicate the residues of β fragments that end in helices and small numbers mark β residues in the loops. (C) Immobilized GST-β1-861 (lane 1), GST-β1-288 (lane 2), GST-β1-353 (lane 3), GST-β1-457 (lane 4), and GST-β348-861 (lane 5) were incubated with recombinant karyopherin α. Bound fractions were analyzed by SDS/PAGE and Coomassie blue staining. The immobilized GST-tagged fragments are marked by asterisks.
To test whether a fragment of karyopherin β containing loops a′ and a–c (Fig. 4 A and B) would interact with karyopherin α in vivo, we expressed a doubly tagged karyopherin β fragment (residues 288-457 linked to GST at the N terminus and protein A at the C terminus) from a galactose inducible promoter in yeast. Four hours after galactose induction, yeast cytosol was prepared and passed over IgG Sepharose. Most of the cytosolic karyopherin α was coisolated with the in vivo-expressed chimeric protein (Fig. 5). We conclude that a karyopherin β fragment containing the acidic loops a′ and a plus loops b and c (Fig. 4B) was able to bind karyopherin α. It remains to be tested whether a karyopherin β fragment containing only loops a′ and a would be sufficient to bind karyopherin α.
Figure 5.
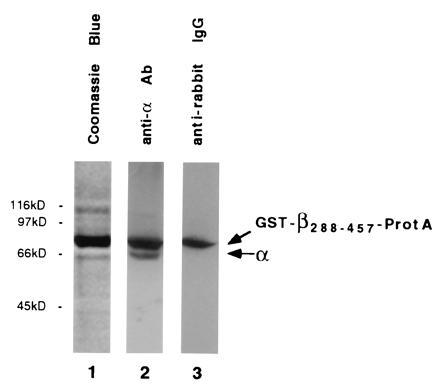
In vivo association of karyopherin α with in vivo-expressed β288-457. Following galactose induction for 4 h, GST-β288-457-ProtA was isolated from yeast cytosol by IgG Sepharose chromatography. The bound fraction was analyzed by SDS/PAGE and Coomassie blue staining (lane 1) or ECL Western blotting using anti-α antibodies and protein A-horseradish peroxidase conjugates (lane 2) or anti-rabbit IgG-HRP conjugates (lane 3).
Ran-GTP and Karyopherin α Share Overlapping Binding Sites on Karyopherin β.
We tested GST-tagged full-length karyopherin β and its various fragments for binding to Ran-GTP in an overlay assay (Fig. 6). As expected, full-length karyopherin β bound Ran-GTP (lane 1). The fragment β1-288 did not bind (lane 2); β1-353 bound weakly (lane 3), and β1-457 bound strongly (lane 4). This indicated that N-terminal fragments containing the two acidic loops a and a′ (Fig. 4B) yielded only weak Ran-GTP binding and that those containing the additional downstream loops b and c yielded strong binding. Surprisingly, a C-terminal fragment containing loops b and c did not bind to Ran-GTP, suggesting that loops b and c were not sufficient for binding to Ran-GTP. A β fragment, β288-457 (lane 6), containing all four loops bound Ran-GTP about as well as full-length β (lane 1) or β1-457 (lane 4). Together these data suggest that maximal binding of Ran-GTP to karyopherin β’s binding site requires all four loops. However, it is possible that either or both of the terminal loops a′ or c are not part of this site. Nevertheless, it strongly suggest that karyopherin α and Ran-GTP share overlapping binding sites on karyopherin β comprising either one or both of the acidic loops (see Discussion).
Figure 6.
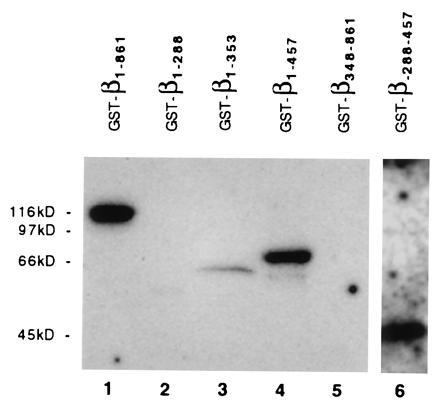
Ran-GTP is bound by β1-861, β1-357, β1-457, and β288-457, but not by β1-288 and β348-861. Equal amounts of the fusion proteins GST-β1-861 (lane 1), GST-β1-288 (lane 2), GST-β1-353 (lane 3), GST-β1-457 (lane 4), GST-β348-861 (lane 5), and inclusion bodies containing GST-β288-457 (lane 6) were separated by SDS-PAGE and subjected to a Ran-[α-32P]GTP ligand blot according to Coutavas et al. (32). Labeled proteins were visualized by autoradiography.
Overexpression of Karyopherins α1-61 and β288-457 Inhibits Growth and Nuclear Import.
Both karyopherins α and β are essential for viability in yeast, and overexpression of α1-61 or β288-457 in yeast cells resulted in sequestering of karyopherin β or α (Figs. 3 and 5), respectively, in complexes with the truncated proteins. We therefore investigated the effects of expression of these fragments on cell growth and localization of a reporter nuclear protein. Cells expressing either fragment grew normally in glucose, when the GAL1 promoter is repressed. In liquid culture, following a shift to galactose, the cells continued to grow, albeit more slowly than control cells expressing protein A alone (Fig. 7A). However, cells expressing α1-61 or β288-457 failed to form colonies when streaked out on galactose-containing plates (Fig. 7B).
Figure 7.
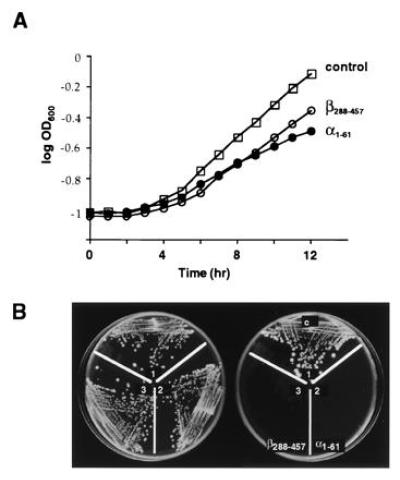
Expression of α1-61 and β288-457 in yeast inhibit cell growth. (A) Cells were grown overnight in dextrose and then shifted to 2% galactose at zero time. The growth rates of cells (control) expressing protein A or of cells expressing α1-61-ProtA or GST-β288-457-ProtA, all from a galactose inducible promoter, were determined by measurement of the optical density of the cultures at 600 nm. (B) Cells were also streaked on SC-ura plates containing 2% glucose (Left) or 2% galactose (Right).
We used histone H2B fused to the naturally GFP (H2B-GFP) as a nuclear reporter protein (28). Its subcellular distribution could be directly observed with the fluorescence microscope in cells expressing H2B-GFP alone (Fig. 8A1) or in combination with either α1-61 (Fig. 8A2) or β288-457 (Fig. 8A3), all under the control of the GAL1 promoter. The reporter protein was primarily localized to the nucleus when it was expressed by itself (Fig. 8A Upper), while it accumulated in the cytoplasm after coexpression of either α1-61 (Fig. 8A2) or β288-457 (Fig. 8A3). These data suggest that either of these peptides is able to inhibit nuclear import of the nuclear reporter protein.
Figure 8.
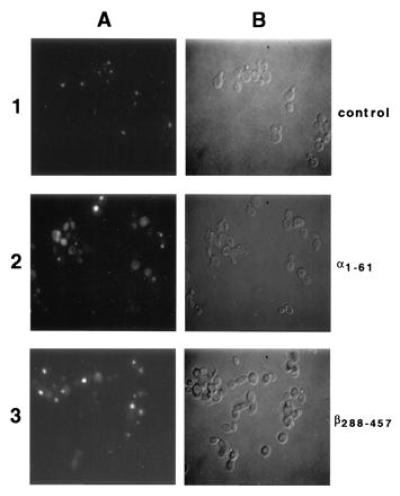
Expression of α1-61 and β288-457 in yeast inhibit nuclear import of a coexpressed nuclear reporter protein. Cells were grown overnight in dextrose, shifted to 2% galactose for 10 h, and then directly examined by fluorescence microscopy (A) or Nomarski optics (B). Shown are cells expressing the galactose inducible fusion protein consisting of the naturally GFP and histone H2B (28) and protein A (1), α1-61-ProtA (2), or GST-β288-457-ProtA (3). All micrographs were exposed for the same length of time.
DISCUSSION
Karyopherin α’s Binding Site for Karyopherin β.
The liquid-phase binding assays using various immobilized karyopherin α fragments and recombinant full-length karyopherin β in the mobile phase (Fig. 1) indicated that a fragment, α1-61, was sufficient for high affinity binding to karyopherin β, suggesting that α1-61 contains the major determinants for contact with karyopherin β. Moreover, a 10-fold excess of α1-61 competitively released some, but not all, of the karyopherin β from an in vivo-formed heterodimeric karyopherin α/β complex (Fig. 2).
However, there are at least two important caveats that should be considered with regard to the type of mapping experiments reported here. First, the liquid-phase binding assays that were used for mapping may detect binding determinants only of high but not of low affinity. Second, additional high affinity binding determinants may have been missed because of improper folding of a given truncated karyopherin fragment. Hence, our experiment may not have identified all of the high affinity binding determinants and may have missed low affinity binding determinants altogether. Furthermore, the possibility that there are additional binding determinants for karyopherin β beyond residue 61 of karyopherin α is consistent with two other sets of results. First, a temperature-sensitive mutant of yeast karyopherin α, in which the highly conserved Ser116, located within a basic cluster, is changed to Phe, inhibits docking of NLS substrate at the nuclear envelope (33). Second, mapping data on a mammalian homologue of karyopherin α have shown that a karyopherin α mutant lacking the first 49 residues still functions in import into nuclei of digitonin permeabilized cells, albeit with reduced efficiency (9, 24). These latter data in particular argue strongly against a proposal (23, 25) that determinants of α for binding to β are exclusively localized to the first 50 or so residues of karyopherin α.
The overexpression in yeast of α1-61 from a galactose-inducible promoter provided in vivo evidence that α1-61 contains major determinants for binding to karyopherin β. Cell fractionation showed that most of the cytosolic karyopherin β was bound to the overexpressed α1-61 (Fig. 3) rather than to native karyopherin α. Furthermore, overexpression of α1-61 inhibited cell growth (Fig. 7) and nuclear protein import (Fig. 8). As these cells contained karyopherins α and β, both full-length, in addition to the α1-61 fragment, these results along with the comparable results for β288-457 demonstrate that the interaction between karyopherins α and β is required for their activity in nuclear import.
Ran-GTP and Karyopherin α Bind to Overlapping Sites on Karyopherin β.
Liquid-phase binding assays using various immobilized fragments of karyopherin β and full-length recombinant karyopherin α in the mobile phase indicated that two acidic clusters of karyopherin β are principal determinants for binding to karyopherin α (Fig. 4). Each of these two acidic clusters might be part of exposed loops (a′ and a, see Fig. 4B) that connect repetitive antiparallel amphipathic alpha helices, termed HEAT repeats (31). Thus, it appears that the contacts between karyopherins α and β are primarily between clusters of basic residues in karyopherin α with complementary clusters of acidic residues in karyopherin β. Similar acidic clusters have been shown to constitute the major determinants for binding to mammalian karyopherin α (26).
Overlay binding assays using fragments of karyopherin β that were separated by SDS/PAGE, transferred to nitrocellulose, and subsequently probed with Ran-GTP indicated that binding determinants for Ran-GTP and karyopherin α overlap. There was weak binding of Ran-GTP to an N-terminal fragment of karyopherin β that included both acidic loops a′ and a (Figs. 4B and 6), but there was strong binding when the N-terminal fragment included the downstream loops b and c (Figs. 4B and 6). However, there was no detectable binding of Ran-GTP to a C-terminal karyopherin β fragment that contained loops b and c but not loops a′ and a (Fig. 6). Loops b and c by themselves may only have weak affinity for Ran-GTP. Alternatively, loops b and c may not be correctly folded in this particular karyopherin β fragment. Together these data suggest that acidic loops a′ and a as well as the b and c loops of karyopherin β might cooperate in the binding of Ran-GTP. Most interestingly, loops a, b, and c of karyopherin β were found to exhibit sequence similarity to discontinuous regions of the Ran-GTP binding protein, RanBP1, as well as to RanBP1 homologous domains in Ran-GTP binding nucleoporins (26). Hence, it is likely that loops a, b, and c of karyopherin β constitute the major determinant for binding to Ran-GTP. Thus, loop a would constitute a shared determinant for binding to Ran-GTP or to karyopherin α. Studies with mammalian recombinant proteins have shown that Ran-GTP binds to karyopherin β with higher affinity than does karyopherin α. Hence, the Ran-GTP-mediated dissociation of the karyopherin α/β heterodimer was proposed to proceed via competitive displacement by Ran-GTP of karyopherin α from karyopherin β and the ensuing formation of a karyopherin β/Ran-GTP heterodimer (20, 26).
In vivo expression of a karyopherin β fragment containing loops a′, a, b, and c (β288-457) from a galactose inducible promoter had a dominant negative effect on both cell growth (Fig. 7) and nuclear import of a model substrate (Fig. 8). Fractionation of galactose-induced cells showed that much of the cytosolic karyopherin α was sequestered and bound to β288-457, providing an explanation for the observed inhibition of nuclear import and of cell growth.
The mapping of binding regions in vitro combined with in vivo functional experiments, as shown here for the karyopherin α/β heterodimer, is a useful paradigm for understanding the interactions of complex systems in yeast.
Acknowledgments
We thank especially Erica Johnson and Mike Matunis for critical reading of the manuscript and helpful discussions. Thanks also go to Doris Krämer, John Aitchison, and Oliver Ernst for helpful advice. We thank Monique Floer for providing recombinant Ran (Gsp1), Michael Rexach for antibodies against yeast karyopherins α and β, Jonathan Loeb and Steffen Rupp for the plasmid H2B-GFP, and Dieter Wolf for yeast strain WCG. C.E. was supported by Research Fellowship En301/1-1 from the Deutsche Forschungsgemeinschaft and funds from the Fritz Thyssen Stiftung (Az.212 95010).
Footnotes
Abbreviations: GFP, green fluorescent protein; GST, glutathione S-transferase; NLS, nuclear localization signal; NPC, nuclear pore complex.
References
- 1.Adam S A, Gerace L. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 2.Adam E J H, Adam S A. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chi N C, Adam E J H, Adam S A. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Görlich D, Prehn S, Laskey R A, Hartmann E. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 5.Görlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 6.Görlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Nature (London) 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 7.Enenkel C, Blobel G, Rexach M. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 8.Imamoto N, Tachibana T, Matsubae M, Yoneda Y. J Biol Chem. 1995;270:8559–8565. doi: 10.1074/jbc.270.15.8559. [DOI] [PubMed] [Google Scholar]
- 9.Moroianu J, Blobel G, Radu A. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radu A, Blobel G, Moore M S. Proc Natl Acad, Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weis K, Mattaj I W, Lamond A I. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 12.Melchior F, Paschal B, Evans J, Gerace L. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore M S, Blobel G. Nature (London) 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 14.Moore M S, Blobel G. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paschal B M, Gerace L. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iovine M K, Watkins J L, Wente S R. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraemer D M, Strambio-de-Castillia C, Blobel G, Rout M P. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- 18.Moroianu J, Hijikata M, Blobel G, Radu A. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radu A, Moore M S, Blobel G. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 20.Rexach M, Blobel G. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 21.Floer M, Blobel G. J Biol Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- 22.Nehrbass U, Blobel G. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- 23.Görlich D, Henklein P, Laskey R A, Hartmann E. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 24.Moroianu J, Blobel G, Radu A. Proc Natl Acad Sci USA. 1996;93:7059–7062. doi: 10.1073/pnas.93.14.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weis K, Ryder U, Lamond A I. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 26.Moroianu J, Blobel G, Radu A. Proc Natl Acad Sci USA. 1996;93:6572–6576. doi: 10.1073/pnas.93.13.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinemeyer W, Gruhler A, Möhrle V, Mahe Y, Wolf D H. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 28.Schlenstedt G, Saavedra C, Loeb J D J, Cole C N, Silver P A. Proc Natl Acad, Sci USA. 1995;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yano R, Oakes M, Yamaghishi M, Dodd J A, Nomura M. Mol Cell Biol. 1992;12:5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peifer M, Berg S, Reynolds A B. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 31.Andrade M A, Bork P. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 32.Coutavas E, Ren M, Oppenheim J D, D’Eustachio P, Rush M G. Nature (London) 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- 33.Loeb J D L, Schlenstedt G, Pellman D, Kornitzer D, Silver P A, Fink G. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]


