Abstract
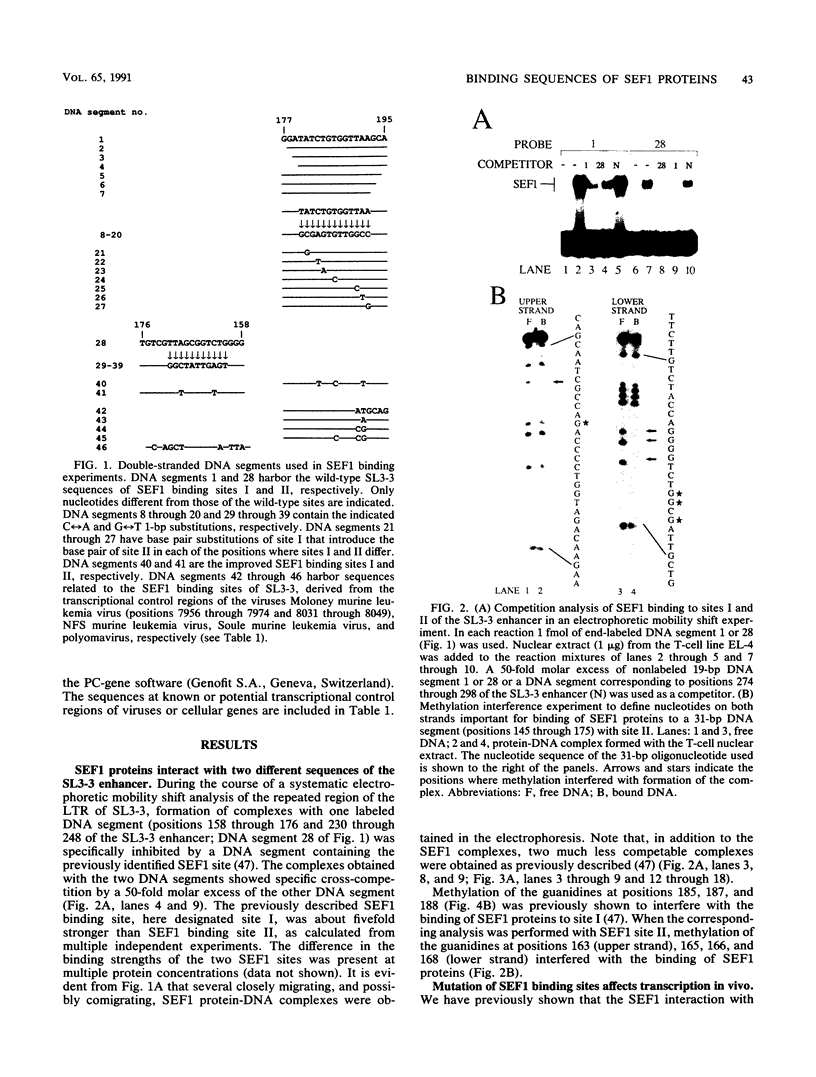
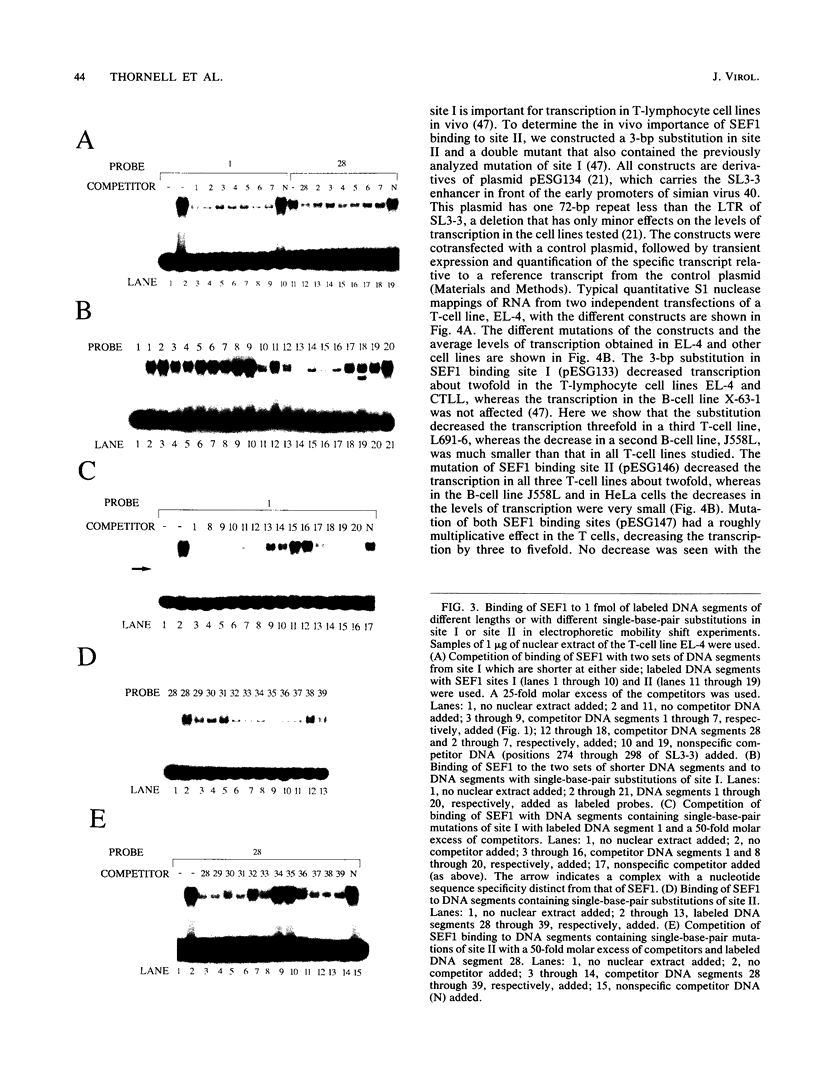
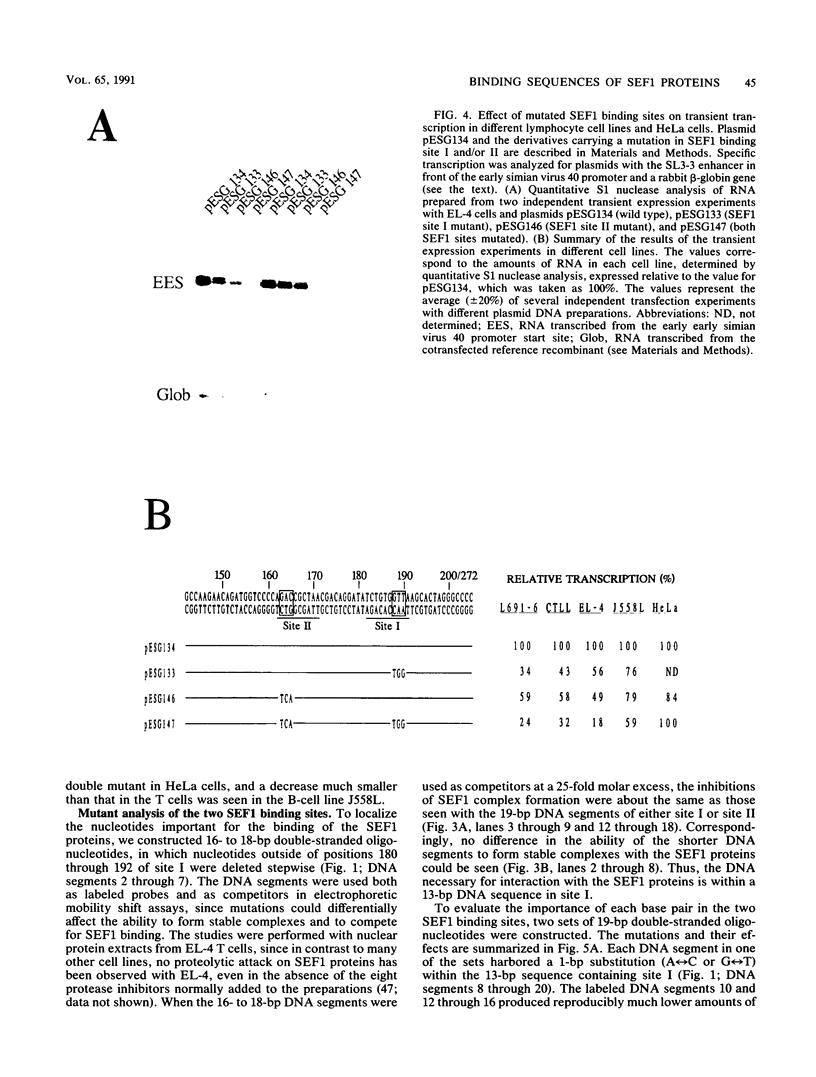
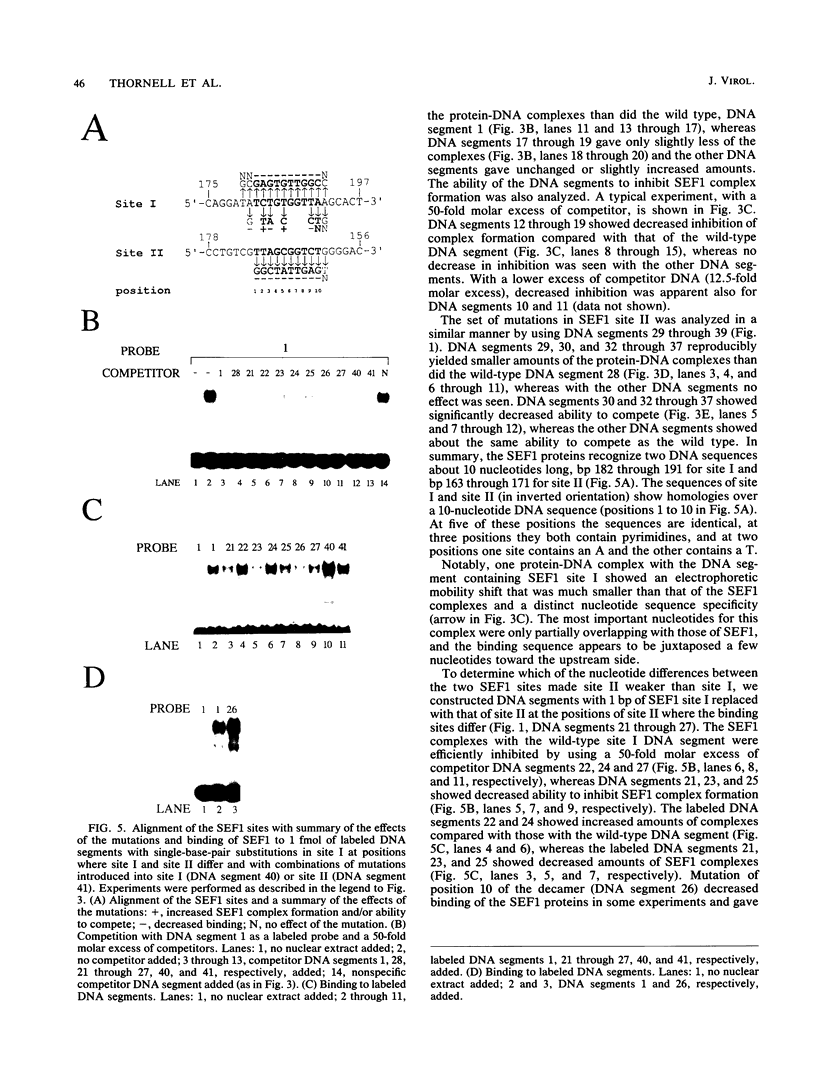
Interactions between SL3-3 enhancer factor 1 (SEF1) proteins and the enhancer of the murine leukemia virus SL3-3 were analyzed. SEF1 proteins were found to interact with two different DNA sequences within the DNA repeat region of the enhancer; these two motifs cooperated in enhancing initiation of transcription in T lymphocytes. Using an electrophoretic mobility shift assay, we identified nucleotides that are important for the SEF1 binding, and we deduced a sequence, 5'-TTTGCGGTTA/T-3' with highly improved binding of SEF1 proteins. We show that many different SEF1 binding sequences exist in the transcription control regions of different viral and cellular genes. The results indicate a general role of SEF1 proteins in T-cell gene expression.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker P. E., Gillis S., Smith K. A. Monoclonal cytolytic T-cell lines. J Exp Med. 1979 Jan 1;149(1):273–278. doi: 10.1084/jem.149.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boral A. L., Okenquist S. A., Lenz J. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J Virol. 1989 Jan;63(1):76–84. doi: 10.1128/jvi.63.1.76-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. J., Holbrook N. J., Mitchell K. F., Vallone C. A., Greengard J. S., Crabtree G. R., Lin Y. A viral long terminal repeat in the interleukin 2 gene of a cell line that constitutively produces interleukin 2. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7284–7288. doi: 10.1073/pnas.82.21.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R., Imagawa M., Imbra R. J., Bockoven J. R., Karin M. Multiple cis- and trans-acting elements mediate the transcriptional response to phorbol esters. Nature. 1987 Oct 15;329(6140):648–651. doi: 10.1038/329648a0. [DOI] [PubMed] [Google Scholar]
- Clark S. P., Kaufhold R., Chan A., Mak T. W. Comparison of the transcriptional properties of the Friend and Moloney retrovirus long terminal repeats: importance of tandem duplications and of the core enhancer sequence. Virology. 1985 Jul 30;144(2):481–494. doi: 10.1016/0042-6822(85)90288-0. [DOI] [PubMed] [Google Scholar]
- Clevers H., Lonberg N., Dunlap S., Lacy E., Terhorst C. An enhancer located in a CpG-island 3' to the TCR/CD3-epsilon gene confers T lymphocyte-specificity to its promoter. EMBO J. 1989 Sep;8(9):2527–2535. doi: 10.1002/j.1460-2075.1989.tb08390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D. R., Hapel A. J., Young I. G. Cloning and expression of the rat interleukin-3 gene. Nucleic Acids Res. 1986 May 12;14(9):3641–3658. doi: 10.1093/nar/14.9.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Deninger P. L., Esty A., LaPorte P., Hsu H., Friedmann T. The nucleotide sequence and restriction enzyme sites of the polyoma genome. Nucleic Acids Res. 1980 Feb 25;8(4):855–860. [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devare S. G., Reddy E. P., Law J. D., Robbins K. C., Aaronson S. A. Nucleotide sequence of the simian sarcoma virus genome: demonstration that its acquired cellular sequences encode the transforming gene product p28sis. Proc Natl Acad Sci U S A. 1983 Feb;80(3):731–735. doi: 10.1073/pnas.80.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORER P. A. Studies in antibody response of mice to tumour inoculation. Br J Cancer. 1950 Dec;4(4):372–379. doi: 10.1038/bjc.1950.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K., van den Elsen P., Bier E., Maxam A., Terhorst C. A T cell-specific enhancer is located in a DNase I-hypersensitive area at the 3' end of the CD3-delta gene. EMBO J. 1988 Aug;7(8):2401–2407. doi: 10.1002/j.1460-2075.1988.tb03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E. A., Speck N. A., Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990 Feb;64(2):534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E., Li Y., Fredrickson T. N., Hartley J. W., Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989 Jan;63(1):328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundström T., Zenke W. M., Wintzerith M., Matthes H. W., Staub A., Chambon P. Oligonucleotide-directed mutagenesis by microscale 'shot-gun' gene synthesis. Nucleic Acids Res. 1985 May 10;13(9):3305–3316. doi: 10.1093/nar/13.9.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg B., Grundström T. Tissue specific sequence motifs in the enhancer of the leukaemogenic mouse retrovirus SL3-3. Nucleic Acids Res. 1988 Jul 11;16(13):5927–5944. doi: 10.1093/nar/16.13.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe A., Gobet M., Even J., Sherr C. J., Galibert F. Nucleotide sequences of feline sarcoma virus long terminal repeats and 5' leaders show extensive homology to those of other mammalian retroviruses. J Virol. 1983 Jan;45(1):466–472. doi: 10.1128/jvi.45.1.466-472.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imler J. L., Schatz C., Wasylyk C., Chatton B., Wasylyk B. A Harvey-ras responsive transcription element is also responsive to a tumour-promoter and to serum. Nature. 1988 Mar 17;332(6161):275–278. doi: 10.1038/332275a0. [DOI] [PubMed] [Google Scholar]
- Ishimoto A., Adachi A., Sakai K., Matsuyama M. Long terminal repeat of Friend-MCF virus contains the sequence responsible for erythroid leukemia. Virology. 1985 Feb;141(1):30–42. doi: 10.1016/0042-6822(85)90180-1. [DOI] [PubMed] [Google Scholar]
- Jones N. C., Rigby P. W., Ziff E. B. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988 Mar;2(3):267–281. doi: 10.1101/gad.2.3.267. [DOI] [PubMed] [Google Scholar]
- Kaushansky K., O'Hara P. J., Berkner K., Segal G. M., Hagen F. S., Adamson J. W. Genomic cloning, characterization, and multilineage growth-promoting activity of human granulocyte-macrophage colony-stimulating factor. Proc Natl Acad Sci U S A. 1986 May;83(10):3101–3105. doi: 10.1073/pnas.83.10.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S., Martin M. A. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci U S A. 1983 May;80(9):2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P., de Jong R., Uematsu Y., Dembic Z., Ryser S., von Boehmer H., Steinmetz M., Berns A. Transcription of T cell receptor beta-chain genes is controlled by a downstream regulatory element. EMBO J. 1988 Mar;7(3):745–750. doi: 10.1002/j.1460-2075.1988.tb02871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Liou R. S., Boone L. R., Kiggans J. O., Yang D. M., Wang T. W., Tennant R. W., Yang W. K. Molecular cloning and analysis of the endogenous retrovirus chemically induced from RFM/Un mouse cell cultures. J Virol. 1983 Apr;46(1):288–292. doi: 10.1128/jvi.46.1.288-292.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoSardo J. E., Boral A. L., Lenz J. Relative importance of elements within the SL3-3 virus enhancer for T-cell specificity. J Virol. 1990 Apr;64(4):1756–1763. doi: 10.1128/jvi.64.4.1756-1763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Matrisian L. M., Leroy P., Ruhlmann C., Gesnel M. C., Breathnach R. Isolation of the oncogene and epidermal growth factor-induced transin gene: complex control in rat fibroblasts. Mol Cell Biol. 1986 May;6(5):1679–1686. doi: 10.1128/mcb.6.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall S., Peterson C. L., Calame K. A transcriptional enhancer 3' of C beta 2 in the T cell receptor beta locus. Science. 1988 Jul 8;241(4862):205–208. doi: 10.1126/science.2968651. [DOI] [PubMed] [Google Scholar]
- McGrath M. S., Pillemer E., Kooistra D., Weissman I. L. The role of MuLV receptors on T-lymphoma cells in lymphoma cell proliferation. Contemp Top Immunobiol. 1980;11:157–184. doi: 10.1007/978-1-4684-3701-0_5. [DOI] [PubMed] [Google Scholar]
- McKnight S., Tjian R. Transcriptional selectivity of viral genes in mammalian cells. Cell. 1986 Sep 12;46(6):795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- Mermod N., Williams T. J., Tjian R. Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vitro. Nature. 1988 Apr 7;332(6164):557–561. doi: 10.1038/332557a0. [DOI] [PubMed] [Google Scholar]
- Mohit B., Fan K. Hybrid cell line from a cloned immunoglobulin-producing mouse myeloma and a nonproducing mouse lymphoma. Science. 1971 Jan 8;171(3966):75–77. doi: 10.1126/science.171.3966.75. [DOI] [PubMed] [Google Scholar]
- Nedwin G. E., Naylor S. L., Sakaguchi A. Y., Smith D., Jarrett-Nedwin J., Pennica D., Goeddel D. V., Gray P. W. Human lymphotoxin and tumor necrosis factor genes: structure, homology and chromosomal localization. Nucleic Acids Res. 1985 Sep 11;13(17):6361–6373. doi: 10.1093/nar/13.17.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oi V. T., Morrison S. L., Herzenberg L. A., Berg P. Immunoglobulin gene expression in transformed lymphoid cells. Proc Natl Acad Sci U S A. 1983 Feb;80(3):825–829. doi: 10.1073/pnas.80.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen F. S., Crowther R. L., Tenney D. Y., Reimold A. M., Haseltine W. A. Novel leukaemogenic retroviruses isolated from cell line derived from spontaneous AKR tumour. Nature. 1981 Jul 9;292(5819):167–170. doi: 10.1038/292167a0. [DOI] [PubMed] [Google Scholar]
- Riedel N., Hoover E. A., Gasper P. W., Nicolson M. O., Mullins J. I. Molecular analysis and pathogenesis of the feline aplastic anemia retrovirus, feline leukemia virus C-Sarma. J Virol. 1986 Oct;60(1):242–250. doi: 10.1128/jvi.60.1.242-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Haseltine W. A., Lenz J., Ruprecht R., Cloyd M. W. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J Virol. 1985 Sep;55(3):862–866. doi: 10.1128/jvi.55.3.862-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988 Apr;8(4):1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor C. D., Scott M. L., Josephs S. F., Fry K. E., Reitz M. S., Jr Nucleotide sequence of the large terminal repeat of two different strains of gibbon ape leukemia virus. Virology. 1984 Aug;137(1):201–205. doi: 10.1016/0042-6822(84)90025-4. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., van Straaten F., Galleshaw J. A., Verma I. M. Nucleotide sequence of the genome of a murine sarcoma virus. Cell. 1981 Nov;27(1 Pt 2):97–108. doi: 10.1016/0092-8674(81)90364-0. [DOI] [PubMed] [Google Scholar]
- Vogt M., Haggblom C., Swift S., Haas M. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J Virol. 1985 Jul;55(1):184–192. doi: 10.1128/jvi.55.1.184-192.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Imler J. L., Chatton B., Schatz C., Wasylyk C. Negative and positive factors determine the activity of the polyoma virus enhancer alpha domain in undifferentiated and differentiated cell types. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7952–7956. doi: 10.1073/pnas.85.21.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J. H., Davidson I., Ferrandon D., Rosales R., Vigneron M., Macchi M., Ruffenach F., Chambon P. One cell-specific and three ubiquitous nuclear proteins bind in vitro to overlapping motifs in the domain B1 of the SV40 enhancer. EMBO J. 1987 Oct;6(10):3005–3013. doi: 10.1002/j.1460-2075.1987.tb02606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J. H., Davidson I., Macchi M., Rosales R., Vigneron M., Staub A., Chambon P. In vitro binding of several cell-specific and ubiquitous nuclear proteins to the GT-I motif of the SV40 enhancer. Genes Dev. 1987 Oct;1(8):794–807. doi: 10.1101/gad.1.8.794. [DOI] [PubMed] [Google Scholar]
- Yasukawa K., Hirano T., Watanabe Y., Muratani K., Matsuda T., Nakai S., Kishimoto T. Structure and expression of human B cell stimulatory factor-2 (BSF-2/IL-6) gene. EMBO J. 1987 Oct;6(10):2939–2945. doi: 10.1002/j.1460-2075.1987.tb02598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]