Abstract
Background
The tissue distributions and functions of Eph receptors and their ephrin ligands have been well studied, however less is known about their evolutionary history. We have undertaken a phylogenetic analysis of Eph receptors and ephrins from a number of invertebrate and vertebrate species.
Results
Our findings indicate that Eph receptors form three major clades: one comprised of non-chordate and cephalochordate Eph receptors, a second comprised of urochordate Eph receptors, and a third comprised of vertebrate Eph receptors. Ephrins, on the other hand, fall into either a clade made up of the non-chordate and cephalochordate ephrins plus the urochordate and vertebrate ephrin-Bs or a clade made up of the urochordate and vertebrate ephrin-As.
Conclusion
We have concluded that Eph receptors and ephrins diverged into A and B-types at different points in their evolutionary history, such that primitive chordates likely possessed an ancestral ephrin-A and an ancestral ephrin-B, but only a single Eph receptor. Furthermore, ephrin-As appear to have arisen in the common ancestor of urochordates and vertebrates, whereas ephrin-Bs have a more ancient bilaterian origin. Ancestral ephrin-B-like ligands had transmembrane domains; as GPI anchors appear to have arisen or been lost at least 3 times.
Background
The Eph family represents the largest subgroup of receptor tyrosine kinases, with most vertebrate genomes having 14 members [1]. Eph receptors can be divided into two classes based on sequence similarity and ligand binding affinity [2,3]. A-type Eph receptors (EphA1–8,10) bind promiscuously to glycosylphosphatidylinositol (GPI) – linked ephrin-A ligands (ephrin-A1–6), whereas B-type Eph receptors (EphB1–4, 6) bind promiscuously to transmembrane ephrin-B ligands (ephrin-B1–3). The only known exceptions to this are EphA4, which can bind ephrin-B2 and 3 in addition to the ephrin-As [3], and EphB2, which can bind ephrin-A5 in addition to the ephrin-Bs [4].
Eph receptors and ephrin ligands classically function in cell-cell repulsion events. For example, retinal ganglion cell (RGC) axons expressing high levels of Eph receptors are excluded from ephrin-rich regions in the optic tectum and instead map to points where ligand expression is low [5,6]. Eph receptors and ephrins similarly show complementary distributions in the developing hindbrain, where their interactions function to keep different cell populations separate and thus establish segment boundaries [7]. Eph receptors are also expressed by the neural crest (NC) and mediate the exclusion of these cells from territories where complementary ephrin ligands are expressed, thus restricting their migration to stereotypical pathways [8,9].
Eph receptors and ephrins are found throughout the metazoans and the phylogeny of these genes in vertebrates is well established [10,11]. However, the relationship of invertebrate Eph receptors and ephrins to their vertebrate counterparts is not as clear [12]. A number of studies have constructed phylogenies using vertebrate and ecdysozoan (e.g. nematode and insect) sequences, generally resulting in trees with three major clades: one comprised of ecdysozoan Eph receptors/ephrins, a second comprised of vertebrate EphAs/ephrin-As, and a third comprised of vertebrate EphBs/ephrin-Bs [11,13,14]. However, this approach is unsatisfactory because of the large evolutionary gaps between the protostomes and the vertebrates. The increasing availability of genomic data from deuterostomes such as echinoderms, urochordates and cephalochordates presents an opportunity to better resolve the question of how Eph receptors and ephrins diversified.
Results
The amino acid sequences of the full-length Eph receptors could be readily aligned with the ephrin binding domains and the tyrosine kinase domains being most highly conserved and acting as anchors (see Additional file 1). Gaps were introduced principally by inclusion of the human EphA6 and C. intestinalis Eph sequences. The full-length Eph sequences ranged from 10% to 68% identity, whereas the sequences between the ephrin binding domain and the kinase domain ranged from 30 to 60% identity. Trees constructed from full-length sequences, sequences between the Ephrin binding and kinase domains, or from the kinase domain alone all had similar topology. The full-length Ephrin amino acid sequences will also readily align with ClustalW centered on the conserved Ephrin domain (see Additional file 2). The amino terminal sequence of N. vectensis and the amino and carboxyl sequences of D. melanogaster sequence are unique and were removed from the analysis. Gaps were introduced to accommodate the D. melanogaster and C. elegans sequences. The overall identity within the Ephrin sequences was from 10 to 60%, whereas within the Ephrin domain identities ranged from 30 to 60%. Several of the sequences used are predictions made from genomic data and introns exon boundary predictions may be incorrect.
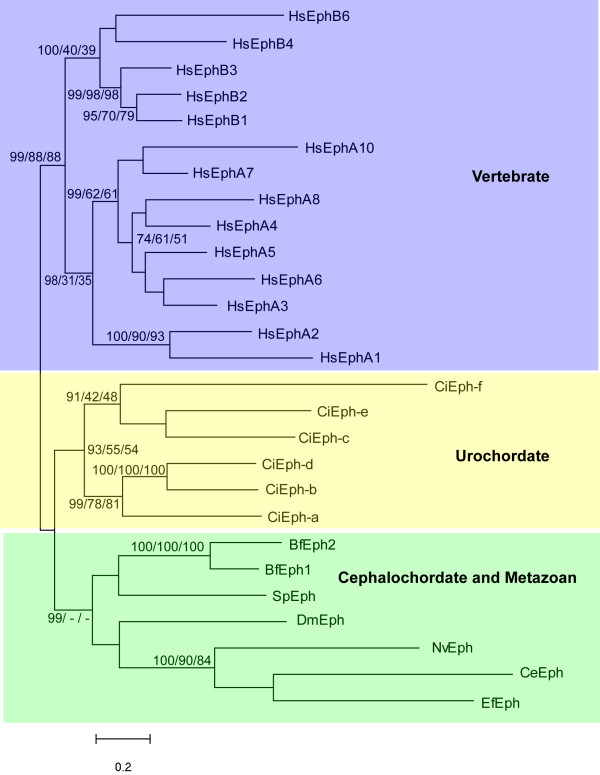
Our phylogenetic tree for invertebrate and vertebrate Eph receptors is comprised of a cephalochordate plus non-chordate clade, a urochordate clade, and a vertebrate clade (Fig. 1). There is weak support for the Urochordate and Cephalochordate/Metazoan grouping, however independent diversification of urochordate, cephalochordate, and ecdysozoan clades is well supported. With this larger data set, there is a distinct vertebrate Eph clade divided into a EphA and EphB groups. Within the EphA clade, there is strong bootstrap support for a subgroup comprised of EphA3–6,8 and another comprised of EphA7 and 10. Similarly, within the EphB clade, there is a distinct group comprised of Eph4,6 and another comprised of Eph1–3.
Figure 1.
Unrooted phylogenetic tree for Eph receptors constructed with the Maximum Likelihood method. Numbers represent percentage bootstrap values for 1000 replicates (Maximum likelihood/Neighbour Joining/Minimum Evolution); unlabelled branches or/- indicates a value less that 50%. Three major clades are present: Vertebrate, Urochordate, and a group containing Cephalochordates, Echinoderms, Ecdysozoans, Coelenterate and Sponges. Using the ecdysozoan branch to root the tree does not change the composition of the major groups. Bf, Brachiostoma floridae; Ce, Caenorhabditis elegans; Ci, Ciona intestinalis; Dm, Drosophila melanogaster; Ef, Ephydatia fluviatilis; Hs, Homo sapiens; Nv, Nematostela vectensis; Sp, Strongylocentrotus purpuratus.
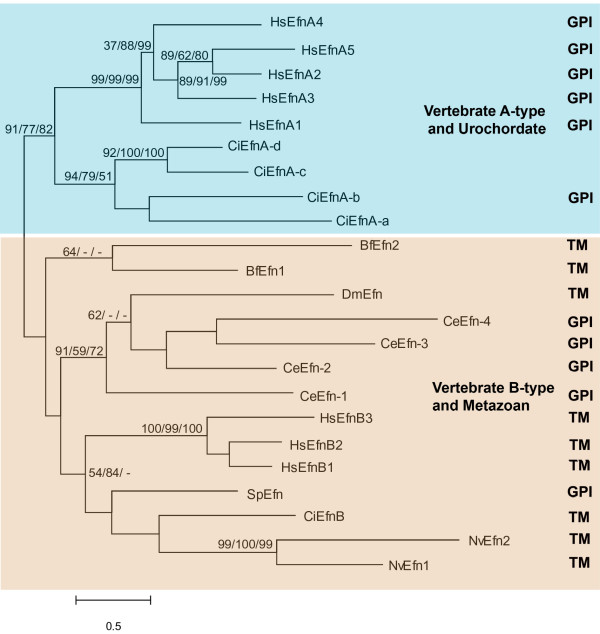
Our phylogenetic tree for ephrin ligands can be broadly divided into two major clades (Fig. 2). One diverse group includes coelenterate, ecdysozoan, echinoderm, cephalochordate, urochordate and vertebrate B ephrins. Within this group there is evidence for independent diversification of cephalochordate, ecdysozoan and deuterostome ephrin ligands. The second major ephrin clade is comprised of a urochordate ephrin-A subclade and a vertebrate ephrin-A subclade. There is good support for this being a monophyletic group in which there have been independent diversifications.
Figure 2.
Phylogenetic tree for ephrin ligands constructed with Maximum Likelihood method. Numbers represent percentage bootstrap values for 1000 replicates (Maximum likelihood/Neighbour Joining/Minimum Evolution); unlabelled branches or/- indicates a value less that 50%. Two major clades are present: Vertebrate A-type and Urochordate ephrins and a group containing Vertebrate B-type, Cephalochordate, Ecdysozoan, Echinoderm, Coelenterate, Sponge and one Urochordate ephrin. Using the ecdysozoan branch to root the tree does not change the composition of the major groups. Efn, ephrin; Bf, Brachiostoma floridae; Ce, Caenorhabditis. elegans; Ci, Ciona intestinalis; Dm, Drosophila melanogaster; Hs, Homo sapiens; Nv, Nematostella vectensis; Sp, Strongylocentrotus purpuratus. Results of predictions for the type of membrane anchorage are aligned with individual branches; TM, transmembrane domains; GPI, glycosylphosphatidylinositol modification. Where no prediction is reported the protein prediction contains neither.
Discussion
The clearest implication from our phylogenetic analysis is that Eph receptors and their ephrin ligands diversified at different times in evolutionary history. Vertebrate EphAs and EphBs do not have invertebrate orthologues, whereas vertebrate ephrin-As and ephrin-Bs have urochordate counterparts. This suggests that the diversification of Eph receptors followed the diversification of ephrins and that the common ancestor of urochordates and vertebrates likely had a single receptor for the two different types of ligand.
The possibility exists that EphA4 and EphB2 have some special relationship to the ancestor of vertebrate Eph receptors, given that these two receptors are uniquely capable of interacting with both ephrin-As and ephrin-Bs [3,4]. Indeed, the most common vertebrate matches for urochordate Eph receptors returned from BLAST searches are EphA4 and EphB2. EphA4 and EphB2 may therefore represent the most ancient vertebrate Eph receptors. By this scenario, the descendants of the ancestral Eph, which was presumably capable of interacting with both types of ligand, became increasingly specific for either ephrin-As or ephrin-Bs, such that EphA4 and EphB2 persist as the only reminders of the ancestral characteristic of dual ligand binding specificity. Our findings, however, fail to support this idea since neither EphA4 nor EphB2 is placed outside of a subclade containing the remaining EphA or EphB receptors. This suggests that vertebrate Eph receptors randomly lost the ability to interact with both ephrin-As and ephrin-Bs as they diversified, which is consistent with the fact that EphA4 cannot bind all ephrin-Bs just as EphB2 cannot bind all ephrin-As. However, the homology of the urochordate Eph receptors to EphA4 and EphB2 remains as a point of support for the idea that the predecessor of vertebrate Eph receptors could bind to both ephrin-As and ephrin-Bs.
Is there evidence that the diversification of Eph receptors in the vertebrates is associated with a diversification of function? The functions performed by Eph receptors and ephrins typically fall into one of several categories including regulation of axon guidance (e.g. formation of visual topographic maps), directed cell migration (e.g. NC cell pathfinding), or tissue segregation (e.g. hindbrain segmentation). Do EphA receptors perform different functions from EphB receptors? A survey of the literature reveals that while some specific roles are unique to each class of receptor, in a broader context both EphAs and EphBs perform a wide range of similar functions. For example, EphAs are uniquely involved in tissue segregation events that are part of somite segmentation [15,16] and limb development [17-20], as well as motor neuron axon guidance [21-27]. On the other hand, both EphAs and EphBs contribute to hindbrain segmentation [7,28-31], segregation of veins from arteries during angiogenesis [32-35], patterning of the inner ear [36,37], directed migration of NC cells [8,9,38,39], and guidance of RGC axons to their targets [39-41]. Thus, it is difficult to ascribe one set of functions to EphAs and another to EphBs. The impressive array of EphAs and EphBs may simply reflect the duplication and diversification of an ancestral receptor to perform subtle variations of the same functions in different cell or tissue types as the body plan become progressively more complex in the lineage from basal chordate to vertebrate.
Another intriguing question is if one of the three prototypical functions performed by Eph receptors can be regarded as ancestral. The surprising discovery of a poriferan Eph [10] indicates that these receptors are not a eumetazoan novelty. Sponges lack nerves, muscles, and true tissues, but do possess a cellular grade of organization in which different cell types are arranged into tissue-like layers such as the porous epidermis (pinacoderm) on the exterior and the lining of flagellated cells on the interior [42,43]. While there has been no research on Eph function in sponges, they may play a role in the positioning of these different cell types during development [44]. In the nematode C. elegans, Eph receptors and ephrins are expressed in adjacent populations of neural precursor cells and when either receptor or ligand function is disrupted, the neural precursors become disorganized and the ventral epidermis fails to close over these cells [45,56]. While the Eph receptors and ephrins are expressed in neural cells, the function performed here is more consistent with a tissue segregation mechanism. On the other hand, defective Eph function in C. elegans also results in axons extending beyond their expected target sites [47], indicating that these receptors can contribute to axon guidance in invertebrates as well. In the insect D. melanogaster, Eph receptors and ephrins function similarly to guide cortical axons to the optic ganglion [48] and restrict interneuronal axons from either crossing the midline or exiting the central nervous system [49]. In the echinoderm S. purpuratus, Eph receptors and ephrins are expressed in non-overlapping ectoderm domains and do not appear to function directly in axon guidance, but rather are involved in patterning domains of ectoderm (Jones, Yaguchi, and Burke, unpublished). In C. intestinalis embryos, Eph and ephrin mediate an early cell fate specification by activation of ERK signalling pathway producing an asymmetric cell division [50]. [51] has speculated that the ancestral function is regulation of cellular movements, but we currently lack sufficient functional data from diverse metazoans to reasonably conclude which functions of Eph receptors can be regarded as ancestral. A better understanding of the interacting signal transduction components of Eph and ephrin signalling pathways may reveal relationships not apparent from considerations of overall function.
A second important implication from our findings is that ephrin-Bs are the more ancient of the two types of vertebrate ephrins. The urochordate and vertebrate ephrin-Bs are actually part of a larger clade that also includes cnidarian, nematode, insect, echinoderm, and cephalochordate ephrins. The urochordate and vertebrate subclade is grouped with one comprised of cephalochordate, echinoderm, and cnidarian ephrins, while the nematode and insect ephrins form an external ecdysozoan clade. The four C. elegans ephrins are paraphyletic with the single D. melanogaster ephrin, perhaps indicating that the common ancestor for nematodes and insects possessed multiple ephrins, some of which were lost in the lineage to D. melanogaster. Although lineage specific expansion of ephrins occur, so an expansion of the C. elegans complement is equally likely.
Our analysis of Eph receptors suggests urochordate Eph receptors are more closely related to vertebrate receptors than the cephalochordate receptors, BfEph1 and BfEph2. Similarly, our analysis of ephrins allies vertebrate A-type ligands with urochordate A-type ligands. These topologies are contrary to the more firmly established relationship of cephalochordates being considered the closest living relatives of vertebrates. Similar results have been reported with other analyses involving small sets of molecules, yet more complete sets confirm the conventional relationships [12,52]. The evolutionary history of Eph receptors and ephrin ligands appears to be distinct from the history of the species, which suggests that functional features of these molecules have constrained their diversification.
The unusual topology of the ephrin-B clade suggests that the ligands possessed by cnidarians, echinoderms, and cephalochordates are more ephrin-B-like than those possessed by ecdysozoans. On the other hand the bootstrap support for the cnidarian, echindoderm, and cephalochordate ephrins grouping with the urochordate and vertebrate ephrin-Bs is low. Therefore, the possibility of a more obvious non-chordate plus cephalochordate subclade existing cannot be excluded. Although there is currently a wealth of genomic information available for an array of invertebrates, there are no Eph receptor or ephrin sequences from lophotrochozoans. This puts a considerable gap into our analysis, the elimination of which might allow for a more clear resolution of ephrin-B phylogeny.
Our analysis suggests that a clade of ephrin-A ligands distinguished by sequence similarity in the ephrin domain arose in a common ancestor of urochordates and vertebrates. Curiously, some of the invertebrate ephrins lack a transmembrane domain and have a putative GPI anchor site, which is the defining feature of the vertebrate A type ephrins. These invertebrate ephrins have an ephrin-B-like receptor binding domain that is the tethered to the cell membrane by a GPI anchor. Predictions of transmembrane domains occur in cnidarians, insects, urochordates, cephalochordates, and vertebrates. Whereas, GPI anchors predictions occur in nematodes, echinoderms, urochordates and vertebrates. This distribution suggests that a transmembrane anchorage of an ephrin with a B-type ligand binding domain is the ancestral state and that GPI anchors have arisen or been lost at least 3 times. The types of membrane anchorage have yet to be confirmed experimentally, but it suggests that the vertebrate ephrin As are a homologous group related by their binding domain and that the mechanism of anchorage is not a reliable defining feature for the group.
Conclusion
We have shown through a phylogenetic analysis that Eph receptors and ephrin ligands followed different paths of evolution. For the receptors, the ancestral Eph receptors of non-chordates and cephalochordates diversified independently in urochordates and vertebrates. For the ligands there were lineage specific diversifications of forms with ephrin-B-like receptor binding domains and ephrin-As appear in the founding lineages of urochordates and vertebrates. Ephrins and Eph receptors therefore diversified at different points in evolution, such that the common ancestor of urochordates and vertebrates possessed a single receptor, but two ligands. Additional data on the functions and signalling pathways employed by Eph receptors and ephrins in diverse metazoans are necessary to more clearly define the evolutionary relationships within this intriguing family of signalling molecules.
Methods
Full length Eph and/or ephrin amino acid sequences for Ephydatia fluviatilis (poriferan), Nematostella vectensis (cnidarian), Caenorhabditis elegans (nematode), Drosophila melanogaster (insect), Strongylocentrotus purpuratus (echinoderm), Branchiostoma floridae (cephalochordate), Ciona intestinalis (urochordate), and Homo sapiens (vertebrate) were retrieved from (53–55, see additional files 1, 2). Sequences were aligned in MEGA 4.0 (56) and assembled into phylogenetic trees using the minimum evolution (ME), maximum parsimony (MP), neighbour joining (NJ), and UPGMA methods. Sequences aligned in BioEdit (57) were assembled into maximum likelihood (ML) trees using the Treefinder (58) program. Trees were created under the Jones-Taylor-Thornton model of amino acid substitution (where appropriate) using 1000 bootstrap replications. In each case, the ME, ML, and NJ trees gave very similar results and so we have included only the ML trees here. GPI modification site predictions were done at (59) and transmembrane domain predictions were made at the TMHMM server (60) (see Additional file 3).
Authors' contributions
DOM did the analysis and prepared the first draft of the paper; RDB conceived of the study, revised, and edited the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Aligned Eph sequences. Aligned Eph amino acid sequence data used in this study.
Aligned Ephrin sequences. Aligned Ephrin amino acid sequence data used in this study.
Accession numbers. A list of accession number for the amino acid sequences used in this study.
Acknowledgments
Acknowledgements
This research was supported by grants to RDB from NSERC and CIHR (Canada). We are grateful to John Taylor and Perry Howard for discussions and help.
Contributor Information
Dan O Mellott, Email: dan.mellott@gmail.com.
Robert D Burke, Email: rburke@uvic.ca.
References
- Murai KK, Pasquale EB. 'Eph'ective signaling: forward, reverse and crosstalk. J Cell Sci. 2003;116:2823–2832. doi: 10.1242/jcs.00625. [DOI] [PubMed] [Google Scholar]
- Eph Nomenclature Committee Unified nomenclature for Eph family receptors and their ligands, the ephrins. Cell. 1997;90:403–404. doi: 10.1016/S0092-8674(00)80500-0. [DOI] [PubMed] [Google Scholar]
- Gale NW, Holland SJ, Valenzuela DM, Flenniken A, Pan L, Ryan TE, Henkemeyer M, Strebhardt K, Hirai H, Wilkinson DG, Pawson T, Davis S, Yancopoulos GD. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron. 1996;17:9–19. doi: 10.1016/S0896-6273(00)80276-7. [DOI] [PubMed] [Google Scholar]
- Himanen JP, Chumley MJ, Lackmann M, Li C, Barton WA, Jeffrey PD, Vearing C, Geleick D, Feldheim DA, Boyd AW, Henkemeyer M, Nikolov DB. Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci. 2004;7:501–509. doi: 10.1038/nn1237. [DOI] [PubMed] [Google Scholar]
- Cheng HU, Nakamoto M, Bergemann AD, Flanagan JG. Complementary gradients in expression and binding of ELF-1 and Mek4 in development of the topographic retinotectal projection map. Cell. 1995;82:371–381. doi: 10.1016/0092-8674(95)90426-3. [DOI] [PubMed] [Google Scholar]
- Monschau B, Kremoser C, Ohta K, Tanaka H, Kaneko T, Yamada T, Handwerker C, Hornberger MR, Loschinger J, Pasquale EB, Siever DA, Verderame MF, Muller BK, Bonhoeffer F, Drescher U. Shared and distinct functions of RAGS and ELF-1 in guiding retinal axons. EMBO J. 1997;16:1258–1267. doi: 10.1093/emboj/16.6.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Mellitzer G, Robinson V, Wilkinson DG. In vivo cell sorting in complementary segmental domains mediated by Eph receptors and ephrins. Nature. 1999;399:267–271. doi: 10.1038/20452. [DOI] [PubMed] [Google Scholar]
- Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, Fraser SE, Bronner-Fraser M. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern of trunk neural crest migration. Curr Biol. 1997;7:571–580. doi: 10.1016/S0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- Smith A, Robinson V, Patel K, Wilkinson DG. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr Biol. 1997;7:561–570. doi: 10.1016/S0960-9822(06)00255-7. [DOI] [PubMed] [Google Scholar]
- Suga H, Koyanagi M, Hoshiyama D, Ono K, Iwabe N, Kuma K, Miyata T. Extensive gene duplication in the early evolution of animals before the parazoan-eumetazoan split demonstrated by G proteins and protein tyrosine kinases from sponge and hydra. J Mol Evol. 1999;48:646–653. doi: 10.1007/PL00006508. [DOI] [PubMed] [Google Scholar]
- Gu J, Gu X. Natural history and functional divergence of protein tyrosine kinases. Gene. 2003;317:49–57. doi: 10.1016/S0378-1119(03)00696-6. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Suga H, Hoshiyama D, Kuraku S, Katoh K, Kubokawa K, Miyata T. Protein tyrosine kinase cDNAs from Amphioxus Hagfish and lamprey: isoform duplications around the divergence of cyclostomes. J Mol Evol. 1999;49:601–608. doi: 10.1007/PL00006581. [DOI] [PubMed] [Google Scholar]
- Satou Y, Sasakura Y, Yamada L, Imai KS, Satoh N, Degnan B. A genomewide survey of developmentally relevant genes in Ciona intestinalis: V. genes for receptor tyrosine kinase pathway and Notch signaling pathway. Dev Genes Evol. 2003;213:254–263. doi: 10.1007/s00427-003-0317-9. [DOI] [PubMed] [Google Scholar]
- Barrios A, Poole RJ, Durbin L, Brennan C, Holder N, Wilson SW. Eph/Ephrin signaling regulates the mesenchymal-to-epithelial transition of the paraxial mesoderm during somite morphogenesis. Curr Biol. 2003;13:1571–1582. doi: 10.1016/j.cub.2003.08.030. [DOI] [PubMed] [Google Scholar]
- Durbin L, Brennan C, Shiomi K, Cooke J, Barrios A, Shanmugalingam S, Guthrie B, Lindberg R, Holder N. Eph signaling is required for segmentation and differentiation of the somites. Genes Dev. 1998;12:3096–3109. doi: 10.1101/gad.12.19.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo M, Piedra ME, Herrara MT, Ros MA, Nieto MA. The expression and regulation of chick EphA7 suggests roles in limb patterning and innervation. Development. 1998;125:4195–4204. doi: 10.1242/dev.125.21.4195. [DOI] [PubMed] [Google Scholar]
- Ganju P, Shigemoto K, Brennan J, Entwistle A, Reith AD. The Eck receptor tyrosine kinase is implicated in pattern formation during gastrulation hindbrain segmentation and limb development. Oncogene. 1994;9:1613–1624. [PubMed] [Google Scholar]
- Patel K, Nittenberg R, D'Souza D, Irving C, Burt D, Wilkinson DG, Tickle C. Expression and regulation of Cek-8 a cell to cell signaling receptor in developing chick limb buds. Development. 1996;122:1147–1155. doi: 10.1242/dev.122.4.1147. [DOI] [PubMed] [Google Scholar]
- Wada N, Tanaka H, Ide H, Nohno T. Ephrin-A2 regulates position-specific cell affinity and is involved in cartilage morphogenesis in the chick limb bud. Dev Biol. 2003;264:550–563. doi: 10.1016/j.ydbio.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Araujo M, Nieto MA. The expression of chick EphA7 during segmentation of the central and peripheral nervous system. Mech Dev. 1997;8:173–177. doi: 10.1016/S0925-4773(97)00147-0. [DOI] [PubMed] [Google Scholar]
- Eberhart J, Barr J, O'Connell S, Flagg A, Swartz ME, Cramer KS, Tosney KW, Pasquale EB, Krull CE. Ephrin-A5 exerts positive or inhibitory effects on distinct subsets of EphA4-positive motor neurons. J Neurosci. 2004;4:1070–1078. doi: 10.1523/JNEUROSCI.4719-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart J, Swartz ME, Koblar SA, Pasquale EB, Krull CE. EphA4 constitutes a population-specific guidance cue for motor neurons. Dev Biol. 2002;247:89–101. doi: 10.1006/dbio.2002.0695. [DOI] [PubMed] [Google Scholar]
- Helmbacher F, Schneider-Manoury S, Topilko P, Tiret L, Charnay P. Targeting of the EphA4 tyrosine kinase receptor affects dorsal/ventral pathfinding of limb motor axons. Development. 2000;127:3313–3324. doi: 10.1242/dev.127.15.3313. [DOI] [PubMed] [Google Scholar]
- Iwamasa H, Ohta K, Yamada T, Ushijima K, Terasaki H, Tanaka H. Expression of Eph receptor tyrosine kinases and their ligands in chick embryonic motor neurons and hindlimb muscles. Dev Growth Differ. 1999;41:685–698. doi: 10.1046/j.1440-169x.1999.00468.x. [DOI] [PubMed] [Google Scholar]
- Kury P, Gale N, Connor R, Pasquale E, Guthrie S. Eph receptors and ephrin expression in cranial motor neurons and the branchial arches of the chick embryo. Mol Cell Neurosci. 2000;15:123–140. doi: 10.1006/mcne.1999.0812. [DOI] [PubMed] [Google Scholar]
- Olivieri G, Miescher GC. Immunohistochemical localization of EphA5 in the adult human central nervous system. J Histochem Cytochem. 1999;47:855–861. doi: 10.1177/002215549904700702. [DOI] [PubMed] [Google Scholar]
- Becker N, Seitanidou T, Murphy P, Mattei MG, Topilko P, Nieto MA, Wilkinson DG, Charnay P, Gilardi-Hebenstreit P. Several receptor tyrosine kinase genes of the Eph family are segmentally expressed in the developing hindbrain. Mech Dev. 1994;47:3–17. doi: 10.1016/0925-4773(94)90091-4. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Gilardi-Hebenstreit P, Charnay P, Wilkinson DG. A receptor protein tyrosine kinase implicated in the segmental patterning of the hindbrain mesoderm. Development. 1992;116:1137–1150. doi: 10.1242/dev.116.4.1137. [DOI] [PubMed] [Google Scholar]
- Henkemeyer M, Marengere LE, McGlade J, Olivier JP, Conlon RA, Holmyard DP, Letwin K, Pawson T. Immunolocalization of the Nuk receptor tyrosine kinase suggests roles in segmental patterning of the brain and axonogenesis. Oncogene. 1994;9:1001–1014. [PubMed] [Google Scholar]
- Xu Q, Alldus G, Holder N, Wilkinson DG. Expression of truncated Sek-1 receptor tyrosine kinase disrupts the segmental restriction of gene expression in the Xenopus and zebrafish hindbrain. Development. 1995;121:4005–4016. doi: 10.1242/dev.121.12.4005. [DOI] [PubMed] [Google Scholar]
- Adams RH, Wilkinson GA, Weiss C, Diella F, Gale NW, Deutsch U, Risau W, Klein R. Roles of ephrinB ligands and EphB receptors in cardiovascular development: demarcation of arterial/venous domains vascular morphogenesis and sprouting angiogenesis. Genes Dev. 1999;13:295–306. doi: 10.1101/gad.13.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantley-Sieders DM, Caughron J, Hicks D, Pozzi A, Ruiz JC, Chen J. EphA2 receptor tyrosine kinase regulates endothelial cell migration and vascular assembly through phosphoinositide 3-kinase-mediated Rac1 GTPase activation. J Cell Sci. 2004;117:2037–2049. doi: 10.1242/jcs.01061. [DOI] [PubMed] [Google Scholar]
- Gerety SS, Wang HU, Chen ZF, Anderson DJ. Symmmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol Cell. 1999;4:403–414. doi: 10.1016/S1097-2765(00)80342-1. [DOI] [PubMed] [Google Scholar]
- Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/S0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- Bianchi LM, Liu H. Comparison of ephrin-A ligand and EphA receptor distribution in the developing inner ear. Anat Rec. 1999;254:127–134. doi: 10.1002/(SICI)1097-0185(19990101)254:1<127::AID-AR16>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Pickles JO, Claxton C, Van Heumen WR. Complementary and layered expression of Ephs and ephrins in developing mouse inner ear. J Comp Neurol. 2002;449:207–216. doi: 10.1002/cne.10231. [DOI] [PubMed] [Google Scholar]
- Helbling PM, Tran CT, Brandli AW. Requirement for EphA receptor signaling in the segregation of Xenopus third and fourth arch neural crest. Mech Dev. 1998;78:63–79. doi: 10.1016/S0925-4773(98)00148-8. [DOI] [PubMed] [Google Scholar]
- Feldheim DA, Nakamoto M, Osterfield M, Gale NW, DeChiara TM, Rohatgi R, Yancopoulos GD, Flanagan JG. Loss-of-function analysis of EphA receptors in retinotectal mapping. J Neurosci. 2004;24:2542–2550. doi: 10.1523/JNEUROSCI.0239-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindges R, McLaughlin T, Genoud N, Henkemeyer M, O'Leary DD. EphB forward signaling controls directional branch extension and arborization required for dorsal-ventral retinotopic mapping. Neuron. 2002;35:475–487. doi: 10.1016/S0896-6273(02)00799-7. [DOI] [PubMed] [Google Scholar]
- Walkenhorst J, Dutting D, Handwerker C, Huai J, Tanaka H, Drescher U. The EphA4 receptor tyrosine kinase is necessary for the guidance of nasal retinal ganglion cell axons in vitro. Mol Cell Neurosci. 2000;16:365–375. doi: 10.1006/mcne.2000.0878. [DOI] [PubMed] [Google Scholar]
- Leys SP, Mackie GO, Reiswig HM. The biology of glass. Advances in Marine Biology. 2007;52:1–145. doi: 10.1016/S0065-2881(06)52001-2. [DOI] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Liubicich D, Hinman VF, Gauthier M, Gongora M, Green K, Worheide G, Leys SP, Degnan BM. Developmental expression of transcription factor genes in a demosponge: insights into the origin of metazoan multicellularity. Evolution & Development. 2006;8:150–173. doi: 10.1111/j.1525-142X.2006.00086.x. [DOI] [PubMed] [Google Scholar]
- Degnan BM, Leys SP, Larroux C. Sponge development and antiquity of animal pattern formation. Integrative and Comparative Biology. 2005;45:335–341. doi: 10.1093/icb/45.2.335. [DOI] [PubMed] [Google Scholar]
- George SE, Simokat K, Hardin J, Chisholm AD. The VAB-1 Eph receptor tyrosine kinase functions in neural and epithelial morphogenesis in C. elegans. Cell. 1998;92:633–643. doi: 10.1016/S0092-8674(00)81131-9. [DOI] [PubMed] [Google Scholar]
- Chin-Sang ID, George SE, Ding M, Moseley SL, Lynch AS, Chisholm AD. The ephrin VAB-2/EFN-1 functions in neuronal signaling to regulate epidermal morphogenesis in C. elegans. Cell. 1998;92:633–643. doi: 10.1016/S0092-8674(00)81131-9. [DOI] [PubMed] [Google Scholar]
- Mohamed AM, Chin-Sang ID. Dev Biol. 290:164–176. doi: 10.1016/j.ydbio.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Dearborn R, Jr, He Q, Kunes S, Dai Y. 2002 Eph receptor tyrosine kinase-mediated formation of a topographic map in the Drosophila visual system. J Neurosci. 2006;22:1338–1349. doi: 10.1523/JNEUROSCI.22-04-01338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T, Brand AH. Dephrin a transmembrane ephrin with a unique structure prevents interneuronal axons from exiting the Drosophila embryonic CNS. Development. 2002;129:4205–4218. doi: 10.1242/dev.129.18.4205. [DOI] [PubMed] [Google Scholar]
- Picco V, Hudson C, Yasuo H. Ephrin-Eph signalling drives the asymmetric division of notochord/neural precursors in Ciona embryos. Development. 2007;134:1491–1497. doi: 10.1242/dev.003939. [DOI] [PubMed] [Google Scholar]
- Drescher U. Eph family functions from an evolutionary perspective. Curr Opin Genet Dev. 2002;12:397–402. doi: 10.1016/S0959-437X(02)00316-7. [DOI] [PubMed] [Google Scholar]
- Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, Lander ES, Thorndyke M, Nakano H, Kohn AB, Heyland A, Moroz LL, Copley RR, Telford MJ. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- NCBI, Protein Home http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein
- NCBI Genome Biology http://www.ncbi.nlm.nih.gov/Genomes/
- JGI Eukaryotic Genomes http://genome.jgi-psf.org/
- MEGA, Molecular Evolutionary Genetic Analysis http://www.megasoftware.net/
- Bioedit, Biological Sequence Alignment Editor http://www.mbio.ncsu.edu/BioEdit/bioedit.html
- Treefinder http://www.treefinder.de/
- GPI Lipid anchor project http://mendel.imp.ac.at/gpi/gpi_server.html
- TMHMM Server http://www.cbs.dtu.dk/services/TMHMM/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Aligned Eph sequences. Aligned Eph amino acid sequence data used in this study.
Aligned Ephrin sequences. Aligned Ephrin amino acid sequence data used in this study.
Accession numbers. A list of accession number for the amino acid sequences used in this study.