Abstract
Retrovirus RNA is synthesized as a single primary transcript that is differentially processed by RNA splicing. Three species of viral RNA (spliced env, spliced src, and unspliced full-length RNA) are produced in chicken embryo fibroblasts infected with Rous sarcoma virus, an avian retrovirus. The env and src mRNAs are synthesized by the alternative use of two 3' splice sites. The mechanism by which balanced splicing at the two sites is maintained was investigated in this report. Mutants that increase or decrease splicing at one of the two 3' splice sites were analyzed for the effect on splicing at the other site. The two splice sites differed in their response. Mutations that caused a specific increase in the level of spliced env mRNA were associated with reciprocal changes in the levels of src mRNA but with no change in overall splicing. In contrast, mutants in which the src 3' splice site was inactivated demonstrated a decreased overall level of spliced RNA but had little or no effect on the level of spliced env mRNA. Mutations that caused specific increases in the level of spliced src mRNA had variable effects on env mRNA levels. Deletion of regions in gag, which was previously shown to contain a cis-acting negative regulator of splicing, resulted in a corresponding increase of both spliced viral mRNAs and a decrease in unspliced RNA, suggesting that this element suppressed both env and src splicing. Several models are discussed which are possible mechanisms for regulation of alternative splicing of Rous sarcoma virus RNA, but none of these models appear to be consistent with all of the data.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G., Nevins J. R. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 1988 Jul;7(7):2107–2116. doi: 10.1002/j.1460-2075.1988.tb03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Arrigo S., Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988 Nov;8(11):4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beldjord C., Lapoumeroulie C., Pagnier J., Benabadji M., Krishnamoorthy R., Labie D., Bank A. A novel beta thalassemia gene with a single base mutation in the conserved polypyrimidine sequence at the 3' end of IVS 2. Nucleic Acids Res. 1988 Jun 10;16(11):4927–4935. doi: 10.1093/nar/16.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Chang L. J., Stoltzfus C. M. Cloning and nucleotide sequences of cDNAs spanning the splice junctions of Rous sarcoma virus mRNAs. J Virol. 1985 Mar;53(3):969–972. doi: 10.1128/jvi.53.3.969-972.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Feinberg M. B., Jarrett R. F., Aldovini A., Gallo R. C., Wong-Staal F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986 Sep 12;46(6):807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- Ficht T. A., Chang L. J., Stoltzfus C. M. Avian sarcoma virus gag and env gene structural protein precursors contain a common amino-terminal sequence. Proc Natl Acad Sci U S A. 1984 Jan;81(2):362–366. doi: 10.1073/pnas.81.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. D., Katz R. A., Skalka A. M., Maniatis T. The role of branchpoint and 3'-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991 Feb;5(2):211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- Glenney J. R., Jr, Glenney P. The microvillus 110K cytoskeletal protein is an integral membrane protein. Cell. 1984 Jul;37(3):743–751. doi: 10.1016/0092-8674(84)90410-0. [DOI] [PubMed] [Google Scholar]
- Hackett P. B., Varmus H. E., Bishop J. M. The genesis of Rous sarcoma virus messenger RNAs. Virology. 1981 Jul 30;112(2):714–728. doi: 10.1016/0042-6822(81)90316-0. [DOI] [PubMed] [Google Scholar]
- Hidaka M., Inoue J., Yoshida M., Seiki M. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988 Feb;7(2):519–523. doi: 10.1002/j.1460-2075.1988.tb02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L. S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol Cell Biol. 1984 Nov;4(11):2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Kotler M., Skalka A. M. cis-acting intron mutations that affect the efficiency of avian retroviral RNA splicing: implication for mechanisms of control. J Virol. 1988 Aug;62(8):2686–2695. doi: 10.1128/jvi.62.8.2686-2695.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990 Feb;10(2):696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Hauber J., Fenrick R., Cullen B. R. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature. 1988 Sep 8;335(6186):181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Sodroski J., Goh W. C., Rosen C., Dayton A., Terwilliger E., Haseltine W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature. 1986 May 22;321(6068):412–417. doi: 10.1038/321412a0. [DOI] [PubMed] [Google Scholar]
- Sompayrac L. M., Danna K. J. Efficient infection of monkey cells with DNA of simian virus 40. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7575–7578. doi: 10.1073/pnas.78.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M., Fogarty S. J. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect the accumulation of unspliced RNA. J Virol. 1989 Apr;63(4):1669–1676. doi: 10.1128/jvi.63.4.1669-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
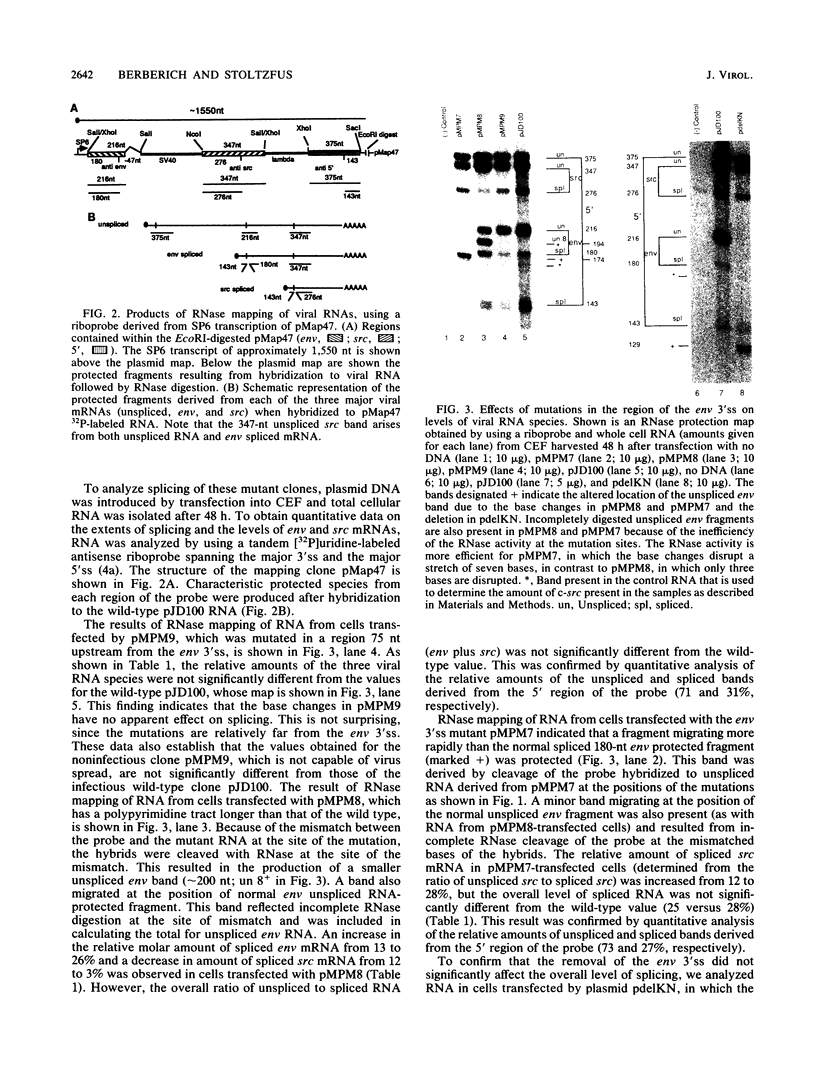
- Stoltzfus C. M., Lorenzen S. K., Berberich S. L. Noncoding region between the env and src genes of Rous sarcoma virus influences splicing efficiency at the src gene 3' splice site. J Virol. 1987 Jan;61(1):177–184. doi: 10.1128/jvi.61.1.177-184.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M. Synthesis and processing of avian sarcoma retrovirus RNA. Adv Virus Res. 1988;35:1–38. doi: 10.1016/s0065-3527(08)60707-1. [DOI] [PubMed] [Google Scholar]
- Strohman R. C., Moss P. S., Micou-Eastwood J., Spector D., Przybyla A., Paterson B. Messenger RNA for myosin polypeptides: isolation from single myogenic cell cultures. Cell. 1977 Feb;10(2):265–273. doi: 10.1016/0092-8674(77)90220-3. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., Parker R. C., Varmus H. E., Bishop J. M. Transduction of a cellular oncogene: the genesis of Rous sarcoma virus. Proc Natl Acad Sci U S A. 1983 May;80(9):2519–2523. doi: 10.1073/pnas.80.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen V. L., Spritz R. A. mRNA precursor splicing in vivo: sequence requirements determined by deletion analysis of an intervening sequence. Proc Natl Acad Sci U S A. 1985 May;82(9):2885–2889. doi: 10.1073/pnas.82.9.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]