Abstract
Pulmonary adenoma susceptibility 1 (Pas1), located on chromosome 6, is the major locus affecting inherited predisposition to lung tumor development in mice. We have fine mapped the Pas1 locus to a region of ≈0.5 megabases by using congenic strains of mice, constructed by placing the Pas1 region of chromosome 6 from A/J mice onto the genetic background of C57BL/6J mice. Systematic characterization of Pas1 candidates establishes the Las1 (lung adenoma susceptibility 1) and Kras2 (Kirsten rat sarcoma oncogene 2) genes as primary candidates for the Pas1 locus. Clearly, Kras2 affects lung tumor progression only, and Las1 is likely to affect lung tumor multiplicity.
Lung cancer is the leading cause of cancer death in men and women in the United States (1). Although lung cancer is largely induced by smoking, there is strong evidence for genetic susceptibility and gene–environment interactions in its development (2–5). However, genetic heterogeneity and enormous variation in exposure levels to environmental agents make it difficult to identify lung cancer susceptibility loci in humans. Thus, inbred mouse models offer an effective means of identifying candidate lung cancer susceptibility loci (6–14). Inbred strains of mice have different susceptibilities to spontaneous and carcinogen-induced lung tumor formation (6, 7). The A/J strain is the most susceptible to lung tumorigenesis, whereas the C3H and C57BL/6J strains are among the most resistant (6, 7). Linkage study using (A/J × C3H/HeJ)F2 and (A/J × C57BL/6J)F2 mice has demonstrated that pulmonary adenoma susceptibility 1 (Pas1) is the major lung tumor susceptibility locus in mice, which has been mapped to the distal region of chromosome 6 and accounts for ≈50% of the phenotypic variance (9, 10). Here we provide definitive evidence to support the candidacy of both Las1 (lung adenoma susceptibility 1) and Kras2 (Kirsten rat sarcoma oncogene 2) as the Pas1 genes on mouse chromosome 6. We are not aware of the identification of any other candidates for this quantitative trait locus (QTL) in either mouse models or humans since the initial mapping of the Pas1 QTL in 1993 (9).
Materials and Methods
Construction of Congenic Strains. Inbred A/J and C57BL/6J mice were purchased from The Jackson Laboratory. The basic breeding scheme in the study was to put an ≈26.1-centimorgan fragment of chromosome 6, encompassed by D6MIT54 and D6MIT373 markers from the lung tumor-susceptible A/J strain, onto the genetic background of lung tumor-resistant C57BL/6J mice. A/J mice were crossed initially to C57BL/6J mice. F1 progeny were backcrossed to C57BL/6J mice to produce the first backcross generation (N2). The N2 generation heterozygous for the chromosome region of interest was then backcrossed again to C57BL/6J mice to produce the N3 generation. This process was repeated for a total of seven backcrosses. At the N5 generation, additional microsatellite markers on chromosomes 9, 10, 17, and 19 (D9MIT75, D9MIT355, D9MIT35, D10MIT106, D10MIT2, D10MIT126, D17MIT246, D17MIT23, D17MIT50, D19MIT36, D19MIT10, and D19MIT89) were screened to obtain the optimal breeders that harbor the least amount of the non-Pas1 donor genome. At N8, 132 male substrains containing different chromosomal regions of interest were generated. These individual substrains were then each crossed to three C57BL/6J females to produce the N9 generation. After selection, an average of 5–12 N9 congenic mice were generated from each subcongenic strain.
Genotyping Using Polymorphic Markers. For selecting mice on the basis of their genotypes throughout the Pas1 region on chromosome 6, the following markers were used: D6MIT54, D6MIT52, D6MIT59, D6MIT57, D6MCO10, D6MCO11, D6MIT15, and D6MIT373. All of the mouse microsatellite primers were purchased from Research Genetics (Huntsville, AL). The forward primer was end labeled with [32P]ATP, and 30 cycles of PCR were performed at 94°C for denaturation, 55°C for annealing, and 72°C for extension. Denaturing 8% polyacrylamide gels were used for resolution of the radiolabeled PCR products, followed by autoradiography.
Lung Tumorigenesis in Congenic Mice. Five-week-old N9 mice were given a single i.p. injection of urethane (1 mg/g of body weight) in 0.2 ml of PBS. All animals were killed by CO2 asphyxiation 7.5 months after urethane initiation. A portion of lung tumors plus normal tissue was removed and flash frozen in liquid nitrogen. The remaining portion was fixed in Tellyesniczky's solution and examined with the aid of a dissecting microscope to count and size the tumors. Tumor volumes were determined by measuring the three-dimensional size of each tumor and by using the average of the three measurements as the diameter. The radius (diameter/2) was determined, and the total tumor volume was calculated by the formula volume = (4/3)πr3, where r is the radius. Two-way ANOVA was used to determine the difference in both the number and sizes of lung tumors between control and congenic groups.
Northern Blot Analysis and Semiquantitative RT-PCR. Total RNAs were prepared from mouse lung tissues by using TRIzol reagent (Life Technologies, Rockville, MD). Poly(A)+ RNAs were purified from the total RNAs with a MicroPoly(A)Pure kit (Ambion, Austin, TX). A 2-μg aliquot of each poly(A)+ RNA was separated on a 1% agarose gel containing 2% formaldehyde and transferred to nylon membrane. The blots were hybridized with a random-primed 32P-labeled cDNA probe in ExpressHyb hybridization solution (Clontech) at 68°C, washed with 0.1× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS at 50–65°C, and exposed for autoradiography at –80°C. For semi-quantitative RT-PCR, first-strand cDNAs were synthesized by using SuperScript 2 (Life Technologies) with random primer and 1 μg of poly(A)+ RNAs or 3 μg of total RNAs described above. Primer sequences were 5′-GACCAAAGCCGAGCGACTGCGGC and 3′-TCGAAGAAGTAGTTCTGTGGC (for Las1), and 5′-TGACATCCGTAAAGACCTCTATGCC and 3′-AAGCAC TTGCGGTGCACGATGGAG (for β-actin). All reactions involved initial denaturation at 94°C for 3 min, followed by 30–35 cycles at 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec (for Las1), and 21 cycles at 94°C for 30 sec, 68°C for 30 sec, and 72°C for 30 sec (for β-actin) on a PTC-100 programmable thermal controller (MJ Research, Cambridge, MA).
Colony-Formation Assay and Athymic Mouse Tumorigenicity Assay. To obtain the entire sequence of cDNAs, we performed rapid amplification of cDNA ends by using a Marathon cDNA amplification kit (Clontech). The entire Las1 cDNAs for both A/J and C57BL/6J were cloned into the pcDNA3.1(+) vector (Invitrogen) with the restriction sites for HindIII and EcoRI. Various mouse lung tumor cells were used in this experiment. LM1 is a metastatic cell line derived from A/J (15). MC4 and MC14 cells were from chemical-induced lung tumors in B6C3 F1 (15). These lines and NIH 3T3 cells (mouse fibroblast cell line) were obtained from either the American Type Culture Collection or the University of Colorado (Denver). For colony-formation assay, LM1 cells were seeded at 1.5 × 106 per 10-cm dish and transiently transfected with 4 μg of the constructed and empty vectors with Lipofectamine (Invitrogen). The transfected cells were cultured in the presence of 1 mg/ml G418 for 2 weeks. The cells that survived were fixed with 10% formalin and stained with 0.125% crystal violet. The colonies (≥1 mm) were counted. We repeated at least three independent experiments. For athymic mouse tumorigenicity, female athymic BALB/c nude mice aged 4–6 weeks were purchased from Charles River Breeding Laboratories. We injected 10 million cells s.c. into each flank of nude mice. Four animals were used per sample. We monitored the health of animals three times a week and measured the size of tumors weekly for 6 weeks. Tumor volume was calculated as described. We also confirmed the expression of Las1 for several tumors resected from nude mice by RT-PCR.
Immunocytochemistry. pcDNA3.1(+) and N-myc-tagged Las1 expression vectors were constructed from the A/J and C57BL/6J alleles, individually, to identify the localization of Las1 in cells. NIH 3T3 cells transiently transfected with pcDNA3.1(+) and N-myc-tagged Las1 were replated on multiwell chamber slides (Beckton Dickinson). The cells were then fixed with 4% paraformaldehyde in PBS and made permeable with 0.1% Triton X-100 in PBS for 3 min. Cells were covered with 3% BSA-containing blocking solution for 1 h at room temperature. The cells were then incubated with mouse anti-myc antibody (Oncogene Science; 1:50 dilution in blocking solution)for 1 h at room temperature. Anti-myc antibody was stained with goat anti-mouse secondary antibody conjugated to rhodamine (1:250 dilution) for 1 h and viewed with an Eclipse E600 microscope (Nikon).
Lung Tumorigenesis on F1 Heterozygous Kras2-Deficient Mice. Six-week-old male 129/Sv-K-ras+/– (Kras2-knockout), A/J female, and C57BL/6J female mice were paired to develop breeding colonies for production of (A/J × 129/Sv-K-ras+/–)F1 mice and (C57BL/6J × 129/Sv-K-ras+/–)F1 mice. Each F1 mouse was genotyped for the presence of the Kras2-targeted mutation and was randomized into groups according to the Kras2 genotypes and carcinogen treatments. For groups treated with urethane, animals were given a single i.p. injection of urethane (1 mg/g of body weight) in 0.2 ml of PBS. For methylnitrosourea (MNU) treatment groups, all animals were given a single i.p. injection of MNU (50 mg/kg of body weight) in 0.2 ml of PBS. Animals from all eight groups were killed by CO2 asphyxiation 18–20 weeks after treatment with carcinogens. For each mouse, portions of tumors plus normal lung tissue were frozen in liquid nitrogen. The remaining tissue and tumors were fixed in Tellyesniczky's solution overnight and then treated with 70% ethanol. The tumor number and sizes were measured.
Kras2 Activity Assays. Mouse lung tumors from Kras2+/– of A/J or C57BL/6J mice were obtained from the lung tumor bioassays. These tumors were homogenized in lysis buffer [PBS containing 5 mM MgCl2, 1 mM DTT, 0.2 mM PMSF, 2 μg/ml leupeptin, 5 μg/ml aprotinin, and 1 mM benzamidine (pH 7.5)] and cleared by centrifugation. Nonidet P-40 was then added to 1% after centrifugation. Protein concentrations were determined by Bradford assay (Bio-Rad) and equalized before incubation with 40 μg of GST–Raf Ras binding domain, precoupled to glutathione agarose. Half as much lysate was used in the pulldown from C57BL/6J tumors. After 2 h of mixing, beads were washed and bound proteins were eluted with SDS sample treatment buffer. Bound Kras2 protein was detected by Western blotting with α-KRas2 F234 antibody (Santa Cruz Biotechnology).
Statistical Analysis. To compare the tumor multiplicity and load genotype/genotype (Kras2+/–/Kras2+/+) ratios between (A/J × 129/Sv-K-ras+/–)F1 strains and (C57BL/6J × 129/Sv-K-ras+/–)F1 strains, tumor numbers and loads were log transformed, and t tests were applied by using the genotype–genotype differences of the log-transformed values. To facilitate log transformation, multiplicity values of zero were assigned a value of 0.5 and load values of zero were assigned a value of 0.001. These reassignments would produce conservative P values.
Results
Congenic Strains. We constructed congenic and subcongenic strains carrying various portions of the Pas1 locus on distal chromosome 6 to fine map the candidate region. In the Celera mouse genome map (CDS3.9), the Pas1 locus is within the D6MIT54 and D6MIT373 markers and is located at contigs GA_x54KRFPKN04 and GA_x6K02T2NTSL, respectively, which define a physical distance of ≈36 megabases (Mb). Pas1 congenic strains were constructed by placing this ≈36-Mb chromosome 6 region from the A/J (donor) mice onto the genetic background of the resistant C57BL/6J (recipient) mice by using a total of nine generations of backcrosses. Nine generations of backcross mating resulted in strains heterozygous for the locus of interest and an overall genome that is ≈99.81% C57BL/6J (16). After seven generations of backcrosses, 20 male N7 congenic mice that carried the A/J donor region (defined by markers D6MIT54 and D6MIT373) were further backcrossed to C57BL/6J females. Mice with crossovers at various segments throughout the Pas1 region were selected to develop the N8 generation. A total of 132 male congenic and subcongenic strains (N8) contained various Pas1 subregions. All mice were genotyped with eight microsatellite markers (D6MIT54, D6MIT52, D6MIT59, D6MIT57, D6MCO10, D6MCO11, D6MIT15, and D6MIT373) to define the chromosomal segments from the A/J mice onto the C57BL/6J background for each subcongenic strain.
Lung Tumorigenesis of Congenic Mice. Lung tumor bioassays were performed by using the N9 generation of congenic and subcongenic mice with urethane as the carcinogen. Lung tumor multiplicity and total tumor volume in congenic mice showed a significant difference between the Pas1 congenic strain and its control strain (Fig. 1A). An average of 6.0 tumors per mouse with a total tumor volume of 1.91 mm3 was observed in the congenic strain (AB), whereas an average of 0.7 tumor per mouse with a total tumor volume of 0.18 mm3 was found in the control mice (BB; Fig. 1A; P < 0.0001). Replacement of a C57BL/6J Pas1 allele with an A/J Pas1 allele increased lung tumor susceptibility by ≈8.6-fold in tumor number and ≈10-fold in total tumor volume. As a result, the Pas1 QTL was narrowed to an ≈0.5-Mb physical region defined by markers D6OSU6 (145.03 Mb) and D6OSU12 (145.53 Mb).
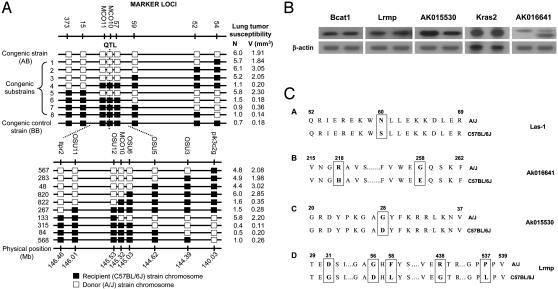
Fig. 1.
Characterizations of the Pas1 locus. (A) Substitution mapping of the Pas1 QTL for mouse lung tumor susceptibility with the use of a set of congenic strains. The open boxes represent a chromosome fragment from the donor strain (A/J), and the filled boxes represent a chromosome fragment from the recipient strain (C57BL/6J). Eight microsatellite markers (top) were used to allelotype the 26.1-centimorgan region containing the Pas1 locus. AB is a congenic strain in which the entire chromosomal region between markers D6MIT54 (54) and D6MIT373 (373) has been substituted into the recipient C57BL/6J strain from the donor A/J strain. Congenic substrains 1–8 carry various donor (A/J) fragments, as shown. BB is the control congenic strain in which no substitution was found in the entire region. (B) Expression of Pas1 candidate genes in mouse lung. Total RNAs were isolated from A/J and C57BL/6J normal lung tissues. Expression levels of five candidate genes were tested by using RT-PCR and Northern blot analysis. For each autoradiograph, Upper shows individual candidate genes and Lower shows β-actin control. For each candidate gene, the left lane shows the expression level in C57BL/6J lung, and the right lane shows the expression level in A/J lung. Bcat1, Lrmp, AK015530, and AK016641 were tested by RT-PCR. Kras2 was tested by Northern blot analysis. (C) Functional polymorphisms of Pas1 candidate genes. (1) Sequence analysis of the Las1 gene revealed a functional polymorphism at codon 60 between lung tumor susceptible/intermediate (A/J, SWR/J, BALB/cJ, 129/J, CBA/J, and SM/J; upper) and resistant strains (C57BL/6J, DBA/2J, SJL/J, C3H/HeJ, AKR/J, and Mus spretus; lower). An amino acid alignment of the codon 52–69 region of Las1 is shown, with the asparagine to serine alteration at codon 60 (boxed). (2) Sequence variations of AK016641 between the A/J and C57BL/6J strains. AK016641 contains two functional polymorphisms at codon 218 (Arg to His) and codon 258 (Gly to Glu) and an alternative splicing transcript without exon 5 (found only in the A/J strain). (3) AK015530 had a polymorphism at codon 28, resulting in a change of Gly to Asp. (4) Lrmp contains the following five functional polymorphisms: codon 31 (Asp to Gly), codon 56 (Gly to Asp), codon 58 (Phe to Leu), codon 438 (Arg to Gly), and codon 537 (Pro to Leu).
Sequence and Expression Analyses of Candidate Pas1 Genes. There are 12 putative genes, detected by a search of the National Center for Biotechnology Information (Sept. 26, 2003), within the ≈0.5-Mb Pas1 region (Table 1). Among these genes, six [Lrmp (17), Bcat1 (18), Las1 (145.25 Mb), AK015530, Kras2, and AK016641] were confirmed to be expressed in lung tissues by RT-PCR and Northern blot analyses (data not shown). Consistent with our previous study (19), Northern blot analysis showed that Kras2 4B expression level in A/J mouse lung was 36% higher than it was in the C57BL/6J strain, whereas the AK015530 expression level in A/J mouse lung was 58% lower than it was in C57BL/6J mouse lung (Fig. 1B). AK016641 contained an elevated frequency of an alternative-splicing transcript in A/J (Fig. 1B). No differential expression was found for Bcat1, Lrmp, and Las1 between lung tissues of A/J and C57BL/6J strains (Figs. 1B and 3C). Next, sequence analyses were performed to detect functional polymorphisms between susceptible and resistant strains. For Lrmp, Bcat1, Kras2, AK016641, and AK015530, the entire ORFs were sequenced. For Las1, additional 5′ and 3′ rapid amplification cDNA ends methods were used to obtain the entire ORF. The mouse Las1 sequence and its comparison with those of rat and human are shown in Fig. 2. As shown in Fig. 1C, four transcripts (AK016641, AK015530, Lrmp, and Las1) showed functional polymorphisms between the A/J and C67BL/6J strains. AK016641 had two functional polymorphisms at codon 218 (Arg to His), codon 258 (Gly to Glu), and an alternative splicing transcript without exon 5 that was found only in the A/J strain. AK015530 had a polymorphism at codon 28, resulting in a change of Gly to Asp. Lrmp had five functional polymorphisms: codon 31 (Asp to Gly), codon 56 (Gly to Asp), codon 58 (Phe to Leu), codon 438 (Arg to Gly), and codon 537 (Pro to Leu). Las1 showed one functional polymorphism at codon 60 (Asn to Ser). Therefore, five genes (AK016641, AK015530, Lrmp, Las1, and Kras2) contained either functional polymorphisms or differential expression between A/J and C57BL/6J and were further characterized as candidates for Pas1 gene(s).
Table 1. Candidate genes located in the Pas1 QTL region encompassed by markers D60SU6 and D60SU12.
| Gene symbol |
Amino acid-changing polymorphisms |
||||||
|---|---|---|---|---|---|---|---|
| Location | Gene ID | Description | Codon | A/J | C57BL/6J | Derived by | |
| 145,053,958-145,144,990 | mCG13310 | Bcatl | Branched chain aminotransferase 1, cytosolic | No* | |||
| 145,189,964-145,249,134 | mCG13301 | Lrmp | Lymphoid-restricted membrane protein | 31 | GAC | GGC | Direct sequencing |
| 56 | GGC | GAC | |||||
| 58 | TTC | TTG | |||||
| 438 | AGG | GGG | |||||
| 537 | CCG | CTG | |||||
| 145,248,888-145,257,579 | mCG13308 | Las1 | 60 | AAT | AGT | Direct sequencing | |
| 145,285,432-145,291,225 | mCG13311 | AK015530‡ | Growth hormone-inducible soluble protein | 28 | GGC | GAC | Direct sequencing |
| 145,290,331-145,324,539 | mCG13312 | Kras2 | Kirsten rat sarcoma oncogene 2 | No* | |||
| 145,340,836-145,341,323 | mCG1027072 | Similar to 40S ribosomal protein S25 | ND† | ||||
| 145,377,523-145,383,294 | mCG1027183 | ND† | |||||
| 145,388,589-145,390,646 | mCG1027184 | ND† | |||||
| 145,391,857-145,392,891 | mCG115945 | Lactate dehydrogenase pseudogene | ND† | ||||
| 145,403,203-145,415,908 | mCG1027185 | ND† | |||||
| 145,460,150-145,460,954 | mCG13304 | 60S ribosomal protein pseudogene | ND† | ||||
| 145,465,863-145,503,064 | mCG13305 | AK01664§ | Intermediate filament-related, alternatively splicing | 218 | CGC | CAC | Direct sequencing |
| 258 | GGA | GAA | |||||
No functional polymorphisms were identified in coding region.
Not determined because pseudogene, ribosomal protein, or transcripts were not detected by RT-PCR.
Corresponds to Riken cDNA 4930469P12.
Corresponds to Riken cDNA 4933403M22.
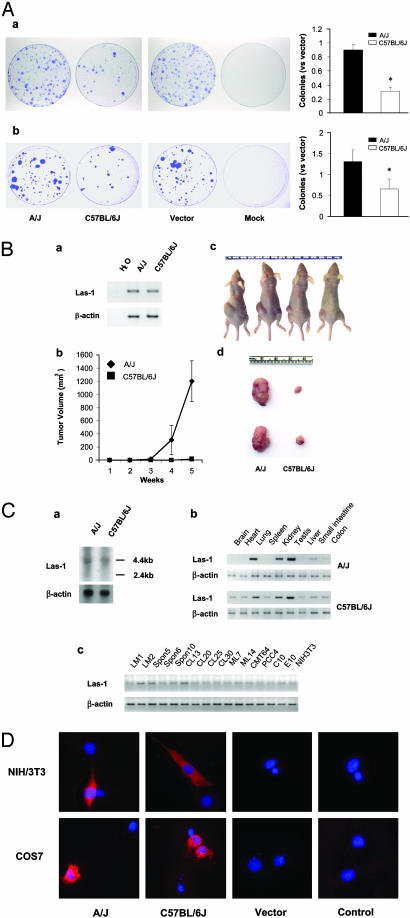
Fig. 3.
Characterization of the Las1 gene as a candidate Pas1 gene. (A) Colony-formation assay. Inhibition of colony formation by transfected Las1 in the LM1 (a) and MC14 (b) cell lines is shown. LM1 and MC14 cells of the same passage were transfected under identical conditions with 4 μg of purified plasmid DNA of Las-1-A/J pcDNA3.1, Las-1-C57BL/6J pcDNA3.1, or pcDNA3.1 vector alone. First, 1.5 × 106 cells were seeded into each of the four 10-cm dishes and incubated in appropriate medium plus G418 (50 μg/ml) for 14 days. The cells were then fixed and stained, colonies >1 mm in diameter were counted, and the proportions against empty vector (±SE) were plotted. *, P < 0.05, compared with vector-transfected cells. (B) Athymic mouse tumorigenicity assays of Las1-transfected tumor cells. (a) RT-PCR of unique sequences carried by the transfected Las1 clones. A/J, LM1 cells transfected with Las-1-A/J pcDNA3.1 clone; C57BL/6J, LM1 cells transfected with Las-1-C57BL/6J pcDNA3.1 clone. (b) Measurement of the tumor size. (c and d) Inhibition of nude mouse tumor development (left sides of mice, Las-1-A/J pcDNA3.1-transfected cells; right sides of mice, Las-1-C57BL/6J pcDNA3.1-transfected cells). (C) Expression of Las1 in mouse tissues and cell lines. (a) Northern blot analysis was used to determine the expression of Las1 in A/J and C57BL/6J lung. (b and c) Total RNA was prepared from different mouse tissues and cell lines. RT-PCR analysis was used to determine the expression of Las1 in mouse multiple organs (b) and cell lines (c). β-Actin was used as an internal control. (D) Subcellular localization of Las1. Myc-tagged Las1 plasmids were transfected into NIH 3T3 and COS7 cells and visualized by immunofluorescent staining with rhodamine (red). Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). A/J, Las-1-A/J pcDNA3.1-transfected cells; C57BL/6J, Las-1-C57BL/6J pcDNA3.1-transfected cells; Vector, pcDNA3.1 vector-transfected cells; Control, untransfected negative control cells.
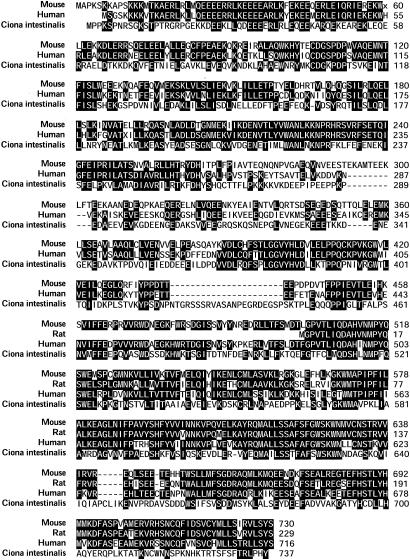
Fig. 2.
Cluster alignment of mouse, rat, human, and Ciona intestinalis Las1 protein sequences. Identical residues and residues identical in at least two species are shown with black backgrounds. In mouse protein, codon 60 (x) encodes an asparagine (AAT) in A/J mice and a serine (AGT) in C57BL/6J mice. The human protein sequence (67% identities and 81% similarities) is based on the predicted human Las1 cDNA sequence. A search of the National Center for Biotechnology Information protein database using the mouse protein sequence revealed a rat homologous protein, encoded by the National Center for Biotechnology Information-predicted gene LOC297720 (84% identities and 92% similarities). The mouse Las1 protein is homologous also to the C. intestinalis protein axonemal p83.9 (GenBank accession no. BAB88834; 33% identities and 52% similarities).
Effect of Pas1 Candidates on the Growth of Mouse Lung Tumor Cells. To evaluate further the effects of Pas1 candidates on cell proliferation, we carried out a series of transfection experiments to determine whether these candidates could promote or inhibit growth of the mouse lung tumor cell lines LM1, MC7, and MC14 (see ref. 22 for detailed descriptions of these cells). Among the five genes (AK016641, AK015530, Lrmp, Las1, and Kras2) tested, only Las1 showed differential effect on cell growth between the A/J and C57BL/6J alleles (data not shown for genes with negative results). As shown in Fig. 3Aa, transfection of Las-1-A/J pcDNA3.1 into LM1 cells produced ≈90% colonies vs. pcDNA3.1 control vector, whereas transfection of Las-1-C57BL/6J pcDNA3.1 produced ≈30% colonies vs. pcDNA3.1 control vector, indicating that Las-1-C57BL/6J pcDNA3.1 significantly inhibited anchorage-dependent cell growth (P < 0.0001; Fig. 3Aa). Similar inhibitory effects were also observed in MC7 (data not shown) and MC14 (Fig. 3Ab) cells. Clearly, Las1 derived from the C57BL/6J allele can suppress tumor cell growth in vitro. We also tested the ability of the C57BL/6J-derived Las1 to inhibit tumor development of LM1 cells in nude mice. As shown in Fig. 3B, after 5 weeks of inoculation, large tumors developed in all four mice injected with Las-1-A/J pcDNA3.1-transfected cells, but only small tumors developed in the four mice injected with transformed LM1 cells carrying Las-1-C57BL/6J pcDNA3.1 (P < 0.05; Fig. 3B b–d). Expression of Las1 was also confirmed in the resected tumors by RT-PCR (Fig. 3Ba). These results indicate that the C57BL/6J-derived Las1 can inhibit the tumorigenic potential of the LM1 cells in vivo.
Strain-Distribution Pattern and Cellular Localization of Las1. Las1 has a 2,193-bp ORF and consists of 730 amino acids; its estimated molecular mass is 84.8 kDa, and it is homologous to axonemal p83.9 (C. intestinalis, Fig. 2). Northern blotting showed that Las1 mRNA hadasizeof ≈4 kb and the same expression level between A/J and C57BL/6J (Fig. 3Ca). RT-PCR data also revealed no differential expression in a total of 12 different strains examined, comprising 6 resistant (C57BL/6J, DBA/2J, SJL/J, C3H/HeJ, AKR/J, and Mus spretus), 4 intermediate (BALB/c, 129/J, CBA/J, and SM/J), and 2 susceptible (A/J and SWR/J) strains (data not shown). In multiple-organ expression panels, Las1 was expressed at higher levels in lung, kidney, and testis in both A/J and C57BL/6J strains (Fig. 3Cb). Moreover, in 16 mouse cell lines (15), LM2, Spon5, CL-13H, and CL20 showed relatively high levels of Las1 expression, whereas some lines, such as LM1, CMT64, and PCC4, showed extremely low expression (Fig. 3Cc). There appears to be a close correlation between functional polymorphism of Las1 and lung tumor susceptibility in various mouse strains. We sequenced the entire ORF of the Las1 gene in 12 mouse strains, comprising 6 resistant, 4 intermediate, and 2 susceptible strains. The 12 strains of mice fall into two genotypes according to their sequence alteration in the 60th codon of the Las1 gene. The susceptible/intermediate groups, including six strains (A/J, SWR/J, BALB/c, 129/J, CBA/J, and SM/J), have an AAT at codon 60 encoding an asparagine, whereas the six resistant strains (C57BL/6J, DBA/2J, SJL/J, C3H/HeJ, AKR/J, and M. spretus) have an AGT at codon 60 encoding a serine (data not shown). To assess the possible function of the Las1 gene, the cellular localization of the Las1 product in NIH 3T3, COS7, and LM1 cells was investigated by transient transfection of N-terminal myc-tagged Las1 vectors derived from A/J and C57BL/6J alleles into NIH 3T3 cells. Fig. 3D shows the cytoplasmic distribution of the Las1 gene product in NIH 3T3 cells. Similar results were obtained when we transfected those plasmids into COS7 and LM1 cells (data not shown).
Looking into Las1 homology between mouse and other species, Las1 products revealed a high degree of conservation from C. intestinalis to human (Fig. 2). Las1 protein is 67% identical and 81% similar to the derived human LAS1 protein (similar to human hypothetical protein FLJ10921, 30.41 Mb). A search of the National Center for Biotechnology Information (Sept. 26, 2003) protein database using a mouse protein sequence revealed a homologous rat protein, encoded by National Center for Biotechnology Information-predicted gene LOC297720 (84% identities and 92% similarities). The mouse Las1 protein is homologous also to a C. intestinalis protein axonemal p83.9 (accession no. BAB88834; 33% identities and 52% similarities; Fig. 2). Axonemes are highly organized microtubule-based structures present in diverse types of cells that perform motile, sensory, and developmental functions in organisms from protists to humans. These functions are consistent with observed cellular distribution.
Kras2 Alleles Contribute to Differential Lung Tumor Progression. Although no functional polymorphisms were detected in the coding sequence of the Kras2 gene between A/J mice and C57BL/6J mice, we further characterized its candidacy for Pas1 because of previously observed allele-specific expression and mutation of Kras2 in mouse lung tumors from hybrid mice, such as (A/J × C67BL/6J)F1 mice or (A/J × C3H/HeJ)F1 mice, in which as much as a 20-fold higher expression of Kras2 mRNA from the A/J allele was observed (20). A mouse lung tumor bioassay was conducted by using heterozygous Kras2-deficient mice (K-ras+/–). The animals were paired to develop breeding colonies for production of (A/J × K-ras+/–)F1 and (C57BL/6J × K-ras+/–)F1 mice. Either wild-type or heterozygous Kras2-knockout and wild-type mice were subjected to lung tumorigenesis assays using two lung carcinogens: urethane and MNU. Treatment of both heterozygous (A/J × K-ras+/–)F1- and (C57BL/6J × K-ras+/–)F1-deficient mice with urethane or MNU produced 4–5 times more tumors per mouse than did treatment of wild-type mice, indicating that no allelic difference was found in promoting lung tumor multiplicity as a result of heterozygous Kras2 deficiency (Fig. 4A). In contrast, the allelic effects on tumor load (or tumor size) showed significant differences; a nearly 50-fold increase in tumor volume was observed in heterozygous (A/J × K-ras+/–)F1-deficient mice when compared with wild-type controls, whereas only ≈8-fold increase in tumor size was observed in heterozygous (C57BL/6J × K-ras+/–)F1-deficient mice when compared with wild-type controls (Fig. 4B). These results indicate that the remaining A/J or C57BL/6J Kras2 allele in K-ras+/– mice has a significant differential effect on lung tumor progression (Fig. 4B; P < 0.001). Thus, the Kras2 allele derived from A/J mouse strains confers a significantly higher susceptibility to lung tumor progression than does the C57BL/6J Kras2 allele.
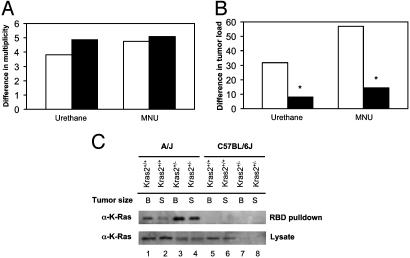
Fig. 4.
Kras2 allelic effects on chemically induced lung tumorigenesis. (A) Allelic effects on tumor multiplicity. In A/J (open bars) or C57BL/6J (filled bars) hybrid mouse groups, the remaining A/J or C57BL/6J Kras2 allele in Kras2+/– mice has no significant differential effect on lung tumor multiplicity: 3.82-fold (A/J Kras2+/– mice and A/J Kras2+/+ mice) vs. 4.88-fold (C57BL/6J Kras2+/– mice and C57BL/6J Kras2+/+ mice; urethane-treated group) or 4.76-fold vs. 5.10-fold (MNU-treated group) between the two Kras2 alleles. (B) Allelic effects on tumor volume. In A/J or C57BL/6J hybrid groups, the remaining A/J or C57BL/6J Kras2 allele in Kras2+/– mice has a significant differential effect on lung tumor progression: 31.73-fold (A/J Kras2+/– mice and A/J Kras2+/+ mice) vs. 8.04-fold (C57BL/6J Kras2+/– mice and C57BL/6J Kras2+/+ mice; urethane-treated group) or 56.85-fold vs. 14.47-fold (*, P < 0.001) between the two Kras2 alleles. (C) Activated Kras2 protein in mouse lung tumors from (A/J × K-ras+/–)F1 and (C57BL/6J × K-ras+/–)F1 mice. Kras2 activity in large and small tumors from Kras2+/+ and Kras2+/– mice was presented that corresponds to A/J and C57BL/6J alleles. B and S correspond to large (>4mm in diameter) and small (<4 mm in diameter) tumors, respectively. Note that half the amount of lysate was used in Ras-binding domain (RBD) pulldowns from A/J tumors (lanes 1–4) compared with C57BL/6J tumors (lanes 5–8).
The Activation State of Kras2 in Lung Tumors from Kras2-Deficient Mice. The mechanism for the Kras2 alleles in lung tumor progression appears to be related to the observed differential mRNA expression of the activated Kras2 alleles in lung tumors from the A/J strain of mice (20). Interestingly, there was considerably higher active Kras2 protein expressed in lung tumors from mice containing onlyanA/J Kras2 allele compared with mice with only a C57BL/6J Kras2 allele, as determined by an assay that used the Ras-binding domain of c-Raf to specifically recognize GTP-bound Kras2 (26) (Fig. 4C). In fact, we were unable to detect any active Kras2 protein in lung tumors from (C57BL/6J × K-ras+/–)F1 mice (Fig. 4C). This result was seen despite the use of half the amount of lysate in lung tumors from (A/J × K-ras+/–)F1 mice. Thus, mice containing only the C57BL/6J Kras2 allele expressed less activated Kras2 protein in a fashion that associates closely with its resistance to chemically induced lung tumorigenesis.
Discussion
The Pas1 locus has been associated with susceptibility to mouse lung tumor development after carcinogen treatment (8–14). We have demonstrated here that a semidominant A/J allele contributed to ≈8.6-fold differences in lung tumor multiplicity and ≈10-fold differences in tumor load between the AB and BB mice. High-resolution mapping using mouse congenic strains has permitted us to narrow the Pas1 QTL to ≈0.5 Mb as defined by markers D6OSU6 and D6OSU12. Further characterization of candidate genes by expression analyses, functional polymorphism searches, and colony-formation assay led us to focus on Las1 and Kras2 as primary candidates for Pas1.
The polymorphism of Las1 at codon 60 is consistent with lung tumor susceptibility in mice and the putative Pas1 allele described (6–14). Sequence analysis of Las1 and its products revealed a high degree of conservation from C. intestinalis to mammals, which suggests that they may play a fundamental role in cellular proliferation and differentiation. Clearly, mice retaining an A/J Kras2 allele in heterozygous Kras2-deficient mice are more susceptible to lung tumor progression than those retaining C57BL/6J Kras2 allele, which provides functional evidence that Kras2 is at least one of the Pas1 genes. Because Kras2 is found to affect only the size of lung tumors, it is likely that Las1 plays an important role in lung tumor multiplicity between the susceptible and resistant strains of mice. The exact relationship between Las1 and Kras2 in promoting tumor multiplicity and tumor progression requires further investigation.
This study also provides evidence that the Kras2 gene is one of the Pas1 genes responsible for lung tumor progression. Two lines of data support this concept. One is the finding that mice retaining an A/J Kras2 allele in heterozygous Kras2-deficient mice are more susceptible to lung tumor progression than those retaining a C57BL/6J Kras2 allele, providing functional evidence that Kras2 is at least one of the Pas1 genes. The other is the finding that there was higher active Kras2 protein expressed in lung tumors from mice containing only an A/J Kras2 allele compared with mice with only a C57BL/6J Kras2 allele. There was virtually no GTP-bound Kras2 protein detected in lung tumors from (C57BL/6J × K-ras+/–)F1 mice. Thus, the genetic basis for the increased lung tumor progression seen in (A/J × K-ras+/–)F1 mice is due to the higher levels of GTP-bound Kras2 protein. We believe that higher active Kras2 protein associated with the A/J Kras2 allele is due, at least in part, to the previously observed allele-specific expression and mutation of Kras2 in mouse lung tumors from hybrid mice (20). Interestingly, the differential expression of RNA or protein seems to occur more frequently during tumorigenesis, suggesting that some of the transcription factors may be made available during the tumorigenic process (20).
Identification of these two primary candidates, Las1 and Kras2, for lung cancer susceptibility is extremely timely and has an immediate application to humans. Three recent reports have demonstrated that the human PAS1 locus, located in the human 12p12 chromosome region and tightly linked to the KRAS2 gene, is involved in susceptibility to human lung adenocarcinoma (21–23). A population-based case-control study showed that the KRAS2/RsaI polymorphism is associated with risk and prognosis in both Italian and Japanese lung adenocarcinoma patients, indicating that the human PAS1 mutation may be in this locus (21). More recently, another case-control study demonstrated that the D12S1034 locus, 800–1,350 kb proximal to the KRAS2 locus, is involved in susceptibility to human lung adenocarcinoma (22). The potential role of Las1 in human lung cancer should be pursued. The significance of the present study is the finding that human Las1 and Kras2 alleles may predispose some individuals to lung cancer.
Acknowledgments
We thank Dr. William J. Lemon for statistical assistance. This work was supported by National Institutes of Health Grants R01 CA58554 (to M.Y.), R01 CA78797 (to Y.W.), and R01 GM62694 (to K.-L.G.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Mb, megabases; MNU, methylnitrosourea; QTL, quantitative trait locus.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY423542 and AY423543).
References
- 1.Minna, J. D., Roth, J. A. & Gazdar, A. F. (2002) Cancer Cell 1 49–52. [DOI] [PubMed] [Google Scholar]
- 2.Sellers, T. A., Bailey-Wilson, J. E., Elston, R. C., Wilson, A. F., Elston, G. Z., Ooi, W. L. & Rothschild, H. (1990) J. Natl. Cancer Inst. 82 1272–1279. [DOI] [PubMed] [Google Scholar]
- 3.Sellers, T. A., Potter, J. D. & Folsom, A. R. (1991) Genet. Epidemiol. 8 199–208. [DOI] [PubMed] [Google Scholar]
- 4.Sellers, T. A., Elston, R. C., Atwood, L. D. & Rothschild, H. (1992) Cancer (Philadelphia) 69 86–91. [DOI] [PubMed] [Google Scholar]
- 5.Rudier, H. W. (1991) in Ecogenetics: Genetic Predisposition to the Toxic Effects of Chemicals, ed. Grandjean, P. (Chapman & Hall, New York), pp. 205–216.
- 6.You, M. & Bergman, G. (1998) Hematol. Oncol. Clin. North Am. 12 1037–1053. [DOI] [PubMed] [Google Scholar]
- 7.Shimkin, M. B. & Stoner, G. D. (1975) Adv. Cancer Res. 21 1–58. [DOI] [PubMed] [Google Scholar]
- 8.Malkinson, A. M. & Beer, D. S. (1983) J. Natl. Cancer Inst. 70 931–936. [PubMed] [Google Scholar]
- 9.Gariboldi, M., Manenti, G., Canzian, F., Falvella, F. S., Radice, M. T., Pierotti, M. A., Della Porta, G., Binelli, G. & Dragani, T. A. (1993) Nat. Genet. 3 132–136. [DOI] [PubMed] [Google Scholar]
- 10.Festing, M. F., Yang, A. & Malkinson, A. M. (1994) Genet. Res. 64 99–106. [DOI] [PubMed] [Google Scholar]
- 11.Devereux, T. R., Wiseman, R. W., Kaplan, N., Garren, S., Foley, J. F., White, C. M., Anna, C., Watson, M. A., Patel, A., Jarchow, S., et al. (1994) Mamm. Genome 5 749–755. [DOI] [PubMed] [Google Scholar]
- 12.Manenti, G., Falvella, F. S., Gariboldi, M., Dragani, T. A. & Pierotti, M. A. (1995) Genomics 29 438–444. [DOI] [PubMed] [Google Scholar]
- 13.Lin, L., Festing, M. F., Devereux, T. R., Crist, K. A., Christiansen, S. C., Wang, Y., Yang, A., Svenson, K., Paigen, B., Malkinson, A. M. & You, M. (1998) Exp. Lung Res. 24 481–497. [DOI] [PubMed] [Google Scholar]
- 14.Festing, M. F., Lin, L., Devereux, T. R., Gao, F., Yang, A., Anna, C. H., White, C. M., Malkinson, A. M. & You, M. (1998) Genomics 53 129–136. [DOI] [PubMed] [Google Scholar]
- 15.McDoniels-Silvers, A. L., Herzog, C. R., Tyson, F. L., Malkinson, A. M. & You, M. (2001) Exp. Lung Res. 27 297–318. [DOI] [PubMed] [Google Scholar]
- 16.Markel, P. D., Bennett, B., Beeson, M., Gordon, L. & Johnson, T. E. (1997) Genome Res. 7 92–99. [DOI] [PubMed] [Google Scholar]
- 17.Behrens, T. W., Jagadeesh, J., Scherle, P., Kearns, G., Yewdell, J. & Staudt, L. M. (1994) J. Immunol. 153 682–690. [PubMed] [Google Scholar]
- 18.Benvenisty, N., Leder, A., Kuo, A. & Leder, P. (1992) Genes Dev. 6 2513–2523. [DOI] [PubMed] [Google Scholar]
- 19.Wang, Y., You, M. & Wang, Y. (2001) Exp. Lung Res. 27 255–267. [DOI] [PubMed] [Google Scholar]
- 20.Chen, B., Johanson, L., Wiest, J. S., Anderson, M. W. & You, M. (1994) Proc. Natl. Acad. Sci. USA 91 1589–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dragani, T. A., Hirohashi, S., Juji, T., Kawajiri, K., Kihara, M., Ono-Kihara, M., Manenti, G., Nomoto, T., Sugimura, H., Genka, K., et al. (2000) Cancer Res. 60 5017–5020. [PubMed] [Google Scholar]
- 22.Yanagitani, N., Kohno, T., Sunaga, N., Kunitoh, H., Tamura, T., Tsuchiya, S., Saito, R. & Yokota, J. (2002) Carcinogenesis 23 1177–1183. [DOI] [PubMed] [Google Scholar]
- 23.Manenti, G., De Gregorio, L., Pilotti, S., Falvella, F. S., Incarbone, M., Ravagnani, F., Pierotti, M. A. & Dragani, T. A. (1997) Carcinogenesis 18 1917–1920. [DOI] [PubMed] [Google Scholar]