Abstract
Aposematism is the association, in a prey organism, of the presence of a warning signal with unprofitability to predators. The origin of aposematism is puzzling, because of its predicted low probability of establishment in a population due to the prey's increased conspicuousness. Aposematism is a widespread trait in invertebrate taxa, but, in vertebrates, it is mostly evident in amphibians, reptiles, and fishes. Poison frogs (Dendrobatidae) are one of the most well known examples of the co-occurrence of warning coloration and toxicity. This monophyletic group of mostly diurnal leaf-litter Neotropical anurans has both toxic/colorful and palatable/cryptic species. Previous studies suggested a single origin of toxicity and warning coloration, dividing the family in two discrete groups of primitively cryptic and more derived aposematic frogs. Recent molecular phylogenetic analyses using mostly aposematic taxa supported this conclusion and proposed a single tandem origin of toxicity and conspicuous warning coloration. By using expanded taxon and character sampling, we reexamined the phylogenetic correlation between the origins of toxicity and warning coloration. At least four or five independent origins of aposematism have occurred within poison frogs; by using simulations, we rejected hypotheses of one, two, or three origins of aposematism (P < 0.002). We also found that diet specialization is linked with the evolution of aposematism. Specialization on prey, such as ants and termites, may have evolved independently at least two times.
Warning signals may inform a predator that the intended prey is toxic, unpalatable, or generally not worth the predator's effort. The association of unprofitability with a warning signal, such as bright or conspicuous coloration, is known as aposematism. Its evolutionary origin has posed a conundrum since the time of Wallace and Darwin (1). Although aposematism evolves as a predator deterrent, its chance of establishment in a population is predicted to be low, because it would lead to an increased probability of predation (2). Aposematism exists in many invertebrates, fishes, amphibians, snakes, and birds (3). Models proposed to explain the origin of aposematism (e.g., individual selection versus kin selection, gregariousness, greenbeard selection; refs. 4–7) treat trait evolution at only the population level. In contrast, a phylogenetic perspective can provide evidence for the likelihood of historical patterns of trait evolution (e.g., does toxicity always evolve before conspicuousness?), but this approach has rarely been examined (4, 8, 9).
Well supported and well sampled phylogenies are fundamental for comparative biology, and reliable inferences should likely be derived from them (8, 10). Ancestral character states, and their order of appearance (i.e., character mapping), can be mistakenly reconstructed if taxon sampling is not comprehensive (11). Although most analyses of aposematism perform tests for predator deterrence based on hypotheses of current utility only, aposematism can be defined within a historical context as the consecutive or prior occurrence of unpalatability, relative to conspicuousness (8). The addition of a phylogenetic framework in these tests would facilitate the identification of the sequences of evolutionary transformations in these traits.
Poison frogs (Dendrobatidae) are a well supported monophyletic group (±210 species) distributed in tropical South and Central America (12). The family includes both aposematic and cryptic species, all of which are diurnal, with Aromobates nocturnus being the one exception (13). Some species (primarily Dendrobates, Phyllobates, and Epipedobates) are brightly colored and possess toxic, lipophilic skin alkaloids (14). Some of these substances are of biomedical importance (15), and their source is probably dietary (16). Other species (e.g., Colostethus, Mannophryne, and Nephelobates) are cryptic and nontoxic, lacking lipophilic skin alkaloids, as far as is known (13, 17, 18). Based on the assumption that structurally complex biochemical compounds are difficult to evolve, the possession of alkaloids was believed to have originated once in dendrobatids (13). Phylogenetic analyses of characters other than DNA (13, 19) also proposed a single origin of toxicity. These findings were supported by an analysis of DNA sequences (20), although other smaller datasets (21) had suggested the possibility of convergence. Here we show, based on a more comprehensive taxon sample, that the association of conspicuous bright coloration and toxicity appeared not once, but several times, within poison frogs.
Aposematic species (Dendrobates, Phyllobates, and some Epipedobates) eat mostly ants, termites, and mites (22–24). Some Dendrobates exclusively eat ants or mites and reject other available prey (22). The majority of cryptic species (Colostethus) eat diverse prey (24), mostly everything available that is the right size. Natural history and ecological studies suggest that higher degrees of toxicity and toxin diversity are directly associated with a specialized diet (16), which has been assumed to have evolved only once within dendrobatids (22, 24). Here we present evidence that diet specialization has occurred more than once, and is tightly associated with the multiple origins of conspicuousness and toxicity.
Materials and Methods
Phylogeny Estimation. We sampled a broadly representative group of cryptic and aposematic dendrobatids. We sequenced 56 samples, of which six were outgroups [Bufo variegatus (Bufonidae), Centrolene grandisonae (Centrolenidae), Pseudacris regilla (Hylidae), Pseudis paradoxa (Pseudidae), Telmatobius niger, and Adenomera sp. (Leptodactylidae); for brevity, these samples are not shown on the phylogenetic trees]. The remaining 50 samples represented 38 species of dendrobatids from most species groups of Colostethus. Taxonomically, we include Minyobates within Dendrobates, and Phobobates in Epipedobates (20). These data included 2,298 characters from the entire 12S, tRNA-val, and almost all of the 16S mitochondrial genes. These data were combined with 22 dendrobatid sequences from GenBank, most of which consist of 900 bases that are completely overlapped by our data. Specimen museum numbers, collection localities, lists of primers used, and GenBank accession nos. can be seen in Tables 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org.
Preliminary alignment was done with clustalx (25). The ambiguously aligned regions were realigned under various parameters of clustalx, and were finally adjusted by eye to produce a parsimonious alignment; thus, informative sites were minimized. Hypothesized secondary structure diagrams from the Comparative RNA web site (www.rna.icmb.utexas.edu) were consulted to optimize alignments. We analyzed four configurations of the data under parsimony by using paup* (26); a combination of both datasets; (i) including and (ii) excluding regions of doubtful alignment (248 base pairs); (iii) a reduced-character dataset with only the GenBank regions common to all taxa (900 base pairs); and (iv) a reduced-taxon dataset using only our sequences. All shorter GenBank sequences were omitted, as were as the unalignable characters. Likelihood and Bayesian analyses (27) were performed by using configuration ii. By using a hierarchical test of models (28) the best fitting model was found to be GTR+G+I, and parameters for this model were estimated during the likelihood search. In the Bayesian analysis, model parameters were also estimated during the run with starting default values of the 10 Markov chains with 0.2 as the value of the exponential prior. Four independent mrbayes runs were executed; each used a random starting tree for 1 million generations and sampled every 10 generations, resulting in 100,000 sampled trees. We determined whether the Markov chains had reached stationarity by examining plots of likelihood scores of sampled trees against generation time. Log-likelihood scores for the sampled trees stabilized after 60,000 generations. The first 10,000 trees sampled were discarded, and the posterior probabilities were estimated with the remaining trees. The posterior probability of each bipartition was calculated by using a majorityrule consensus tree of the retained trees. A clade was considered significantly supported if its posterior probability was ≥95%. All four Bayesian runs produced concordant results. Parsimony and likelihood analyses yielded almost identical trees.
Hypothesis Testing. For testing hypotheses of multiple origins of aposematism, we compared the optimal tree (alternative hypothesis) to trees constrained to represent null hypotheses of one, two, three, and four origins of conspicuous coloration. Sequence evolution parameters were estimated by using maximum likelihood under the GTR+G+I model. We used parametric bootstrapping procedures (29) to evaluate 500 simulated datasets generated by using seq-gen 1.2.5 (30) for each test. Because the GenBank sequences were shorter than our sequences (i.e., 900 vs. 2,298 base pairs), only the regions in common (configuration iii above) were used. Thus, the bootstrapping tests are more conservative; with fewer data, the null hypothesis is more difficult to reject (31)
Phylogenetic analysis with no topological constraints indicated five origins of bright coloration. This tree was compared with other topologies that were constrained to have various number of origins (null hypotheses). For the hypothesis of a single origin, all brightly colored species were constrained to be an exclusive clade. For the hypothesis of two origins, all possible constraints using pairwise combinations of the five brightly colored clades were evaluated, and the shortest resulting tree (i.e., the one least likely to reject the null hypothesis) was used (one constraint for Dendrobates plus Phyllobates and a second for all other brightly colored species). The constraints for three and four origins were determined in a similar fashion by using combinations of three and four clades.
To infer the presence of aposematism, a correlation between conspicuous coloration and toxicity was tested by using the concentrated changes test in macclade 4.0 (32). The reconstructed changes of the two traits (conspicuous coloration and presence of toxins) on the optimal tree were compared with a null distribution from 10,000 simulated datasets. Because there is some disagreement as to whether certain species are “conspicuously” colored, we repeated the correlated changes tests under three criteria: (i) the presence of obvious bright coloration on the exposed surfaces; (ii) a published table of coloration scores (33) for 21 species, with interpolation of additional species based on our field experience with these animals in life, by using field notes and photographs; and (iii) a traditional taxonomic definition, in which species of Dendrobates, Phyllobates, Allobates, Cryptophyllobates, and Epipedobates are scored as conspicuous, and species of Colostethus are scored as cryptic (see Table 1). The different criteria affected three species (Allobates femoralis, Epipedobates boulengeri, and Epipedobates sp. F), but the results of the correlation tests were the same regardless of which definition of conspicuous coloration was used.
Results and Discussion
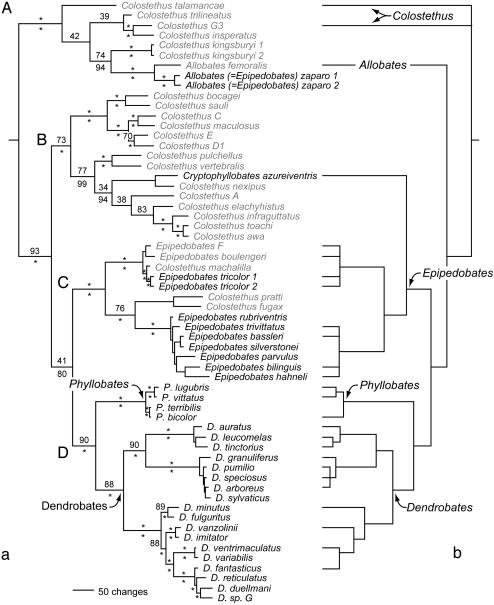
The results of parsimony, likelihood, and Bayesian analyses were highly concordant, and four well supported clades were identified (Fig. 1a, clades A–D), each with conspicuously colored species. On the parsimony topology (Fig. 1), the most-parsimonious reconstruction shows five independent origins of warning coloration (species names are bold). A less-parsimonious reconstruction of six changes requires three origins and three losses. On the likelihood and Bayesian topologies (Fig. 2) the reconstruction also requires five origins (shown as gray boxes); a less-parsimonious reconstruction under the assumption of no losses requires seven origins.
Fig. 1.
(a) Phylogeny of poison frogs. Species names in black refer to conspicuous and (as far as is known) toxic species. Names in gray refer to cryptic and nontoxic species. The tree shown is based on the parsimony analysis, but the likelihood tree (Fig. 2) is almost identical. The sister-group relationship of clades C and D was recovered in all three types of analyses. In the parsimony analysis, the sister-group relationship of clades B and C was equally supported, but this topology was not the best estimate under likelihood or Bayesian analysis. Neither alternative (clade C + D vs. clade B + C) is strongly supported by bootstrap proportions or Bayesian posterior probabilities. Parsimony bootstrap proportions are above each branch, and Bayesian posterior probabilities are below. *, a value of ≥95. (b) Previous molecular phylogeny of poison frogs (33). The difference between a and b is due to the degree of taxon sampling.
Fig. 2.
The likelihood topology, with gray boxes representing the species names shown in black on Fig. 1. The column of photos on the left shows representative cryptic and nontoxic species, and the column on the right shows conspicuous and toxic species (the toxicity of A. zaparo is unknown). The ant icons indicate two origins of specialized diet, and a possible third origin is indicated by a question mark.
With the exception of Cryptophyllobates azureiventris, for which no data on toxins are available, each group of conspicuously colored species includes species with toxic skin alkaloids (see Table 1 and ref. 14). The correlated changes test indicates that the independent appearances of conspicuous coloration and skin alkaloids are significantly correlated (P < 0.001), even though data on toxins are missing for many of the cryptic species. Thus, we conclude that aposematism (i.e., an association between defense and warning coloration) evolved de novo four (and probably five) times; this result is independent of the three criteria used to characterize species as conspicuous.
By using parametric bootstrapping, we tested whether the tree requiring five origins of aposematism (null hypothesis) could be distinguished from a tree with fewer origins. The null hypothesis was rejected at P < 0.002 for each hypothesis of one, two, or three origins. In each case, the observed tree-length difference was >10 steps longer than the greatest difference from the simulated data. We could not reject the four origins of aposematism (P = 0.782), indicating that the two observed origins of aposematism in the Epipedobates group within clade C (Fig. 1) cannot be statistically distinguished from one origin. However, the reduced dataset of 900 base pairs used for the simulations diminishes the power of these tests. More extensive analyses, including more Epipedobates species and more complete sequences, would be needed to distinguish between four or five independent origins of aposematism within the poison frogs.
The hypothesis of multiple origins of aposematism is more robust than previous hypotheses, because we sampled a broad array of cryptically colored species from throughout the taxonomic diversity of the family. If only aposematic species are analyzed, almost no differences exist between our tree and that of a previous study (20) (compare Fig. 1 a and b). Also, the current taxonomy is misleading, with respect to evolutionary relationships, because it relied on bright coloration as a diagnostic character. Colostethus, a group of inconspicuous species, is paraphyletic with respect to Allobates, Cryptophyllobates, and Epipedobates, with the latter being a polyphyletic assemblage of independent aposematic lineages. Previously, C. azureiventris (34) was considered to be an Epipedobates. The extensive polyphyly of Epipedobates can be reduced by transferring Epipedobates zaparo (clade A) to Allobates; thus, the name is Allobates zaparo (new combination). However, Colostethus remains grossly paraphyletic; reducing the degree of polyphyly of Epipedobates by recognizing Allobates and Cryptophyllobates is but a transitional solution toward a classification of dendrobatids based solely on monophyletic taxa. Notwithstanding these taxonomic changes, the remaining Epipedobates species in clade C are not monophyletic. Resolution of this clade is crucial for the Linnean taxonomy of dendrobatids, because it contains the type species of Epipedobates (tricolor) and may also contain the type species of Colostethus (latinasus), based on morphological similarity (17). Given the current lack of molecular data on the position of Colostethus latinasus, a wholesale revision of poison frog taxonomy is beyond the scope of this paper.
A striking feature of the multiple origins is that they occur on different time scales, indicating recurring origins through evolutionary history. Aposematism had a single ancient origin at the base of clade D (Dendrobates plus Phyllobates) and was not lost in any descendants in this clade. Alkaloid data are available for most species in this clade, and all are toxic. The other origins of aposematism are much more recent. For example, sequence divergence (uncorrected p-distance) between the cryptically colored Colostethus machalilla and its toxic, bright red sister species Epipedobates tricolor (Fig. 2) is a mere 0.9–1.0%. The latter species is the natural source of epibatidine, an alkaloid that is an opioid analgesic (18). In contrast, divergence in the same genes is 1.7–5.7% among five well differentiated and strikingly variable and brightly colored species of the Dendrobates pumilio group (Fig. 1). Such low genetic divergence is not necessarily inconsistent with extreme color divergence, because color pattern in some model organisms is controlled by very few genes (35). Nonetheless, the low genetic divergence suggests the microevolutionary lability of defensive signals and toxicity in these frogs.
A significant correlation between evolutionary change in degree of conspicuous coloration and degree of toxicity in poison frogs was demonstrated (33) under the hypothesis of a single origin of aposematism, and this correlation holds under a hypothesis of multiple origins. However, we found no significant correlation between the appearance of each trait under the single-origin hypothesis (correlated changes test; P = 0.975). However, as stated above, the independent appearances of conspicuous coloration and toxicity under the multiple-origins hypothesis are significantly correlated (P < 0.001). A correlation between coloration and toxicity is not surprising when one considers distantly related groups such as coral snakes, monarch butterflies, and nudibranch molluscs. However, the presence of this correlation within a group of closely related species invites a consideration of possible mechanisms for the origin of aposematism.
Accumulating evidence suggests that poison frogs sequester toxins from their diet. Most brightly colored and toxic dendrobatids (Dendrobates, Phyllobates, and Epipedobates) are dietary specialists on ants, termites, or mites. Thus, they eat many and smaller prey, whereas the cryptic and nontoxic species (Colostethus) have generalized diets of few and larger prey (22, 24). Specialized phenotypic traits in diet-specialized dendrobatids (e.g., narrow head and tongue) are recognized as adaptations for foraging on tiny prey (36). The aposematic species have probably evolved narrow diets to maximize the accumulation of toxins from diet (16). Experiments have shown that at least two poison frog species derive alkaloids from ants, and that Dendrobates auratus can sequester certain alkaloids (allopumiliotoxins and izidines) from food supplements (37). When raised on fruit flies, captive Phyllobates, Dendrobates, and Epipedobates tricolor lose much of their toxicity (38, 39). Nonetheless, despite the demonstration of alkaloid sequestration, the source of the most biologically active alkaloids; i.e., batrachotoxins, histrionicotoxins, epibatidine, and others, remains unknown (18).
Based on the available information, our phylogeny demonstrates at least two, and perhaps three, independent origins of dietary specialization (Fig. 2). One origin of diet specialization is in the ancestor of clade D (Phyllobates and Dendrobates), in which all species are ant, termite, or mite specialists (24). A second origin is within clade C (Epipedobates and Colostethus), in which some species have generalist diets (including the brightly colored E. tricolor), but E. parvulus and its relatives are ant specialists (24). A possible third origin is in clade A, in which most species, including A. femoralis, have generalist diets. However, the limited data (40) indicate that A. zaparo, its brightly colored sister species (Fig. 1), eats mostly ants. Although there is a clear phylogenetic correlation between bright coloration and toxicity (as demonstrated by the correlated changes test), data sufficient to test the association of diet are lacking for most species.
Under the single-origin hypothesis, dietary specialization and foraging ecology were predicted to be key evolutionary factors in the diversification of poison frogs (22, 24, 36). Under this multiple-origins hypothesis, the evolutionary association between diet and aposematism may be more complex. Although one large clade (D) displays an ancient origin of aposematism, most of the origins are relatively recent and involve one or a few species, suggesting that this homoplasy is dynamic and recurring. The specialization on different prey types (ants, termites, or mites), which may explain the great diversity of alkaloids, suggests selection for specialization per se (41), rather than commitment to a particular food resource. For example, E. tricolor is toxic but is not specialized on ants or termites (L.A.C., unpublished data), which suggests that this species might sequester toxins from unrecognized sources, such as larger-prey items.
The association of dietary specialization and sequestration of toxic defensive compounds in aposematic organisms is not novel to frogs. For example, two unrelated lineages of aposematic papilionid butterflies sequester aristocholic acid compounds from pipevines (Aristolochiaceae) (42). Longitarsus leaf beetles (Chrysomelidae) have evolved the sequestration of pyrrolizidine alkaloids multiple times (43). However, dendrobatid frogs are unique among vertebrates in their recurring associations of coloration, toxicity, and diet specialization. This observation suggests an as-yet-unidentified physiological mechanism in the ancestor of poison frogs that allowed sequestration of toxic compounds.
Fragmentary but exciting evidence suggests other behavioral traits that may be associated with aposematism and dietary specialization. In contrast to almost all other frogs, both cryptic and aposematic dendrobatids are diurnal, rather than nocturnal, with the apparent exception of A. nocturnus (13). This change to a diurnal habit, in which visual signals would be favored, may have facilitated repeated adaptive shifts toward novel foraging ecology, dietary specialization, toxicity, and bright coloration. Also, high aerobic and low anaerobic metabolic capacity have been found in the few aposematic dendrobatid species studied (44). In contrast, cryptic species of other leaf-litter frogs (Eleutherodactylus) that co-occur with these dendrobatids have low aerobic and high anaerobic capacity (44), and are not dietary specialists on ants (24). This physiological trait in some dendrobatids probably favored a recurring association of these traits. More data about metabolic rate in dendrobatids are needed within this phylogenetic framework.
Ecological specialization is a widespread evolutionary outcome in many animal systems (41). It is commonly stated that a specialization should derive from a generalized, plesiomorphic trait. This finding appears to be true in the case at hand; the traits of conspicuousness, unpalatability, and narrow diet are derived from crypticity, palatability, and a generalized diet, respectively. In the phylogeny presented here, these derived traits are not statistically independent, and probably reinforce each other, promoting evolutionary specialization. The appearance of toxicity may generally precede the appearance of diet specialization and warning coloration. Evidence for this possibility comes from three cases. First, A. nocturnus, the putative sister species of all dendrobatids, has a noxious, mercaptan-like odor, despite the lack of alkaloids (13) and its cryptic coloration. Second, although the few Colostethus sampled for lipophilic alkaloids show no traces (14), Colostethus inguinalis has tetrodotoxin (45), a water-soluble (rather than lipophilic) toxin otherwise unknown in dendrobatids. These two cases suggest parallel but isolated origins of defense in obviously cryptic species. Third, certain species are not brightly colored, but have either flash coloration or contrasting patterns on concealed surfaces; these species also have some degree of alkaloid toxicity (A. femoralis, E. boulengeri, and its sister species, E. sp. F), and are closely related to brightly colored species. These species may represent microevolutionary cases of dynamic intermediate conditions between the crypticpalatable and conspicuous-toxic extremes. However, the data are meager and much work at the population level is needed to confirm this hypothesis.
At the other extreme, all conspicuous species (Dendrobates, Phyllobates, and most Epipedobates) surveyed possess diverse and often abundant toxins (14); i.e., no Batesian mimics are known. Furthermore, although the degree of diet specialization among the toxic species varies, the diets of the least specialized toxic species are still narrower than those of the cryptic species (22, 24).
In summary, a more comprehensive phylogeny reveals the multiple appearances of this complex of traits (visual signals, toxicity, narrow diet, and, perhaps, higher metabolic rate), which suggests parallel and correlated evolutionary trends toward specialization. These multiple occurrences may indicate directional selection for the acquisition of toxins from dietary components, which likely led to aposematic coloration and feeding specializations.
Supplementary Material
Acknowledgments
We thank César Paz y Miño and Paola Leone of the Human Molecular Genetics Lab of the Pontificia Universidad Católica del Ecuador, who provided laboratory facilities and advice to J.C.S.; David Kizirian of the Los Angeles County Museum for support of field work; Ulrich Müller for the use of the automated sequencer; Alisha Holloway for laboratory assistance; Mike Ryan, Ulrich Müller, Rafe Brown, Cat Darst, Beckie Symula, Nicole Gerardo, Ben Evans, Mike Singer, Ted Townsend, Larry Gilbert, and Jim Bull for comments on the manuscript; and other members of the Cannatella–Bull–Hillis laboratories for discussion. Comments from one reviewer were particularly helpful in clarifying points of the manuscript. This work was supported by the Pontificia Universidad Católica del Ecuador Research Fund, National Science Foundation Grant 9981631 (to D.C.C.), and a National Science Foundation Integrative Graduate Education and Research Traineeship training grant.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY364538–AY364580).
See commentary on page 12533.
References
- 1.Mallet, J. & Joron, M. (1999) Annu. Rev. Ecol. Syst. 30 201–233. [Google Scholar]
- 2.Yachi, S. & Higashi, M. (1998) Nature 394 882–884. [Google Scholar]
- 3.Komárek, S. (1998) Mimicry, Aposematism, and Related Phenomena in Animals and Plants. Bibliography 1800–1990 (Vesmir, Prague).
- 4.Guilford, T. (1988) Am. Nat. 131 7–21. [Google Scholar]
- 5.Lindström, L., Alatalo, R. V., Mappes, J., Riipi, M. & Vertainen, L. (1999) Nature 397 249–251. [Google Scholar]
- 6.Servedio, M. R. (2000) Evolution (Lawrence, Kans.) 54 751–763. [DOI] [PubMed] [Google Scholar]
- 7.Fisher, R. A. (1958) The Genetical Theory of Natural Selection (Dover, New York).
- 8.Härlin, C. & Härlin, M. (2003) Evol. Ecol. 17 197–212. [Google Scholar]
- 9.Sillén-Tullberg, B. (1988) Evolution (Lawrence, Kans.) 42 293–305. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. (1985) Am. Nat. 125 1–15. [Google Scholar]
- 11.Pollock, D. D., Zwickl, D. J., McGuire, J. A. & Hillis, D. M. (2002) Syst. Biol. 51 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford, L. S. & Cannatella, D. C. (1993) Herpetol. Monogr. 7 94–117. [Google Scholar]
- 13.Myers, C. W., Paolillo, A. & Daly, J. W. (1991) Am. Mus. Novit. 3002 1–20. [Google Scholar]
- 14.Daly, J. W., Garraffo, H. M. & Spande, T. F. (1999) in Alkaloids: Chemical and Biological Perspectives, ed. Pelletier, S. W. (Elsevier, London), Vol. 13, pp. 1–153. [Google Scholar]
- 15.Daly, J. W., Garraffo, H. M., Spande, T. F., Decker, M. W., Sullivan, J. P. & Williams, M. (2000) Nat. Prod. Rep. 17 131–135. [DOI] [PubMed] [Google Scholar]
- 16.Daly, J. W., Kaneko, T., Wilham, J., Garraffo, H. M., Spande, T. F., Espinosa, A. & Donnelly, M. A. (2002) Proc. Natl. Acad. Sci. USA 99 13996–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coloma, L. A. (1995) Misc. Publ. Mus. Nat. Hist. Univ. Kansas 87 1–72. [Google Scholar]
- 18.Daly, J. W. (1998) J. Nat. Prod. 61 162–172. [DOI] [PubMed] [Google Scholar]
- 19.Myers, C. W., Daly, J. W., Garraffo, H. M., Wisnieski, A. & Cover, J. (1995) Am. Mus. Novit. 3144 1–21. [Google Scholar]
- 20.Clough, M. E. & Summers, K. (2000) Biol. J. Linn. Soc. 70 515–540. [Google Scholar]
- 21.Vences, M., Kosuch, J., Lötters, S., Widmer, A., Jungfer, K. H., Kohler, J. & Veith, M. (2000) Mol. Phylogenet. Evol. 15 34–40. [DOI] [PubMed] [Google Scholar]
- 22.Toft, C. A. (1995) Herpetologica 51 202–216. [Google Scholar]
- 23.Toft, C. A. (1981) J. Herpetol. 15 139–144. [Google Scholar]
- 24.Caldwell, J. P. (1996) J. Zool. (London) 240 75–101. [Google Scholar]
- 25.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swofford, D. L. (2002) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 27.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17 754–755. [DOI] [PubMed] [Google Scholar]
- 28.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14 817–818. [DOI] [PubMed] [Google Scholar]
- 29.Swofford, D. L., Olsen, G. J., Waddell, P. J. & Hillis, D. M. (1996) in Molecular Systematics, eds. Hillis, D. M., Moritz, C. & Mable, B. K. (Sinauer, Sunderland, MA), pp. 407–514.
- 30.Rambaut, A. & Grassly, N. C. (1997) Comput. Appl. Biosci. 13 235–238. [DOI] [PubMed] [Google Scholar]
- 31.Huelsenbeck, J. P., Hillis, D. M. & Jones, J. (1996) in Molecular Zoology: Advances, Strategies, and Protocols, eds. Ferraris, J. D. & Palumbi, S. R. (Wiley, New York), pp. 19–45.
- 32.Maddison, D. R. & Maddison, W. P. (2001) macclade (Sinauer, Sunderland, MA).
- 33.Summers, K. & Clough, M. E. (2001) Proc. Natl. Acad. Sci. USA 98 6227–6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lötters, S., Jungfer, K. H. & Widmer, A. (2000) Jahresh. Ges. Naturkd. Württemb. 156 233–243. [Google Scholar]
- 35.Koch, P. B., Behnecke, B. & ffrench-Constant, R. H. (2000) Curr. Biol. 10 591–594. [DOI] [PubMed] [Google Scholar]
- 36.Vences, M., Glaw, F. & Böhme, W. (1998) Zool. Anz. 236 217–230. [Google Scholar]
- 37.Daly, J. W., Secunda, S., Garraffo, H. M., Spande, T. F., Wisnieski, A. & Cover, J. F., Jr. (1994) Toxicon 32 657–663. [DOI] [PubMed] [Google Scholar]
- 38.Daly, J. W., Garraffo, H. M., Jain, P., Spande, T. F., Snelling, R. R., Jaramillo, C. & Rand, A. S. (2000) J. Chem. Ecol. 26 73–85. [Google Scholar]
- 39.Daly, J. W., Secunda, S. I., Garraffo, H. M., Spande, T. F., Wisnieski, A., Nishihira, C. & Cover, J. F., Jr. (1992) Toxicon 30 887–898. [DOI] [PubMed] [Google Scholar]
- 40.Almendáriz, A. (1987) Revista Politécnica, Quito 12 144–177. [Google Scholar]
- 41.Futuyma, D. J. & Moreno, G. (1988) Annu. Rev. Ecol. Syst. 19 207–233. [Google Scholar]
- 42.Nishida, R. (2002) Annu. Rev. Entomol. 47 57–92. [DOI] [PubMed] [Google Scholar]
- 43.Dober, S. (2001) Basic Appl. Ecol. 2 15–26. [Google Scholar]
- 44.Taigen, T. & Pough, F. H. (1985) Am. Zool. 25 987–997. [Google Scholar]
- 45.Daly, J. W., Gusovsky, F., Myers, C. W., Yotsu-Yamashita, M. & Yasumoto, T. (1994) Toxicon 32 279–285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.