Abstract
Notch receptors and the amyloid precursor protein are type I membrane proteins that are proteolytically cleaved within their transmembrane domains by a presenilin (PS)-dependent γ-secretase activity. In both proteins, two peptide bonds are hydrolyzed: one near the inner leaflet and the other in the middle of the transmembrane domain. Under saturating conditions the substrates compete with each other for proteolysis, but not for binding to PS. At least some Alzheimer's disease-causing PS mutations reside in proteins possessing low catalytic activity. We demonstrate (i) that differentially tagged PS molecules coimmunoprecipitate, and (ii) that PS N-terminal fragment dimers exist by using a photoaffinity probe based on a transition state analog γ-secretase inhibitor. We propose that γ-secretase contains a PS dimer in its catalytic core, that binding of substrate is at a site separate from the active site, and that substrate is cleaved at the interface of two PS molecules.
Mutations in the amyloid precursor protein (APP), presenilin (PS) 1, and PS2 genes comprise the known causes of autosomal dominant familial Alzheimer's disease (FAD; www.alzforum.org). All of these mutations affect the metabolism of APP, a type I transmembrane (TM) protein and precursor of the Aβ peptides that aggregate in senile plaques characteristic of Alzheimer's disease (1, 2). The APP ectodomain is released after either BACE (β-secretase)-mediated or TACE (α-secretase)-mediated cleavage to generate two membrane-associated C-terminal fragments (CTFs) (C99 and C83, respectively). C99 and C83 are then substrates for γ-secretase, an enzymatic activity(s) residing in a multiprotein complex that generates Aβ (from C99) and p3 (from C83). Recent discoveries present strong evidence that the catalytic center of γ-secretase resides within a complex of two PS fragments [N-terminal fragment (NTF) and CTF] generated after cleavage within the cytoplasmic loop (3–5). Proteolysis by γ-secretase releases Aβ peptides that are predominantly 40 aa long (Aβ40). Shorter and longer species of 38–43 aa (6) are also released, but at a lower frequency (7). Of these, the longer amyloidogenic Aβ42/43 fragments are thought to be central to the disease process. γ-Secretase also releases the APP intracellular domain (AICD) by cleaving near the inner leaflet of the TM domain (2). AICD is rapidly degraded, but a small amount has been shown to be translocated to the nucleus (8–11).
Another substrate for γ-secretase is the family of Notch receptors (1, 2, 12), which directly relay signals from the cell surface to the nucleus via regulated intramembrane proteolysis triggered by type I TM ligands. Upon ligand binding, Notch undergoes an ectodomain-shedding extracellular cleavage. The C-terminal product of this event is an intermediate that undergoes further proteolysis within the TM domain to release the Notch intracellular domain (NICD). NICD release and translocation to the nucleus are essential for signal transduction in vitro (12) and in vivo (13, 14). As in APP, an additional peptide bond is hydrolyzed within the middle of the Notch TM domain, releasing an NTF (Nβ) (15).
Analyses of Aβ or Nβ fragments identify a variety of γ-secretase cleavage sites located roughly at the middle of the TM domain (16–20). NICD and AICD formation is strictly associated with cleavage near the inner leaflet; the P1′ position of the cleavage site (S3 in Notch, ε in APP) is located three to five amino acids N terminal to the stop translocation signal (21–24). This distribution of sites is hard to reconcile with a single enzyme given that both substrate and enzyme are likely constrained within the lipid bilayer. Furthermore, some FAD mutations in PS behave as partial loss-of-function mutations in relation to NICD and AICD release (25–27) but as gain-of-function mutations in relation to Aβ42 cleavage. The distribution of missense FAD mutations is more consistent with many FAD mutations being loss-of-function alleles (28). However, because nonsense mutations have not been associated with FAD, missense mutations may share some unique properties, including perhaps partial activity. Several models can be postulated to explain this conundrum. First, it has been suggested that Notch and APP may be cleaved by different protein assemblies (29–31). Second, the proximity of S3 and ε sites to the cytoplasmic face of the membrane may imply that a soluble enzyme associated with PS mediates this cleavage, whereas the true “γ-secretase” cleaves within the membrane (32). Third, a single multimeric enzyme may mediate cleavage at both sites in both substrates.
Materials and Methods
Cell Lines and Plasmids. PS1/PS2-deficient mouse embryonic fibroblasts (MEFs) and controls have been described (33). MEFs and human embryonic kidney (HEK) 293 cells were grown at 37°C and 5% CO2 in DMEM supplemented with 10% FBS. MEFs were transiently transfected by using FuGene (Roche Diagnostics) transfection reagent, and transient transfections of HEK293 cells were performed as described (21, 34, 35). Harvesting and analysis of transfected cells were generally performed 40–48 h after transfection. The plasmid pCS2+Notch ΔE (M1726V), referred to henceforth as NΔE, and NLNR were as described (34). NΔE produces a truncated Notch protein that allows analysis of S3 cleavage independent of ligand regulation and upstream cleavage events. pCS2+C99 contains the C-terminal 100 aa of APP fused to its leader peptide and inserted into the pCS2+ vector. The C99–6MT vector was made by cloning C99 in-frame with a 3′ 6-Myc tag sequence in the pCS2+MT vector. All Notch constructs also contained a 3′ 6-Myc tag, which facilitates protein detection and immunoprecipitation (IP) with the 9E10 antibody. The APP expression vector was a gift from T. Golde (Mayo Clinic, Jacksonville, FL). FAD mutant PS constructs were made by site-directed mutagenesis. All plasmids were sequenced to confirm that only the desired mutations were introduced during cloning.
Inhibitor Assays. Dishes (100 mm) of HEK293 cells were transfected with 10 μg of either NΔE or pCS2+C99. Twenty four hours after transfection multiple dishes were trypsinized, and the cells were pooled and plated onto multiwell dishes for analysis. Inhibitors were solubilized in DMSO, diluted into medium, and then used for both the Notch- and C99-expressing cells.
To analyze the effects of inhibitors on Notch cleavage, cells transfected with NΔE were starved for 1 h in Met-free media and labeled with 35S-Met (40 μCi/ml, Amersham Pharmacia Redivue) for 5 h in the presence or absence of inhibitor. Cells were then lysed and immunoprecipitated as described (34). 35S-labeled samples were visualized with a Molecular Dynamics PhosphorImager. IMAGEQUANT software was used to quantify both uncleaved NΔE and NICD. Results are expressed either as percentage of NICD (NICD/NΔE + NICD) or normalized to an untreated control sample.
To determine Aβ40 levels, fresh medium containing the inhibitors or DMSO was added, and the medium was collected after 5 h of treatment. Aβ40 levels were determined by ELISA (BioSource International, Camarillo, CA). Values for each concentration of inhibitor were determined relative to the untreated (DMSO) control. At least four data points were generated for each concentration of inhibitor. IC50 values were calculated with PRIZM graphing software (GraphPad, San Diego) by using the nonlinear regression analysis algorithm.
Notch-APP Competition. Dishes (60 mm) of HEK293 cells were transfected with 2 μg of pCS2+C99 and 2 μg of Notch-expressing plasmid or empty pCS2+ vector and 1 μg of pSEAP2 to normalize for transfection efficiency. Forty eight hours after transfection fresh medium was added to the plates. Aβ and secreted alkaline phosphatase (SEAP) were allowed to accumulate overnight after which both media and cells were harvested for assay. Aβ levels were assayed by ELISA. SEAP levels in the medium were determined by using the Phospholight luminescence assay (Tropix, Bedford, MA). The ratio of (NICD/NΔE + NICD) was determined by quantitative Western analyses (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org) or 35S labeling as above. The ratio of (NICD/NΔE + NICD) in three independent experiments was determined; the average ratio and the standard error are shown.
IPs and Antibodies. For co-IPs, cells were lysed in co-IP lysis buffer (0.5% Triton X-100/1% Nonidet P-40/150 mM NaCl/2 mM EDTA/50 mM Tris, pH 7.6) as described (36). PS proteins were separated by SDS/PAGE and transferred to nitrocellulose membranes (Amersham Pharmacia) and immunoblotted. Notch was detected with monoclonal 9E10 α-Myc antibody. 9N14 is a polyclonal antibody raised against a peptide antigen composed of residues 1–14 of human PS1. Other antibodies against PS1 have been described: NT1 (monoclonal, provided by P. Mathews, New York University School of Medicine, Orangeburg) (38), 13A11 (monoclonal, provided by P. Seubert, Elan Pharmaceuticals, San Francisco) (37), and R22 (polyclonal) (36).
Results
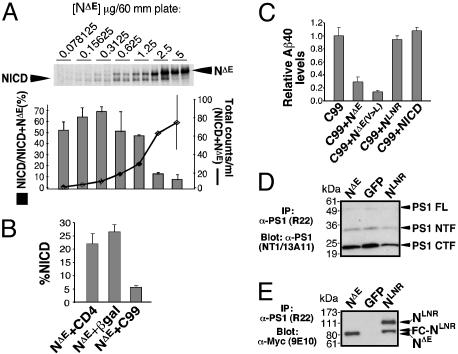
Competition Between S3 Cleavage of Notch and Middle TM Cleavage of APP (Aβ Production). If a single enzyme activity catalyzes proteolysis in the middle of the TM domain (Aβ, Nβ) and near the inner leaflet (S3, ε), Aβ cleavage should act as a specific competitive inhibitor of S3 cleavage and vice versa. However, if the Aβ and S3 cleavages are mediated by different activities, no competition should be observed. Some report that these substrates do not compete (29, 31), whereas others argue that they do (39, 40). A competitive interaction between APP and Notch in cells will be revealed only if the combined substrate concentration is in excess; merely altering the ratio of substrates would not result in competitive inhibition. To determine whether γ-secretase activity can be saturated, we titrated the expression of a truncated Notch 1 protein that does not require ectodomain shedding (NΔE) and determined the ratio of NICD produced (NICD/NICD + NΔE) at different plasmid concentrations (Fig. 1A). Cytosolic β-galactosidase (β-gal) was cotransfected to keep the total amount of transfected plasmid DNA at 5 μg and control for nonspecific effects caused by promoter competition and burden on the translation machinery. Saturation was reached at 1.25 μg of NΔE (Fig. 1A).
Fig. 1.
Notch and APP compete for a limiting activity but not for binding to PS1. (A) Saturation of γ-secretase was demonstrated by transfecting HEK293 cells with a mixture NΔE and cytoβ-gal DNA to a total of 5 μg (two independent experiments were done for each concentration shown). Cells were metabolically labeled overnight with 35S-Met, and cell extracts were immunoprecipitated with 9E10 antibody and separated by SDS/PAGE. The counts in each lane were determined by PhosphorImaging and plotted to show total Notch labeling (line graph) and percentage of NICD (NICD/NΔE + NICD; histograms) as a function of DNA concentration. (B and C) HEK293 cells were transfected with pCS2+C99 and Notch expressing plasmid or control plasmid. Aβ levels were assayed by ELISA. Percentage of NICD was determined by quantitative Western analysis and 35S labeling, which yielded similar results. Results show that APP competes with Notch for the same cleavage activity. NΔE and, more effectively, NΔE(V>L) compete for Aβ production although NLNR and NICD do not. (D) HEK293 cells were transiently transfected with GFP, NΔE, or NLNR, lysed in co-IP buffer, and immunoprecipitated with R22 antibody. PS1 NTF and CTF were detected via Western blotting with NT1 and 13A11 antibodies, respectively. (E) Notch proteins were detected in the same immunoprecipitate by using 9E10. NLNR and NΔE interact with PS1 as assayed by co-IP (equal protein input, data not shown). FC-NLNR, furin-cleaved NLNR; FL, full length.
To assess the possibility of competitive interactions between Notch and APP, NΔE proteolysis was compared in HEK293 cells cotransfected with the type I protein CD4 or β-gal (as nonspecific controls) or C99. NICD production was significantly reduced when NΔE was cotransfected with C99 but remained at 20% when CD4 or β-gal was coexpressed (Fig. 1B). Similarly, production of Aβ40 was reduced when C99 was coexpressed with NΔE but not when coexpressed with NICD, the γ-secretase product and active form of Notch (Fig. 1C). Thus, no product-based inhibition occurs in our experimental system. NLNR is an inactive form of Notch that cannot undergo extracellular S2 cleavage and therefore is not cleaved by γ-secretase (35). As expected, NLNR does not compete with C99 (Fig. 1C). However, NLNR binds to PS as efficiently as NΔE (Fig. 1 D and E) and full-length Notch (36, 41). Lack of competition between NLNR and C99 implies that NLNR binding does not prevent C99 entry into the active site. This result is consistent with the existence of distinct substrate binding and active sites on γ-secretase (42, 43). Importantly, NΔE(V>L) acts as a better inhibitor of Aβ cleavage as might be expected from a protein that still interacts with the active site of the enzyme but is cleaved less efficiently (21) (Fig. 1C).
γ-Secretase Inhibitors Block Notch S3 Cleavage and Aβ Production with the Same Rank Order of Potency. The ability of NΔE(V>L) to compete with C99 strongly implies that the inner leaflet cleavage site competes with the middle membrane cleavage site for a single or two very closely positioned active sites. To investigate this further, we tested the efficacy of multiple inhibitors on both substrates. If a single protease produced NICD and Aβ, then the same compounds should inhibit the proteolysis of both substrates with similar IC50. Indeed, several γ-secretase inhibitors block Aβ production and Notch S3 cleavage: MG132 (34), Calpeptin (35), MD28170 (44), and difluoro ketone peptidomimetics of Aβ42 (44, 45). Although suggestive, these data are insufficient to conclude that a single activity catalyzes the proteolysis of Notch and APP. Most studies did not compare the inhibition of Notch and APP proteolysis in parallel, and in most only a single inhibitor concentration was tested. Two substrates with different affinities for a single protease may not compete equivalently at any given concentration of inhibitor. Thus, any inhibitor at a particular concentration may differentially affect Notch and APP cleavage whether or not a single protease is involved. To determine the inhibition profiles and derive IC50 values for Notch and APP cleavages, a series of titration experiments were carried out in cells expressing either C99 or NΔE, which were analyzed in parallel by transient transfections. Dose–response curves for both Aβ40 and NICD production were generated for seven different inhibitors. Then, the different IC50 values derived for both substrates were plotted against each other (Fig. 6, which is published as supporting information on the PNAS web site). The rank order of potency for both Notch and APP proteolysis is identical for these inhibitors. The values for six of these inhibitors also fall upon a trend line with near unit slope (y = 1.1337x - 0.3373, R2 = 0.9703), showing that not only the rank order, but also the absolute potency is the same for both substrates in cell culture. Considering that the group of inhibitors we tested contains molecules with diverse structures, this correlation is remarkable and consistent with a single catalytic activity for both Aβ and NICD production. Compound 15 did not fall directly upon the trend line generated by the other six compounds because of the observation that at low concentrations, this inhibitor elevated the levels of Aβ40 (Fig. 6), thereby shifting the dose–response curve to produce an elevated IC50 value. This profile is typical for Aβ42, which has often been observed to increase when cells are treated with low levels of several inhibitors, including Compound 15 (46).
PS Could Act as a Dimer Within γ-Secretase. Increased production of Aβ42 at low levels of inhibitor can be explained if γ-secretase is acting in an allosteric manner (47, 48). Allosteric enzymes tend to be multimeric enzymes comprised either of multiples of a single subunit or a complex of several, distinct subunits. γ-Secretase activity is known to require at least four proteins in a complex (49–52) of 250 kDa (53), 500 kDa (54, 55), or >1,000 kDa (54, 56) depending on the detergent used. In addition, evidence that NTF and CTF homodimers can form in yeast was recently presented (57, 58). If such interactions are functionally significant, the simplest complex comprising γ-secretase could consist of two PS molecules at the catalytic site, together with Nicastrin, Aph-1, and Pen-2 at a yet-to-be-determined stoichiometry (49–52, 59, 60), possibly 2:1:1:1 (≈250 kDa) or 2:2:2:2 (>250 kDa). If functional γ-secretase contained a PS dimer, binding of inhibitor to one molecule could cause a change in the specificity of a second PS molecule, which is not yet bound to inhibitor.
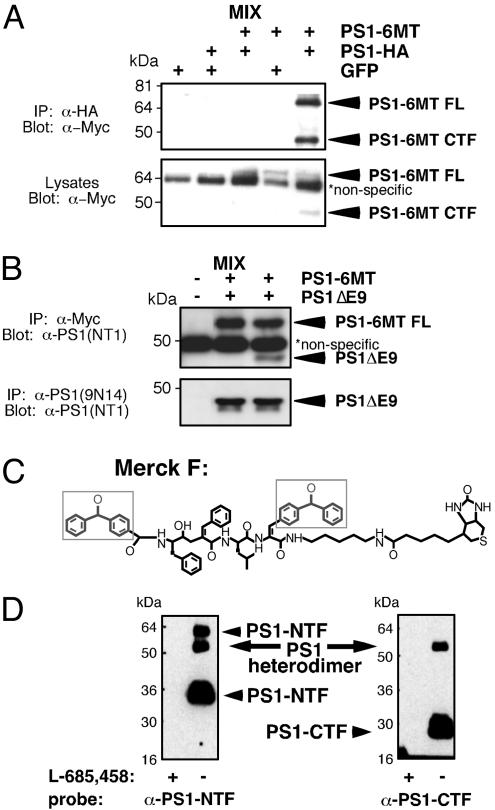
To test PS dimer formation in mammalian cells, we first determined whether differentially tagged PS molecules could undergo co-IP. For these experiments we used PS constructs with C-terminal hemagglutinin (HA) and Myc tags. These PS molecules can reconstitute γ-secretase in PS1/PS2-deficient MEFs although less efficiently than PS1WT (Fig. 7, which is published as supporting information on the PNAS web site). Indeed, the HA-tagged CTF of one PS molecule can immunoprecipitate the Myc-tagged CTF of another (Fig. 2A). Likewise, untagged PS1ΔE9, an active FAD molecule that exists as a single polypeptide (61, 62) coimmunoprecipitated with the Myc-tagged WT PS molecule (Fig. 2B). Importantly, interactions were observed only in lysates from cotransfected cells but not in pooled lysates from separate transfections (mix in Fig. 2 A and B), suggesting that these PS oligomers exist in live cells. Moreover, the PS interactions are apparently strong, as they remain intact under relatively stringent conditions (i.e., using detergents known to disrupt γ-secretase activity).
Fig. 2.
Co-IP and cross-linking analyses suggest dimerization of PS1 molecules within γ-secretase. (A) HEK293 cells were transfected with the indicated constructs, lysed, and immunoprecipitated with a polyclonal α-HA antibody (Covance). Immunoprecipitates and lysates were resolved by SDS/PAGE and immunoblotted with 9E10 antibodies. (B) PS1/PS2-deficient MEFs were transfected with PS1-6MT or PS1ΔE9 or both. They were then lysed, immunoprecipitated with 9E10 or 9N14 antibodies, and analyzed by immunostaining with NT1 antibody. As a control, IPs were also performed by using lysates from singly transfected cells mixed postlysis (labeled MIX). (C) Structure of Merck F (benzophenone photoreactive groups are indicated by boxes). (D) Covalent labeling of PS1 by photoreactive active site-directed γ-secretase inhibitor (Merck F). 3-[(3-Cholamidopropyl)dimethylammonio]-2-hydroxy-1-propane-sulfonate-solubilized HeLa cell membranes were photoactivated in the absence (-) or presence (+) of L-685,458 as a competitor. The sample was diluted with radioimmunoprecipitation assay buffer. Biotinylated proteins were captured with streptavidin-agarose and probed by SDS/PAGE/immunoblotting using α-PS1 antibodies. FL, full length.
To further address the nature of the PS–PS interaction, we performed cross-linking studies on endogenous PS under conditions that maintain γ-secretase activity. We used photoaffinity probes based on the Merck L-685,458 inhibitor that contain two benzophenone groups (Merck F; Fig. 2C and ref. 63). As demonstrated by Esler et al. (43), such inhibitors do not compete for substrate binding but rather interact with the active site. In addition to the expected cross-linking between NTF and CTF units, we observed cross-linking of two NTF subunits (Fig. 2D). Although the high molecular weight band observed could be caused by cross-linking between PS NTF and Aph1, this was ruled out by Western analyses using the α-Aph1 antibody H2D-1 (data not shown; antibody provided by G. Yu, University of Texas Southwestern Medical Center, Dallas; ref. 51). Given that cross-linking is irreversible, the NTF subunits are likely in close proximity within cells supporting a hypothetical catalytic configuration of D2571::D2572 or one NTF presenting substrate to another PS molecule. A D3851::D3852 catalytic pairing or substrate presentation by CTF cannot be ruled out because the positions of the benzophenone groups in Merck F may preclude trapping of the CTF::CTF dimer. Additional biophysical experiments are necessary to determine precisely the structure of the PS oligomer.
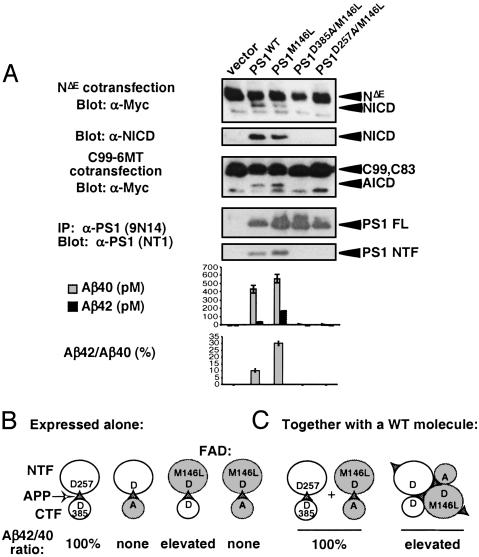
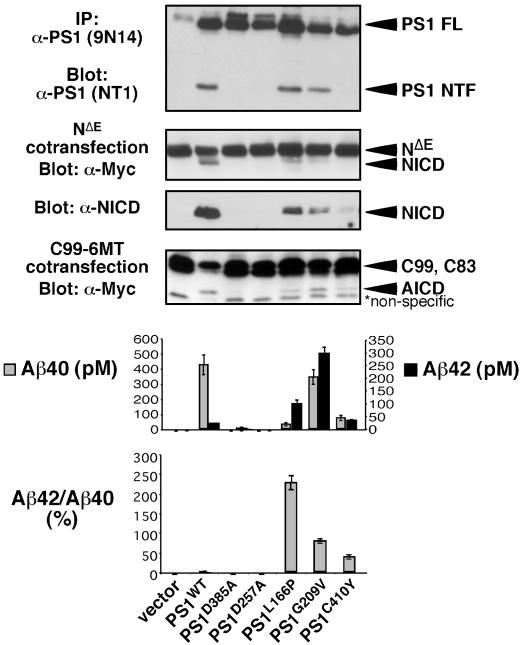
The reported increase in Aβ42 in kindreds with FAD mutations could be caused by PS1FAD interfering with PS1WT in heterozygous individuals (in trans, see also ref. 64) by altering either substrate presentation or active site conformation within a dimer. We characterized the intrinsic activities of several hypoactive FAD mutations [G209V, C410Y (25), and L166P (26)], which were discovered because of their ability to elevate Aβ42/Aβ40 ratios yet are defective in generating NICD (25, 26). These would be candidates for PS FAD molecules that act in trans. When tested for their ability to reconstitute γ-secretase activity in PS1/PS2-deficient MEFs, only the PS1D385A and PS1D257A controls failed to restore any activity. PS1M146L restored activity comparable to that of PS1WT (see Fig. 4A), as did PS1I143T (data not shown). In contrast, PS1L166P, PS1G209V, and PS1C410Y restored only some aspects of γ-secretase activity (Fig. 3): PS1L166P restored WT presenilinase activity but low levels of NICD and AICD. PS1G209V restored WT presenilinase activity and AICD formation but low levels of NICD. PS1C410Y restored weak presenilinase activity and released low levels of both AICD and NICD. Although all restored some Aβ40, PS1L166P and PS1C410Y were only able to restore ≈10% of Aβ40 relative to WT; PS1G209V produced ≈80% of WT Aβ40. However, all FAD mutants tested generated Aβ42 to levels comparable or exceeding that of WT (Figs. 3 and 4A).
Fig. 4.
A strategy for testing trans acting FAD activity. (A) WT and mutant forms of PS1 were transiently transfected into PS1/PS2-deficient MEFs and their associated proteolytic activities were assayed. Results show that mutating Asp-385 and Asp-257 in the FAD mutant PS1M146L abolishes NICD and AICD release, Aβ formation, and endoproteolysis. For Aβ measurements, average of independent triplicates from one representative experiment is shown (n > 10). (B) A schematic showing the expected Aβ42/Aβ40 ratio associated with different PS molecules when expressed alone (i.e., intrinsic activity). (C) Coexpression of the double mutant PS1D385A/M146L with PS1WT is predicted to distinguish between a monomeric (Left) or dimeric (Right) acting PS within the catalytic core of γ-secretase. If PS strictly acts as monomer the loss of Asp-385 should be epistatic to the intramolecular FAD mutation and the Aβ42/Aβ40 ratio should not be altered. However, a dimeric PS at the core of γ-secretase will allow an inactive, cleavage-incompetent PS to alter substrate presentation to and/or the specificity of the WT molecule. In this case, the FAD phenotype will be epistatic to the Asp-385 mutation. FL, full length.
Fig. 3.
Some PS1 FAD mutations are intrinsically hypomorphic for different proteolytic activities. WT and mutant forms of PS1 were transiently transfected into PS1/PS2-deficient MEFs to characterize their associated activities. To determine PS endoproteolysis, lysates of transfected cells were immunoprecipitated with 9N14, resolved by SDS/PAGE, and immunostained with NT1. To assess NICD production, PS1 was cotransfected with NΔE and 9E10 and α-V1744 antibodies (Cell Signaling Technology, Beverly, MA) were used to detect NICD on Western blots. To determine AICD release, PS1 was cotransfected with C99 – 6MT and AICD was visualized with 9E10 antibody. Aβ production was assessed by ELISA on conditioned media from cells cotransfected with PS1 and APP (n = 4) (see Supporting Methods, which is published as supporting information on the PNAS web site). Average of independent triplicates from one representative experiment is shown. FL, full length.
Although these results are consistent with either a PS monomer or dimer at the catalytic center, the cross-linking and co-IP results we report favor a model in which a dimer is involved. One strategy to determine whether FAD proteins can indeed act in trans would be to create truly catalytically inactive molecules capable of elevating Aβ42. Introducing a second site mutation (D257A or D385A) into an FAD mutant PS molecule should render the FAD molecule catalytically inactive. If γ-secretase contains a single PS molecule, PS1D385A/FAD molecules could not alter cellular Aβ42/Aβ40 ratios when coexpressed with PS1WT (Fig. 4C). Conversely, if two PS molecules interacted within γ-secretase, the PS1D385A/FAD may still be able to affect Aβ cleavage despite the intramolecular D385A mutation (Fig. 4C). It should be noted, however, that not all FAD mutations may work in conjunction with a mutation at the catalytic site because of structural interactions between the two mutations. Less of a concern is the possibility that in cells coexpressing PS1D385A/FAD and PS1WT, some cleavage could occur within the loop domain of PS1D385A/FAD, thereby allowing NTFFAD and CTFWT fragments to associate and produce an active PS1FAD molecule. Such “domain swapping” interactions have been ruled out by previous studies (65, 66); overexpression of NTFY115H or CTFC392V can only impact the Aβ42/Aβ40 ratio if expressed with the reciprocal PS fragment but not when overexpressed alone in HEK293 cells. To confirm that no domain swaps can occur in PS1/PS2-deficient cells we coexpressed hypomorphic PS proteins with mutations in their NTF (L166P, G209V, or D257A) with others containing CTF mutations (C410Y or D385A) and assayed for AICD and NICD formation. In no combination were we able to enhance γ-secretase activity (data not shown). Therefore, any FAD effect of PS1D385A/FAD should be the result of transmolecular interactions and not domain swapping.
PS1M146L was selected for the initial experiment because it retains the ability to produce NICD, AICD, and Aβ peptides when expressed in PS1/PS2-deficient MEFs (Fig. 4 A and B). As expected, PS1D385A/M146L and PS1D257A/M146L were unable to reconstitute any γ-secretase activity in these cells (Fig. 4 A and B). When PS1D385A/M146L was transiently coexpressed with PS1WT at 1:1 ratio, however, it could increase the Aβ42/Aβ40 ratio. The magnitude of this effect was found to be variable, and with multiple independent repeats the overall outcome was not statistically significant. This finding may indicate that, when coexpressed in such a system, PS1D385A/M146L and PS1WT association occurs only at a low level and in a transient manner.
Discussion
The major hypothesis emerging from the literature is that PS proteins contain the active site of γ-secretase, whereas Nicastrin, APH-1, and PEN-2 are important for the formation and maturation of the enzymatic complex (2, 67). Here we provide biochemical evidence in support of a single γ-secretase activity involved in Notch and APP proteolysis at both intramembranous sites and propose that this activity resides, at least transiently, in a dimer of PS molecules identified via cross-linking and co-IP experiments. A model in which γ-secretase contains a PS dimer can explain several long-standing and perplexing observations. Many laboratories have observed an increase in cleavage products from minor sites with low inhibitor concentrations; this may occur if only one of two PS molecules in γ-secretase is inhibited. Some small molecules are predicted to act as allosteric modulators, having differential effects on different cleavage sites by promoting a conformation that cleaves preferentially at one site or by altering substrate presentation. This is the most likely mechanism for the shifting of the preferred site from Aβ42 to Aβ38 by nonsteroidal antiinflammatory drugs like ibuprofen (7). To explain how a single active site catalyzes cleavages both near the inner leaflet and in the middle of the membrane in close succession or even simultaneously, it may be necessary to postulate that at least three aspartyl residues can assemble at the dimer interface, forming at least two functional catalytic pairs. Indirect experimental evidence for an aspartyl triad hypothesis comes from the observation that photoaffinity probes can cross-link both NTF-CTF and NTF-NTF dimers but not CTF-CTF dimers (Fig. 3). The dimeric PS core can therefore hydrolyze peptide bonds in the middle of the TM domain by using any combination of up to four aspartyl residues (Fig. 4C). Alternatively, a catalytic dimer may simply not exist; the cross-linking data could also be consistent with substrate presentation by one NTF to the catalytic site on another PS1 molecule. It is interesting to speculate whether other intramembrane cleaving activities also act as oligomers.
Although some model predictions of a dimer mechanism were confirmed experimentally, others were not. The FAD effects of PS1D385A/M146L were not statistically significant; this could be caused by interactions between these two specific mutations that negated a trans effect. Although it is possible that using another PS1D385A/FAD molecule could provide evidence for trans effects on Aβ42 production, formal validation of this mechanistic model awaits advances in crystallography that will permit structural work on purified, active γ-secretase and detailed kinetic analysis of this enzyme.
Finally, the hypothesis that γ-secretase is an allosterically modulated oligomer and the experimental evidence provided here and in the literature indicate that identification of small molecules able to specifically reduce Aβ42 production is a feasible goal.
Supplementary Material
Acknowledgments
We thank Stephen Gardell, Jules Shafer, Ian Ragan, Tim Harrison, Luis Castro, and Alan Nadin (Merck), Andrew Nyborg (Mayo Clinic, Jacksonville, FL), Gene Johnson, and the members of the Kopan lab (past and present) for support and discussion. This work is supported by National Institutes of Health Grant GM55479, Alzheimer's Association Grant RG991516, and Zenith Award ZEN-01-3050 (to R.K.), National Institutes of Health Grants AG17050 (to A.G. and R.K.), and NS41355 (to M.S.W.), and a postdoctoral fellowship from the French Foundation (to S.H.).
Abbreviations: APP, amyloid precursor protein; AICD, APP intracellular domain; NICD, Notch intracellular domain; NTF, N-terminal fragment; CTF, C-terminal fragment; PS, presenilin; FAD, familial Alzheimer's disease; TM, transmembrane; MEF, mouse embryonic fibroblast; HEK, human embryonic kidney; IP, immunoprecipitation; β-gal, β-galactosidase; HA, hemagglutinin.
References
- 1.Selkoe, D. & Kopan, R. (2003) Annu. Rev. Neurosci. 26 565-597. [DOI] [PubMed] [Google Scholar]
- 2.Haass, C. & Steiner, H. (2002) Trends Cell Biol. 12 556-562. [DOI] [PubMed] [Google Scholar]
- 3.Annaert, W. & De Strooper, B. (2002) Annu. Rev. Cell Dev. Biol. 18 25-51. [DOI] [PubMed] [Google Scholar]
- 4.De Strooper, B. & Annaert, W. (2000) J. Cell Sci. 113 1857-1870. [DOI] [PubMed] [Google Scholar]
- 5.Vassar, R. (2002) Adv. Drug Deliv. Rev. 54 1589-1602. [DOI] [PubMed] [Google Scholar]
- 6.Murphy, M. P., Hickman, L. J., Eckman, C. B., Uljon, S. N., Wang, R. & Golde, T. E. (1999) J. Biol. Chem. 274 11914-11923. [DOI] [PubMed] [Google Scholar]
- 7.Weggen, S., Eriksen, J. L., Das, P., Sagi, S. A., Wang, R., Pietrzik, C. U., Findlay, K. A., Smith, T. W., Murphy, M. P., Butler, T., et al. (2001) Nature 414 212-216. [DOI] [PubMed] [Google Scholar]
- 8.Cao, X. & Sudhof, T. C. (2001) Science 293 115-120. [DOI] [PubMed] [Google Scholar]
- 9.Kimberly, W. T., Zheng, J. B., Guenette, S. Y. & Selkoe, D. J. (2001) J. Biol. Chem. 276 40288-40292. [DOI] [PubMed] [Google Scholar]
- 10.Cupers, P., Orlans, I., Craessaerts, K., Annaert, W. & De Strooper, B. (2001) J. Neurochem. 78 1168-1178. [DOI] [PubMed] [Google Scholar]
- 11.Gao, Y. H. & Pimplikar, S. W. (2001) Proc. Natl. Acad. Sci. USA 98 14979-14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mumm, J. S. & Kopan, R. (2000) Dev. Biol. 228 151-165. [DOI] [PubMed] [Google Scholar]
- 13.Struhl, G. & Adachi, A. (1998) Cell 93 649-660. [DOI] [PubMed] [Google Scholar]
- 14.Huppert, S., Le, A., Schroeter, E. H., Mumm, S. J., Saxena, M. T., Milner, A. L. & Kopan, R. (2000) Nature 405 966-970. [DOI] [PubMed] [Google Scholar]
- 15.Okochi, M., Steiner, H., Fukumori, A., Tanii, H., Tomita, T., Tanaka, T., Iwatsubo, T., Kudo, T., Takeda, M. & Haass, C. (2002) EMBO J. 21 5408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama, K., Tomita, T., Shinozaki, K., Kume, H., Asada, H., Saido, T. C., Ishiura, S., Iwatsubo, T. & Obata, K. (1996) Biochem. Biophys. Res. Commun. 227 730-735. [DOI] [PubMed] [Google Scholar]
- 17.Lichtenthaler, S. F., Ida, N., Multhaup, G., Masters, C. L. & Beyreuther, K. (1997) Biochemistry 36 15396-15403. [DOI] [PubMed] [Google Scholar]
- 18.Lichtenthaler, S. F., Wang, R., Grimm, H., Uljon, S. N., Masters, C. L. & Beyreuther, K. (1999) Proc. Natl. Acad. Sci. USA 96 3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tischer, E. & Cordell, B. (1996) J. Biol. Chem. 271 21914-21919. [DOI] [PubMed] [Google Scholar]
- 20.Murphy, M. P., Uljon, S. N., Fraser, P. E., Fauq, A., Lookingbill, H. A., Findlay, K. A., Smith, T. E., Lewis, P. A., McLendon, D. C., Wang, R., et al. (2000) J. Biol. Chem. 275 26277-26284. [DOI] [PubMed] [Google Scholar]
- 21.Schroeter, E. H., Kisslinger, J. A. & Kopan, R. (1998) Nature 393 382-386. [DOI] [PubMed] [Google Scholar]
- 22.Saxena, M. T., Schroeter, E. H., Mumm, J. S. & Kopan, R. (2001) J. Biol. Chem. 276 40268-40273. [DOI] [PubMed] [Google Scholar]
- 23.Gu, Y., Misonou, H., Sato, T., Dohmae, N., Takio, K. & Ihara, Y. (2001) J. Biol. Chem. 276 35235-35238. [DOI] [PubMed] [Google Scholar]
- 24.Sastre, M., Steiner, H., Fuchs, K., Capell, A., Multhaup, G., Condron, M. M., Teplow, D. B. & Haass, C. (2001) EMBO Rep. 2 835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song, W. H., Nadeau, P., Yuan, M. L., Yang, X. D., Shen, J. & Yankner, B. A. (1999) Proc. Natl. Acad. Sci. USA 96 6959-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moehlmann, T., Winkler, E., Xia, X. F., Edbauer, D., Murrelll, J., Capell, A., Kaether, C., Zheng, H., Ghetti, B., Haass, C., et al. (2002) Proc. Natl. Acad. Sci. USA 99 8025-8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, F., Gu, Y., Hasegawa, H., Ruan, X., Arawaka, S., Fraser, P., Westaway, D., Mount, H. & St. George-Hyslop, P. (2002) J. Biol. Chem. 277 36521-36526. [DOI] [PubMed] [Google Scholar]
- 28.Hardy, J. & Crook, R. (2001) Neurosci. Lett. 306 203-205. [DOI] [PubMed] [Google Scholar]
- 29.Chen, F., Yu, G., Arawaka, S., Nishimura, M., Kawarai, T., Yu, H., Tandon, A., Supala, A., Song, Y. Q., Rogaeva, E., et al. (2001) Nat. Cell Biol. 3 751-754. [DOI] [PubMed] [Google Scholar]
- 30.Taniguchi, Y., Karlstrom, H., Lundkvist, J., Mizutani, T., Otaka, A., Vestling, M., Bernstein, A., Donoviel, D., Lendahl, U. & Honjo, T. (2002) Proc. Natl. Acad. Sci. USA 99 4014-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petit, A., St. George-Hyslop, P., Fraser, P. & Checler, F. (2002) Biochem. Biophys. Res. Commun. 290 1408-1410. [DOI] [PubMed] [Google Scholar]
- 32.Wilson, C. A., Doms, R. W., Zheng, H. & Lee, V. M. (2002) Nat. Neurosci. 5 849-855. [DOI] [PubMed] [Google Scholar]
- 33.Herreman, A., Hartmann, D., Annaert, W., Saftig, P., Craessaerts, K., Serneels, L., Umans, L., Schrijvers, V., Checler, F., Vanderstichele, H., et al. (1999) Proc. Natl. Acad. Sci. USA 96 11872-11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopan, R., Schroeter, E. H., Weintraub, H. & Nye, J. S. (1996) Proc. Natl. Acad. Sci. USA 93 1683-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mumm, J. S., Schroeter, E. H., Saxena, M. T., Griesemer, A., Tian, X., Pan, D. J., Ray, W. J. & Kopan, R. (2000) Mol. Cell 5 197-206. [DOI] [PubMed] [Google Scholar]
- 36.Ray, W. J., Yao, M., Nowotny, P., Mumm, J., Zhang, W. J., Wu, J. Y., Kopan, R. & Goate, A. M. (1999) Proc. Natl. Acad. Sci. USA 96 3263-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Podlisny, M. B., Citron, M., Amarante, P., Sherrington, R., Xia, W., Zhang, J., Diehl, T., Levesque, G., Fraser, P., Haass, C., et al. (1997) Neurobiol. Dis. 3 325-337. [DOI] [PubMed] [Google Scholar]
- 38.Malin, S. A., Guo, W. X., Jafari, G., Goate, A. M. & Nerbonne, J. M. (1998) Neurobiol. Dis. 4 398-409. [DOI] [PubMed] [Google Scholar]
- 39.Kimberly, W. T., Esler, W. P., Ye, W., Ostaszewski, B. L., Gao, J., Diehl, T., Selkoe, D. J. & Wolfe, M. S. (2003) Biochemistry 42 137-144. [DOI] [PubMed] [Google Scholar]
- 40.Berezovska, O., Jack, C., Deng, A., Gastineau, N., Rebeck, G. W. & Hyman, B. T. (2001) J. Biol. Chem. 276 30018-30023. [DOI] [PubMed] [Google Scholar]
- 41.Ray, W. J., Yao, M., Mumm, J., Schroeter, E. H., Saftig, P., Wolfe, M., Selkoe, D. J., Kopan, R. & Goate, A. M. (1999) J. Biol. Chem. 274 36801-36807. [DOI] [PubMed] [Google Scholar]
- 42.Annaert, W. G., Esselens, C., Baert, V., Boeve, C., Snellings, G., Cupers, P., Craessaerts, K. & De Strooper, B. (2001) Neuron 32 579-589. [DOI] [PubMed] [Google Scholar]
- 43.Esler, W. P., Kimberly, W. T., Ostaszewski, B. L., Ye, W., Diehl, T. S., Selkoe, D. J. & Wolfe, M. S. (2002) Proc. Natl. Acad. Sci. USA 99 2720-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Strooper, B., Annaert, W., Cupers, P., Saftig, P., Craessaerts, K., Mumm, J. S., Schroeter, E. H., Schrijvers, V., Wolfe, M. S., Ray, W. J., et al. (1999) Nature 398 518-522. [DOI] [PubMed] [Google Scholar]
- 45.Wolfe, M. S., Citron, M., Diehl, T. S., Xia, W., Donkor, I. O. & Selkoe, D. J. (1998) J. Med. Chem. 41 6-9. [DOI] [PubMed] [Google Scholar]
- 46.Wolfe, M. S., Xia, W., Moore, C. L., Leatherwood, D. D., Ostaszewski, B., Rahmati, T., Donkor, I. O. & Selkoe, D. J. (1999) Biochemistry 38 4720-4727. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, L., Song, L., Terracina, G., Liu, Y., Pramanik, B. & Parker, E. (2001) Biochemistry 40 5049-5055. [DOI] [PubMed] [Google Scholar]
- 48.Foote, J. & Schachman, H. K. (1985) J. Mol. Biol. 186 175-184. [DOI] [PubMed] [Google Scholar]
- 49.LaVoie, M. J., Fraering, P. C., Ostaszewski, B. L., Ye, W., Kimberly, W. T., Wolfe, M. S. & Selkoe, D. J. (2003) J. Biol. Chem. 278 37213-37222. [DOI] [PubMed] [Google Scholar]
- 50.Edbauer, D., Winkler, E., Regula, J. T., Pesold, B., Steiner, H. & Haass, C. (2003) Nat. Cell Biol. 5 486-488. [DOI] [PubMed] [Google Scholar]
- 51.Luo, W. J., Wang, H., Li, H., Kim, B. S., Shah, S., Lee, H. J., Thinakaran, G., Kim, T. W., Yu, G. & Xu, H. (2003) J. Biol. Chem. 278 7850-7854. [DOI] [PubMed] [Google Scholar]
- 52.Takasugi, N., Tomita, T., Hayashi, I., Tsuruoka, M., Niimura, M., Takahashi, Y., Thinakaran, G. & Iwatsubo, T. (2003) Nature 422 438-441. [DOI] [PubMed] [Google Scholar]
- 53.Kimberly, W. T., LaVoie, M. J., Ostaszewski, B. L., Ye, W., Wolfe, M. S. & Selkoe, D. J. (2003) Proc. Natl. Acad. Sci. USA 100 6382-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farmery, M. R., Tjernberg, L. O., Pursglove, S. E., Bergman, A., Winblad, B. & Naslund, J. (2003) J. Biol. Chem. 278 24277-24284. [DOI] [PubMed] [Google Scholar]
- 55.Steiner, H., Winkler, E., Edbauer, D., Prokop, S., Basset, G., Yamasaki, A., Kostka, M. & Haass, C. (2002) J. Biol. Chem. 277 39062-39065. [DOI] [PubMed] [Google Scholar]
- 56.Li, Y. M., Lai, M. T., Xu, M., Huang, Q., DiMuzio-Mower, J., Sardana, M. K., Shi, X. P., Yin, K. C., Shafer, J. A. & Gardell, S. J. (2000) Proc. Natl. Acad. Sci. USA 97 6138-6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cervantes, S., Gonzalez-Duarte, R. & Marfany, G. (2001) FEBS Lett. 505 81-86. [DOI] [PubMed] [Google Scholar]
- 58.Hebert, S. S., Godin, C., Tomiyama, T., Mori, H. & Levesque, G. (2003) Biochem. Biophys. Res. Commun. 301 119-126. [DOI] [PubMed] [Google Scholar]
- 59.Kopan, R. & Goate, A. (2002) Neuron 33 321-324. [DOI] [PubMed] [Google Scholar]
- 60.Francis, R., McGrath, G., Zhang, J., Ruddy, D. A., Sym, M., Apfeld, J., Nicoll, M., Maxwell, M., Hai, B., Ellis, M. C., et al. (2002) Dev Cell 3 85-97. [DOI] [PubMed] [Google Scholar]
- 61.Thinakaran, G., Borchelt, D. R., Lee, M. K., Slunt, H. H., Spitzer, L., Kim, G., Ratovitsky, T., Davenport, F., Nordstedt, C., Seeger, M., et al. (1996) Neuron 17 181-190. [DOI] [PubMed] [Google Scholar]
- 62.Steiner, H., Romig, H., Grim, M. G., Philipp, U., Pesold, B., Citron, M., Baumeister, R. & Haass, C. (1999) J. Biol. Chem. 274 7615-7618. [DOI] [PubMed] [Google Scholar]
- 63.Li, Y. M., Xu, M., Lai, M. T., Huang, Q., Castro, J. L., DiMuzio-Mower, J., Harrison, T., Lellis, C., Nadin, A., Neduvelil, J. G., et al. (2000) Nature 405 689-694. [DOI] [PubMed] [Google Scholar]
- 64.Kwok, J. B., Halliday, G. M., Brooks, W. S., Dolios, G., Laudon, H., Murayama, O., Hallupp, M., Badenhop, R. F., Vickers, J., Wang, R., et al. (2003) J. Biol. Chem. 278 6748-6754. [DOI] [PubMed] [Google Scholar]
- 65.Saura, C. A., Tomita, T., Davenport, F., Harris, C. L., Iwatsubo, T. & Thinakaran, G. (1999) J. Biol. Chem. 274 13818-13823. [DOI] [PubMed] [Google Scholar]
- 66.Levitan, D., Lee, J., Song, L. X., Manning, R., Wong, G., Parker, E. & Zhang, L. L. (2001) Proc. Natl. Acad. Sci. USA 98 12186-12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Strooper, B. (2003) Neuron 38 9-12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.