Abstract
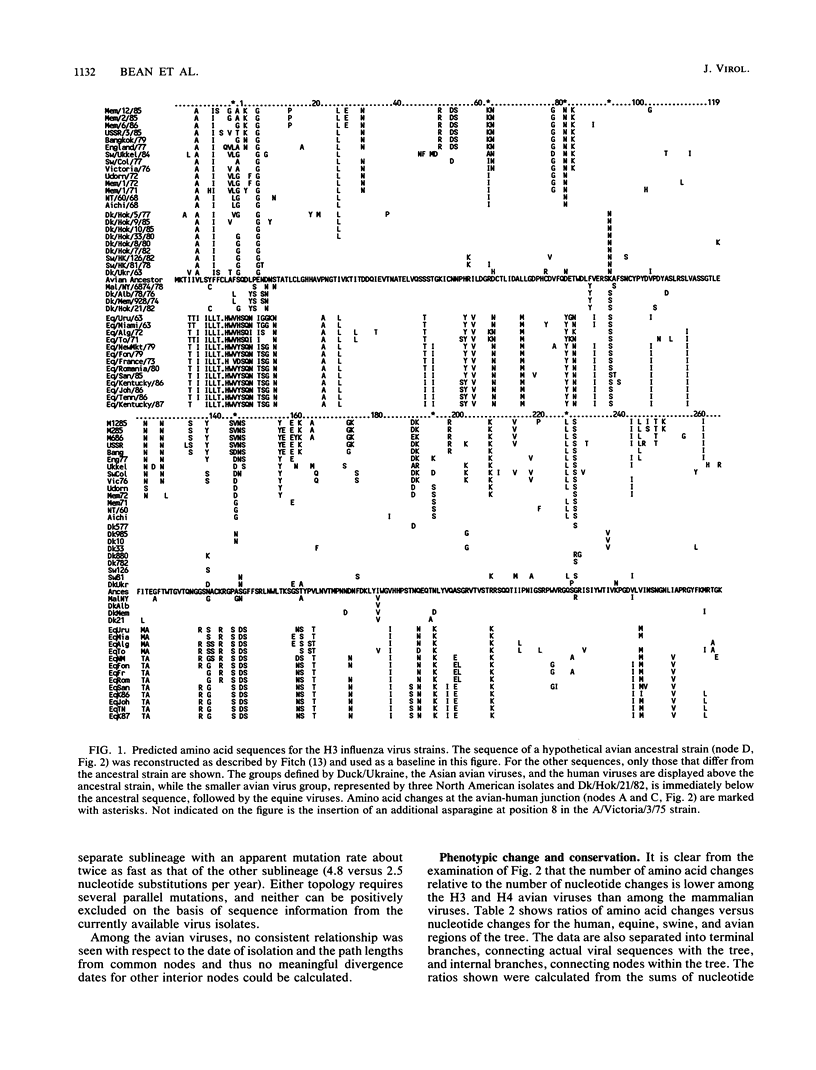
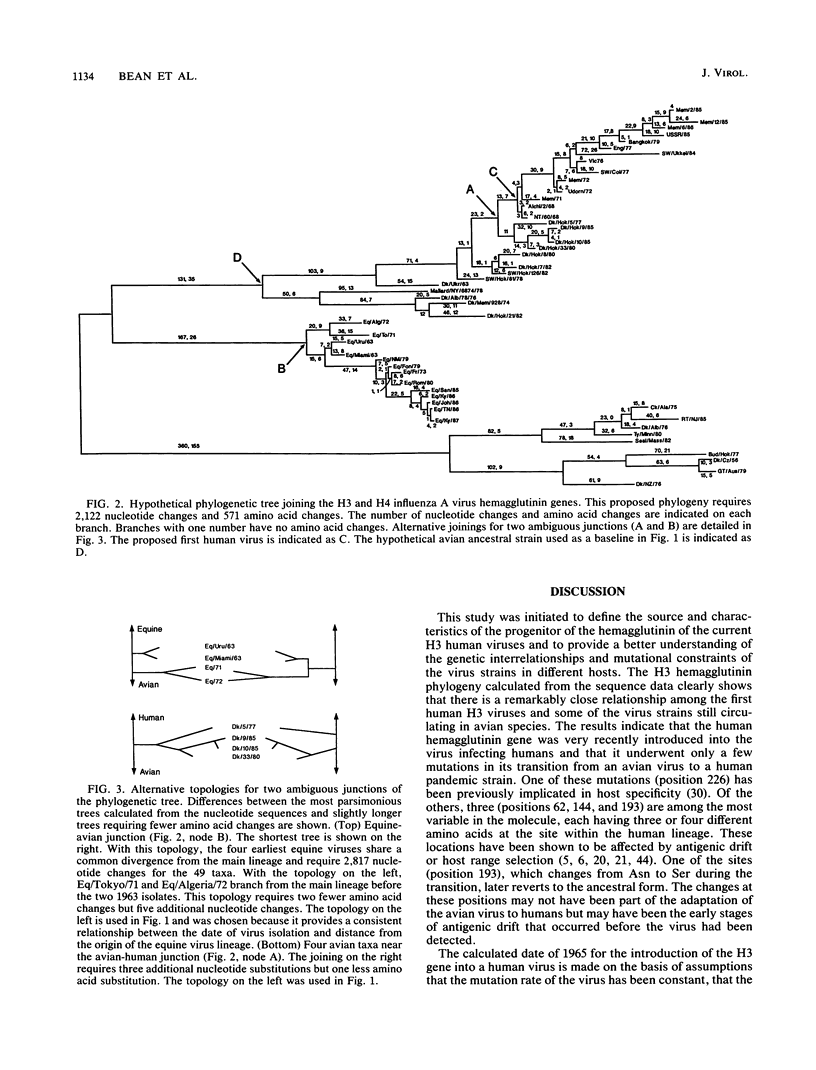
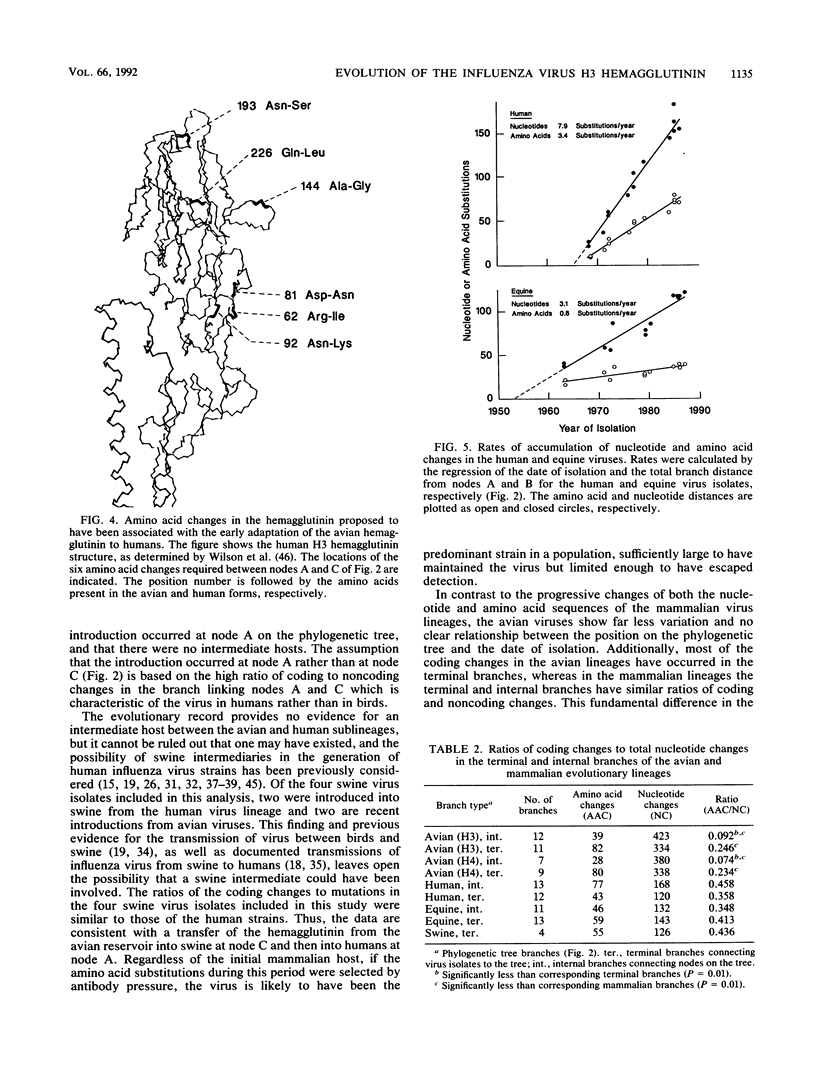
The nucleotide and amino acid sequences of 40 influenza virus hemagglutinin genes of the H3 serotype from mammalian and avian species and 9 genes of the H4 serotype were compared, and their evolutionary relationships were evaluated. From these relationships, the differences in the mutational characteristics of the viral hemagglutinin in different hosts were examined and the RNA sequence changes that occurred during the generation of the progenitor of the 1968 human pandemic strain were examined. Three major lineages were defined: one containing only equine virus isolates; one containing only avian virus isolates; and one containing avian, swine, and human virus isolates. The human pandemic strain of 1968 was derived from an avian virus most similar to those isolated from ducks in Asia, and the transfer of this virus to humans probably occurred in 1965. Since then, the human viruses have diverged from this progenitor, with the accumulation of approximately 7.9 nucleotide and 3.4 amino acid substitutions per year. Reconstruction of the sequence of the hypothetical ancestral strain at the avian-human transition indicated that only 6 amino acids in the mature hemagglutinin molecule were changed during the transition between an avian virus strain and a human pandemic strain. All of these changes are located in regions of the molecule known to affect receptor binding and antigenicity. Unlike the human H3 influenza virus strains, the equine virus isolates have no close relatives in other species and appear to have diverged from the avian viruses much earlier than did the human virus strains. Mutations were estimated to have accumulated in the equine virus lineage at approximately 3.1 nucleotides and 0.8 amino acids per year. Four swine virus isolates in the analysis each appeared to have been introduced into pigs independently, with two derived from human viruses and two from avian viruses. A comparison of the coding and noncoding mutations in the mammalian and avian lineages showed a significantly lower ratio of coding to total nucleotide changes in the avian viruses. Additionally, the avian virus lineages of both the H3 and H4 serotypes, but not the mammalian virus lineages, showed significantly greater conservation of amino acid sequence in the internal branches of the phylogenetic tree than in the terminal branches. The small number of amino acid differences between the avian viruses and the progenitor of the 1968 pandemic strain and the great phenotypic stability of the avian viruses suggest that strains similar to the progenitor strain will continue to circulate in birds and will be available for reintroduction into humans.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Air G. M., Gibbs A. J., Laver W. G., Webster R. G. Evolutionary changes in influenza B are not primarily governed by antibody selection. Proc Natl Acad Sci U S A. 1990 May;87(10):3884–3888. doi: 10.1073/pnas.87.10.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Air G. M. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7639–7643. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean W. J., Jr, Sriram G., Webster R. G. Electrophoretic analysis of iodine-labeled influenza virus RNA segments. Anal Biochem. 1980 Feb;102(1):228–232. doi: 10.1016/0003-2697(80)90343-7. [DOI] [PubMed] [Google Scholar]
- Bean W. J., Threlkeld S. C., Webster R. G. Biologic potential of amantadine-resistant influenza A virus in an avian model. J Infect Dis. 1989 Jun;159(6):1050–1056. doi: 10.1093/infdis/159.6.1050. [DOI] [PubMed] [Google Scholar]
- Both G. W., Sleigh M. J. Conservation and variation in the hemagglutinins of Hong Kong subtype influenza viruses during antigenic drift. J Virol. 1981 Sep;39(3):663–672. doi: 10.1128/jvi.39.3.663-672.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both G. W., Sleigh M. J., Cox N. J., Kendal A. P. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J Virol. 1983 Oct;48(1):52–60. doi: 10.1128/jvi.48.1.52-60.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonagurio D. A., Nakada S., Desselberger U., Krystal M., Palese P. Noncumulative sequence changes in the hemagglutinin genes of influenza C virus isolates. Virology. 1985 Oct 30;146(2):221–232. doi: 10.1016/0042-6822(85)90006-6. [DOI] [PubMed] [Google Scholar]
- Buonagurio D. A., Nakada S., Fitch W. M., Palese P. Epidemiology of influenza C virus in man: multiple evolutionary lineages and low rate of change. Virology. 1986 Aug;153(1):12–21. doi: 10.1016/0042-6822(86)90003-6. [DOI] [PubMed] [Google Scholar]
- Buonagurio D. A., Nakada S., Parvin J. D., Krystal M., Palese P., Fitch W. M. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in NS gene. Science. 1986 May 23;232(4753):980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- Daniels R. S., Skehel J. J., Wiley D. C. Amino acid sequences of haemagglutinins of influenza viruses of the H3 subtype isolated from horses. J Gen Virol. 1985 Mar;66(Pt 3):457–464. doi: 10.1099/0022-1317-66-3-457. [DOI] [PubMed] [Google Scholar]
- Donis R. O., Bean W. J., Kawaoka Y., Webster R. G. Distinct lineages of influenza virus H4 hemagglutinin genes in different regions of the world. Virology. 1989 Apr;169(2):408–417. doi: 10.1016/0042-6822(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Fang R., Min Jou W., Huylebroeck D., Devos R., Fiers W. Complete structure of A/duck/Ukraine/63 influenza hemagglutinin gene: animal virus as progenitor of human H3 Hong Kong 1968 influenza hemagglutinin. Cell. 1981 Aug;25(2):315–323. doi: 10.1016/0092-8674(81)90049-0. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Leiter J. M., Li X. Q., Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman O. T., Bean W. J., Kawaoka Y., Donatelli I., Guo Y. J., Webster R. G. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J Virol. 1991 Jul;65(7):3704–3714. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman O. T., Bean W. J., Kawaoka Y., Webster R. G. Evolution of the nucleoprotein gene of influenza A virus. J Virol. 1990 Apr;64(4):1487–1497. doi: 10.1128/jvi.64.4.1487-1497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann R., Clarke L. D., Mountford R. C., Bachmayer H., Almond J. W. Nucleotide sequence of the haemagglutinin gene of influenza virus A/England/321/77. J Gen Virol. 1983 Jan;64(Pt 1):215–220. doi: 10.1099/0022-1317-64-1-215. [DOI] [PubMed] [Google Scholar]
- Hinshaw V. S., Bean W. J., Jr, Webster R. G., Easterday B. C. The prevalence of influenza viruses in swine and the antigenic and genetic relatedness of influenza viruses from man and swine. Virology. 1978 Jan;84(1):51–62. doi: 10.1016/0042-6822(78)90217-9. [DOI] [PubMed] [Google Scholar]
- Hinshaw V. S., Webster R. G., Bean W. J., Downie J., Senne D. A. Swine influenza-like viruses in turkeys: potential source of virus for humans? Science. 1983 Apr 8;220(4593):206–208. doi: 10.1126/science.6298942. [DOI] [PubMed] [Google Scholar]
- Jou W. M., Verhoeyen M., Devos R., Saman E., Fang R., Huylebroeck D., Fiers W., Threlfall G., Barber C., Carey N. Complete structure of the hemagglutinin gene from the human influenza A/Victoria/3/75 (H3N2) strain as determined from cloned DNA. Cell. 1980 Mar;19(3):683–696. doi: 10.1016/s0092-8674(80)80045-6. [DOI] [PubMed] [Google Scholar]
- Katz J. M., Naeve C. W., Webster R. G. Host cell-mediated variation in H3N2 influenza viruses. Virology. 1987 Feb;156(2):386–395. doi: 10.1016/0042-6822(87)90418-1. [DOI] [PubMed] [Google Scholar]
- Katz J. M., Webster R. G. Antigenic and structural characterization of multiple subpopulations of H3N2 influenza virus from an individual. Virology. 1988 Aug;165(2):446–456. doi: 10.1016/0042-6822(88)90588-0. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Webster R. G. Origin of the hemagglutinin on A/Equine/Johannesburg/86 (H3N8): the first known equine influenza outbreak in South Africa. Arch Virol. 1989;106(1-2):159–164. doi: 10.1007/BF01311048. [DOI] [PubMed] [Google Scholar]
- Kida H., Kawaoka Y., Naeve C. W., Webster R. G. Antigenic and genetic conservation of H3 influenza virus in wild ducks. Virology. 1987 Jul;159(1):109–119. doi: 10.1016/0042-6822(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Kida H., Shortridge K. F., Webster R. G. Origin of the hemagglutinin gene of H3N2 influenza viruses from pigs in China. Virology. 1988 Jan;162(1):160–166. doi: 10.1016/0042-6822(88)90405-9. [DOI] [PubMed] [Google Scholar]
- Kundin W. D. Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature. 1970 Nov 28;228(5274):857–857. doi: 10.1038/228857a0. [DOI] [PubMed] [Google Scholar]
- Laver W. G., Webster R. G. Studies on the origin of pandemic influenza. 3. Evidence implicating duck and equine influenza viruses as possible progenitors of the Hong Kong strain of human influenza. Virology. 1973 Feb;51(2):383–391. doi: 10.1016/0042-6822(73)90437-6. [DOI] [PubMed] [Google Scholar]
- Masurel N., Marine W. M. Recycling of Asian and Hong Kong influenza A virus hemagglutinins in man. Am J Epidemiol. 1973 Jan;97(1):44–49. doi: 10.1093/oxfordjournals.aje.a121483. [DOI] [PubMed] [Google Scholar]
- Naeve C. W., Hinshaw V. S., Webster R. G. Mutations in the hemagglutinin receptor-binding site can change the biological properties of an influenza virus. J Virol. 1984 Aug;51(2):567–569. doi: 10.1128/jvi.51.2.567-569.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Nobusawa E., Nakajima S. Genetic relatedness between A/Swine/Iowa/15/30(H1N1) and human influenza viruses. Virology. 1984 Nov;139(1):194–198. doi: 10.1016/0042-6822(84)90341-6. [DOI] [PubMed] [Google Scholar]
- Nerome K., Ishida M., Oya A., Oda K. The possible origin H1N1 (Hsw1N1) virus in the swine population of Japan and antigenic analysis of the isolates. J Gen Virol. 1982 Sep;62(Pt 1):171–175. doi: 10.1099/0022-1317-62-1-171. [DOI] [PubMed] [Google Scholar]
- Newton S. E., Air G. M., Webster R. G., Laver W. G. Sequence of the hemagglutinin gene of influenza virus A/Memphis/1/71 and previously uncharacterized monoclonal antibody-derived variants. Virology. 1983 Jul 30;128(2):495–501. doi: 10.1016/0042-6822(83)90277-5. [DOI] [PubMed] [Google Scholar]
- Pensaert M., Ottis K., Vandeputte J., Kaplan M. M., Bachmann P. A. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ. 1981;59(1):75–78. [PMC free article] [PubMed] [Google Scholar]
- Rota P. A., Rocha E. P., Harmon M. W., Hinshaw V. S., Sheerar M. G., Kawaoka Y., Cox N. J., Smith T. F. Laboratory characterization of a swine influenza virus isolated from a fatal case of human influenza. J Clin Microbiol. 1989 Jun;27(6):1413–1416. doi: 10.1128/jcm.27.6.1413-1416.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Bürger H., Kistner O., Shortridge K. F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985 Dec;147(2):287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Rohde W., Von Hoyningen V., Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978 Jun 1;87(1):13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- Schultz U., Fitch W. M., Ludwig S., Mandler J., Scholtissek C. Evolution of pig influenza viruses. Virology. 1991 Jul;183(1):61–73. doi: 10.1016/0042-6822(91)90118-u. [DOI] [PubMed] [Google Scholar]
- Shortridge K. F., Cherry A., Kendal A. P. Further studies of the antigenic properties of H3N2 strains of influenza A isolated from swine in South East Asia. J Gen Virol. 1979 Jul;44(1):251–254. doi: 10.1099/0022-1317-44-1-251. [DOI] [PubMed] [Google Scholar]
- Shortridge K. F., Webster R. G., Butterfield W. K., Campbell C. H. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977 Jun 24;196(4297):1454–1455. doi: 10.1126/science.867041. [DOI] [PubMed] [Google Scholar]
- Sleigh M. J., Both G. W., Underwood P. A., Bender V. J. Antigenic drift in the hemagglutinin of the Hong Kong influenza subtype: correlation of amino acid changes with alterations in viral antigenicity. J Virol. 1981 Mar;37(3):845–853. doi: 10.1128/jvi.37.3.845-853.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. L., Katz J. M., Webster R. G. Extensive heterogeneity in the hemagglutinin of egg-grown influenza viruses from different patients. Virology. 1989 Jul;171(1):275–279. doi: 10.1016/0042-6822(89)90538-2. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Yamashita M., Krystal M., Fitch W. M., Palese P. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology. 1988 Mar;163(1):112–122. doi: 10.1016/0042-6822(88)90238-3. [DOI] [PubMed] [Google Scholar]
- Zhdanov V. M., Petrov N. A., Grinev A. A., Iakhno M. A., Isachenko V. A. Pervichnaia struktura gemaggliutinina virusov grippa A (H3N2), izolirovannykh v SSSR v 1985 g. Vopr Virusol. 1989 Mar-Apr;34(2):155–160. [PubMed] [Google Scholar]


