Abstract
Determination of the molecules that regulate astrocyte development has been hindered by the paucity of markers that identify astrocytic precursors in vivo. Here we report that the chondroitin sulfate proteoglycan aggrecan both regulates astrocyte development and is expressed by embryonic glial precursors. During chick brain development, the onset of aggrecan expression precedes that of the astrocytic marker GFAP and is concomitant with detection of the early glial markers GLAST and glutamine synthetase. In co-expression studies, we established that aggrecan-rich cells contain the radial glial markers nestin, BLBP and GLAST and later in embryogenesis, the astroglial marker GFAP. Parallel in vitro studies showed that ventricular zone cultures, enriched in aggrecan-expressing cells, could be directed to a GFAP -positive fate in G5-supplemented differentiation media. Analysis of the chick aggrecan mutant nanomelia revealed marked increases in expression of the astrocyte differentiation genes GFAP, GLAST and GS in the absence of extracellular aggrecan. These increases in astrocytic marker gene expression could not be accounted for by changes in precursor proliferation or cell death, suggesting that aggrecan regulates the rate of astrocyte differentiation. Taken together, these results indicate a major role for aggrecan in the control of glial cell maturation during brain development.
Keywords: aggrecan, astrocyte, nanomelia, gliogenesis, precursors, extracellular matrix
INTRODUCTION
The environment in which neurons, glial cells and their precursors proliferate, migrate, differentiate and survive becomes progressively more complex as the central nervous system (CNS) develops. A key component of this complexity, and one increasingly recognized as a regulator of cell behavior, is the extracellular matrix (ECM). Chondroitin sulfate proteoglycans (CSPGs), the most abundant proteoglycans in the CNS, are a major component of brain ECM. To date, study of CSPG function has focused principally on their roles in late events in brain development, particularly axon outgrowth (Brittis and Silver, 1995; Faissner et al., 1994), and on their importance for axon regeneration in the mature brain (Johnson, 1993; Morgenstern et al., 2002). For example, in studies of the dopaminergic nigrostriatal system and spinal cord, degradation of CSPG glycosaminoglycan side chains by chondroitinase enhances axon regeneration in vivo (Barritt et al., 2006; Bradbury et al., 2002; Cafferty et al., 2007; Moon et al., 2001; Yick et al., 2000), and in related work, release of CSPGs from their anchorage in the extracellular matrix by hyaluronidase treatment has been shown to enhance local sprouting of cut axons (Moon et al., 2003). By contrast, even though CSPGs are also abundant in embryonic brain, their possible contribution to neural patterning and cell type differentiation has received much less attention.
The major CSPGs expressed during brain development are the aggrecan family members neurocan, brevican, versican and aggrecan. These proteoglycans share an N-terminal IgG-like globular domain that binds hyaluronan, a central chondroitin sulfate domain and a C-terminal globular domain that may be important for intracellular trafficking (Domowicz et al., 2000). Previous studies of CSPG expression during CNS development have found that neurocan is expressed by neurons and astrocytes (Margolis and Margolis, 1997), versican is found in oligodendrocyte precursors and neurons (Asher et al., 2002; Lebaron, 1996; Schmalfeldt et al., 2000; Yamagata and Sanes, 2005) and brevican is enriched in cells in the embryonic ventricular zone (Jaworski et al., 1995). To date, no brain-specific developmental phenotypes due to the loss of aggrecan family CSPGs have been reported (Hartmann and Maurer, 2001; Rauch et al., 2005). Perhaps the most striking finding in this regard has been the absence of any obvious effect on early neural development from quadruple gene targeting of neurocan, brevican and tenascin C and R (Brakebusch et al., 2002; Hartmann and Maurer, 2001; Rauch et al., 2005; Zhou et al., 2001)
Among the CSPGs in brain, aggrecan exhibits the most dynamic pattern of developmental gene regulation. In chick embryos, aggrecan mRNA expression brain is detected by embryonic days 6–7 (E6–7), peaks at E14–15 and is nearly extinguished by hatching (Krueger et al., 1992; Li et al., 1996). Biochemical analysis of brain aggrecan has shown that it is distinct from the aggrecan of cartilage, which contains keratan sulfate and has a larger hydrodynamic size, probably due to a higher content of chondroitin sulfate (Krueger et al., 1992; Li et al., 1996). Functional studies of aggrecan to date have focused upon cartilage development. Spontaneously occurring aggrecan mutants, nanomelia in chickens and cmd in mouse, have profound defects in skeletal growth (Kimata et al., 1981; Landauer, 1965; Rittenhouse et al., 1978). Brain defects in aggrecan-deficient lines have not been previously analyzed in part because mutants die at birth. However, one in vitro study of chick forebrain cultures has found that dissociated nanomelic cells on poly-lysine plates form fewer and smaller spontaneous neuronal aggregates than do wild type cells (Domowicz et al., 2003). This observation raises the possibility that, unlike neurocan and brevican, aggrecan may have an important role in early neuronal development.
We report here our observations on the development and cellular expression of brain aggrecan and the functional consequences of aggrecan deficiency studied in the nanomelic chick. Our findings establish that aggrecan is expressed in glial precursors and is required for normal astrocyte differentiation.
MATERIAL AND METHODS
Wild type (wt) fertilized White Leghorn chicken eggs were purchased from Sharp Sales (West Chicago, IL). Fertilized eggs from heterozygous nanomelic (+/nm) crosses were provided by Dr Louis Pierro and Ms Karen Morè of the Department of Animal Genetics, University of Connecticut (Storrs, CT). Eggs were incubated at 37.9 °C at 60% humidity in a Midwest incubator with automatic egg turning and regularly hatched at 21-days of incubation. Conformance of the eggs to a 21-day incubation period to hatching was periodically confirmed. In all experiments, both White Leghorn embryos and embryos of normal progeny from the nanomelic heterozygous crosses (flock mates, fm) were tested as controls. No differences between these two set of controls were found in the levels, distributions and developmental regulation of any messages assayed by in situ hybridization or Northern blotting.
mRNA in situ hybridization
Heads from E3–E19 chick embryos were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) and the harvested brains were processed for whole mount in situ hybridization as described (Grove et al., 1998). For tissue section analysis, brains were sunk in 20% sucrose-10% formalin in PBS, embedded in gelatin and cut at a thickness of 40 microns on a sledge microtome. Sections were mounted on SuperFrost/Plus microscope slides and processed for non-radioactive in situ hybridization. Sections of E12 and younger brain tissue were permeabilized by two 15 minute treatments with a detergent mixture of 1%NP-40, 1%SDS, 0.5% deoxycholate in 1mM EDTA/150 mM NaCl/50 mM Tris-HCl (pH 8). Older tissue was digested for 15 minutes with 1 microgram/ml proteinase K in PTw (PBS with 1% Tween-20). Slides were then rinsed twice in PTw, post-fixed for 15 minutes in 4% paraformaldehyde-PTw, and rinsed in PTw. Hybridizations with digoxigenin (DIG)-labeled riboprobes were conducted overnight at 70 °C in 50% formamide, 5xSSC, 1% SDS, with tRNA, acetylated BSA and heparin added as carriers. Four 30 minute post-hybridization washes were performed at 70 °C in solution X (50% formamide, 2xSSC, 1% SDS). DIG-labeled RNA duplexes were detected with NBT-BCIP histochemistry after incubation with an alkaline phosphatase/anti-DIG Fab conjugate.
Two-color fluorescent in situ hybridization (FISH) was performed on E9 chick brain coronal sections with FITC- and DIG-labeled riboprobes. Sections were permeabilized by treatment with proteinase K (1µg/ml) for 20 minutes. After post-hybridization washes, bound FITC was detected with a peroxidase-conjugated anti-FITC antibody (Perkin Elmer) and incubation with the peroxidase substrate FITC-tyramide. Following inactivation at 70°C, the second probe was localized with a peroxidase-conjugated anti-DIG antibody and Cy5-tyramide (Roche). Fluorescence was studied with a Leica SP2 spectral confocal microscope in the BSD Digital Light Microscopy Core Facility of the University of Chicago Cancer Research Center.
cDNAs
Aggrecan gene expression was analyzed with riboprobes derived from a 690 bp fragment of exon 12 of the chicken aggrecan gene, which encodes the chondroitin sulfate domain of the aggrecan core protein and bears no sequence identity to other aggrecan family members. cDNAs for GLAST/SLC1A3 (nt 1053–1765 of XM425011), BLBP/FABP7 (nt 46–582 of NM205308), nestin/transitin/NES (nt 130–617 of NM205033), FGFR3/cek2 (nt 1164–1491 of NM205509) and PTGS2/COX2 (nt1013–1547 of M64990) were obtained by PCR from a chick E12 brain cDNA using specific primers based on deposited sequence. Brevican/BCAN (2034–2731 of XM423655) and GFAP (621–1146 of XM418091) cDNA fragments were generated using degenerate primers based on the sequences of the mammalian orthologs. The degenerate oligonucleotides were designed with the aid of the Blocks and CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primers) programs (Rose et al., 1998). All isolated PCR products were ligated into the pCRII-dual promoter vector (Invitrogen) for riboprobe synthesis. Glutamine synthetase, proteolipid protein 1, tenascin C and class III beta-tubulin were studied with cDNAs provided by W.-K. Chen (GLUL), J. Thomas (PLP1), R. Chiquet-Ehrismann (TNC) and D. Cleveland (TUBB3).
Northern blots
Total RNA was extracted from wt and nm brains using Trizol reagent (Invitrogen). RNA (10µg per lane) was electrophoresed in a denaturing 1.2% agarose-formaldehyde gel and transferred to Nytran SuperCharge (Schleicher and Schuell) membranes by TurboBlotter rapid downward transfer. Hybridization was performed using QuikHyb solution (Stratagene). Radiolabeled probes were prepared using the Rediprime system (Roche). Following hybridization, the nylon membranes were washed twice with 2X SSC/0.05% SDS for 30 minutes at room temperature and twice with 0.1X SSC/ 0.1% SDS for 30 minutes at 58 °C. Autoradiography was performed overnight at −70°C with Kodak MR2 film. Bands were quantified with the Quantity One image analysis software (Bio-Rad). Blots were stripped by boiling in 0.1X SSC/ 0.1%SDS and re-probed. GAPDH mRNA level was used as the internal standard for loading differences.
Immunostaining and proliferation
After mRNA in situ hybridization, sections were stained with the monoclonal antibody S103L that recognized the core protein of chick aggrecan (Krueger et al., 1990) using a peroxidase conjugated anti-rat IgG secondary antibody. Peroxidase activity was then detected using the ImmunoHisto peroxidase detection kit (Pierce). For immunocytochemistry after mRNA FISH, sections were permeabilized for 40 minutes with a detergent mixture consistent of 1%NP-40, 1%SDS, 0.5% deoxycholate in 1mM EDTA/150 mM NaCl/50 mM Tris-HCl (pH 8) and with proteinase K (1µg/ml) for 7 minutes at room temperature. Anti-GFAP (Chemicon) and anti-BLBP (Chemicon) were used as primary antibodies at a 1:500 dilution. For proliferation assays, BrdU (1.6 mg/100 µl) was applied to the vitelline membrane of E18 eggs 3h prior to tissue harvest. Tissue was then fixed, embedded, sectioned and processed for immunocytochemistry using an anti-BrdU IgG (BD Bioscience) primary antibody and alkaline phosphatase conjugated anti-rabbit IgG (Roche, Germany) secondary antibody. As an alternative method to determine mitotic levels, the mitotic nuclei were identified by immunocytochemistry with anti-phospho-histone H3 (Ser10) polyclonal antibody (pHH3) (Upstate Biotechnology) in paraffin embedded sections from E15 and E18 cerebellum. A FITC-conjugated anti-rabbit IgG was used as secondary antibody and DAPI was utilized to counterstain the nuclei. Positive BrdU and pHH3 mitotic nuclei from two brains were counted in cerebellar white matter using ImageJ software and data was analyzed for statistical significance using the Student’s t-test.
Ventricular zone cultures
Midbrain ventricular zone (VZ) was dissected from E11 chick midbrain and mechanically dissociated into single cells. Cells were plated on Primaria dishes (Becton-Dickinson) in 10% fetal calf serum/DMEM with EGF (40ng/ml) and bFGF (40ng/ml). Some cultures were switched to F12/DMEM media supplemented with G5 and 1.5 µM trans-retinoic acid after seven days in vitro. All cultures were harvested for study nine days after plating. Tissue culture reagents were purchased from Invitrogen.
RESULTS
Aggrecan gene expression in chick embryogenesis
Aggrecan mRNA in brain was first detected by in situ hybridization at E6 (HH st 30). By early E7, aggrecan message, restricted to the ventricular zone (VZ), presented a richly patterned distribution, with lateral and medial longitudinal stripes in hindbrain, tufts in midbrain and diencephalon, and a focus in rostroventral telencephalon marking the prospective olfactory bulb (Fig. 1). Over the next day, AGC brain expression levels increased, revealing rhombomeric territories in hindbrain, arcuate territories in ventral midbrain and bands in forebrain. Superimposed on these patterns were strong rostral/high-caudal/low gradients of AGC levels in telencephalon and, separately, in midbrain (Fig. 1A). By E8, AGC gene expression was no longer restricted to the VZ, but included aggrecan-rich cells in hindbrain and near the mesodiencephalic junction that appeared to be migrating from the VZ deep into the mantle layer (Fig. 2A).
Figure 1.

Aggrecan wholemount gene expression demonstrates that a patterned organization to the chick brain VZ continues after neurogenesis is largely complete. A, Midline cut displays heterogeneities in aggrecan mRNA distributions in the right brain VZ of the E8 chick embryo. Dorsal is to the top and rostral is to the left. Arrow marks the prospective olfactory bulb. B, C, Dorsal view of late E7 (B) and E8 (C) hindbrains oriented with rostral to the top. Arrow in B points to labeling in rhombomere 4. D, Dorsal view of E8 ventral midbrain, with the prospective midbrain tectum resected. Arrows point to arcuate territories in VZ gene expression patterns. Arrowhead identifies dense labeling of the isthmus. di, diencephalon; hb, hindbrain; mb, midbrain; ob, obex; tel, telencephalon;
Figure 2.
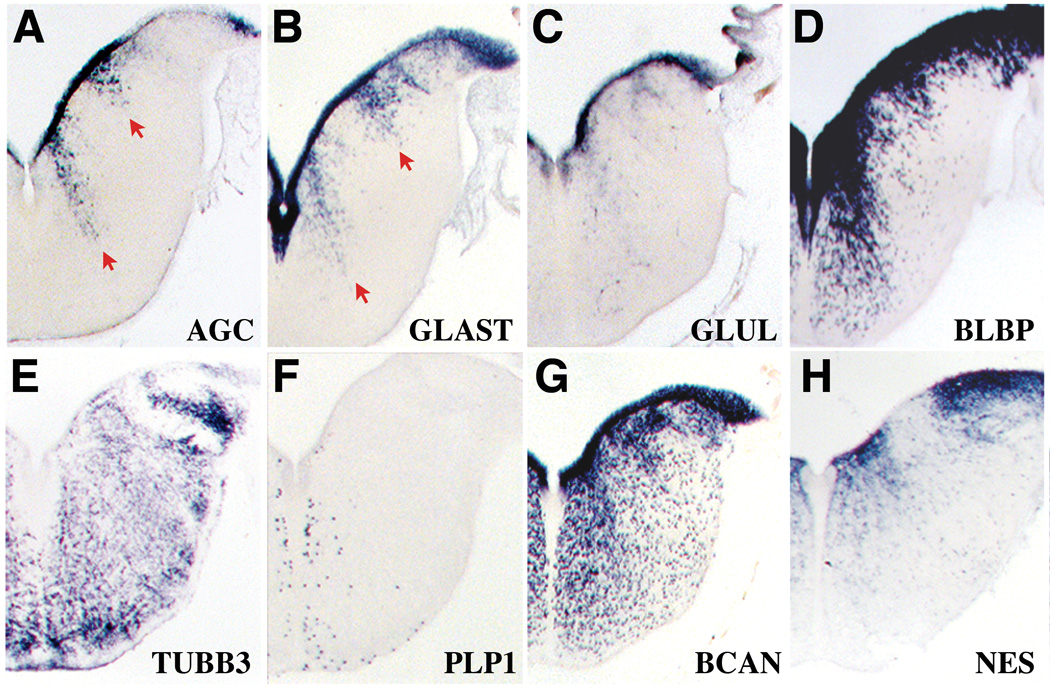
Medial and lateral streams of aggrecan-rich cells in E8 hindbrain cross-sections follow a pattern seen with astroglial markers and not markers of postmitotic neurons or oligodendrocytes. A, Aggrecan gene expression, showing labeled cells (arrows) that appear to be migrating from the VZ. B–D, Similarly positioned streams (but ones differing greatly in their density of labeling) are identified with the astrocyte markers GLAST (B), GLUL (C) and BLBP (D). E, F, Labeling of VZ and medial and lateral streams are not seen with the neuronal marker class III beta-tubulin/TUBB3 (E) or the oligodendrocyte marker PLP1 (F). G, H, Gene expression distributions of the intermediate filament nestin/NES (F) and the aggrecan family member brevican/BCAN (H) are more extensive than those of aggrecan but include key features such as strong VZ labeling. No hybridization signal for the intermediate filament gene GFAP (glial fibrillary acidic protein) was detected at this stage (not shown).
Although the complex pattern of initial AGC gene expression could suggest a role for the proteoglycan in neuronal patterning, the relatively late onset and initial VZ restriction of AGC raises the intriguing possibility of involvement in gliogenesis. An astroglia association was further suggested by comparisons of E8 hindbrain AGC cross-sections with cross-sections hybridized with markers of neurons and glia (Fig. 2). The E8 AGC hindbrain pattern of medial and lateral streams of cells extending from the ventricle (Fig. 2A) was paralleled by the gene expression distributions of the radial glial marker GLAST (high affinity, sodium-dependent L-glutamate/L-aspartate transporter) (Hartfuss et al., 2001; Shibata et al., 1997). Partially overlapping but clearly similar medial and lateral streams were also demonstrated with the glial markers glutamine synthetase (GLUL), brain lipid binding protein (BLBP), FGF receptor 3 (FGFR3) and nestin (NES) (Fig. 2C, D and H; Fig. S1) (Hemken et al., 1997; Lee and Cole, 2000; Napier et al., 1999; Pringle et al., 2003) and another member of the aggrecan family, brevican (Fig 2 G). In contrast, labeling of the VZ and the medial and lateral streams was not seen with markers of postmitotic neurons (class III beta-tubulin, TUBB3; Fig. 2E) or prospective oligodendrocytes (proteolipid protein-1, PLP1; Fig. 2F).
By E12 AGC mRNA was much more widely distributed in the mantle layer. Its pattern of expression frequently reflected nuclear boundaries, but overlapped the distribution of glial markers rather than neuronal markers when the two markers diverged. Fig. 3 documents in serial sections the similarity of expression territories among AGC, the glial enzyme GLUL and tenascin-C (TNC), an extracellular matrix molecule expressed by radial glial cells, astroglial precursors (Bartsch et al., 1995; Yuasa, 2001), and cultured oligodendrocyte precursors (Garwood et al., 2004). AGC expression patterns were not, however, identical to those of the glial mRNA markers, suggesting that aggrecan may identify a subset of these cells. Quantitative analysis of brain mRNA demonstrated that relative AGC levels peak around E15 and decline sharply thereafter (Fig. 4). Comparison of the AGC temporal profile with that of a battery of glial markers suggests that any glial role for AGC would involve glial precursor development. The rise in AGC levels from E8 to E15 closely parallels that of GLAST, and precedes the upregulation of GLUL expression and the expression onset of the mature astroglial marker GFAP. From E18 to E21, when AGC levels are dropping, the glial marker mRNAs GLAST, GLUL and GFAP are attaining their maximal levels of expression, which are maintained in the juvenile chick brain. The low level of hatchling brain AGC may reflect a residual role for aggrecan in glial function. Alternatively, it could reflect a shift in the cellular expression of AGC mRNA to neurons. In support of the latter possibility, postnatal expression of aggrecan in the mouse has been associated with perineuronal nets and neuronal nuclei (Carulli et al., 2007; Dino et al., 2006; Matthews et al., 2002; McRae et al., 2007; Popp et al., 2003) and we have preliminary evidence for a neuronal pattern of AGC gene expression in the hatchling hindbrain (not presented).
Figure 3.
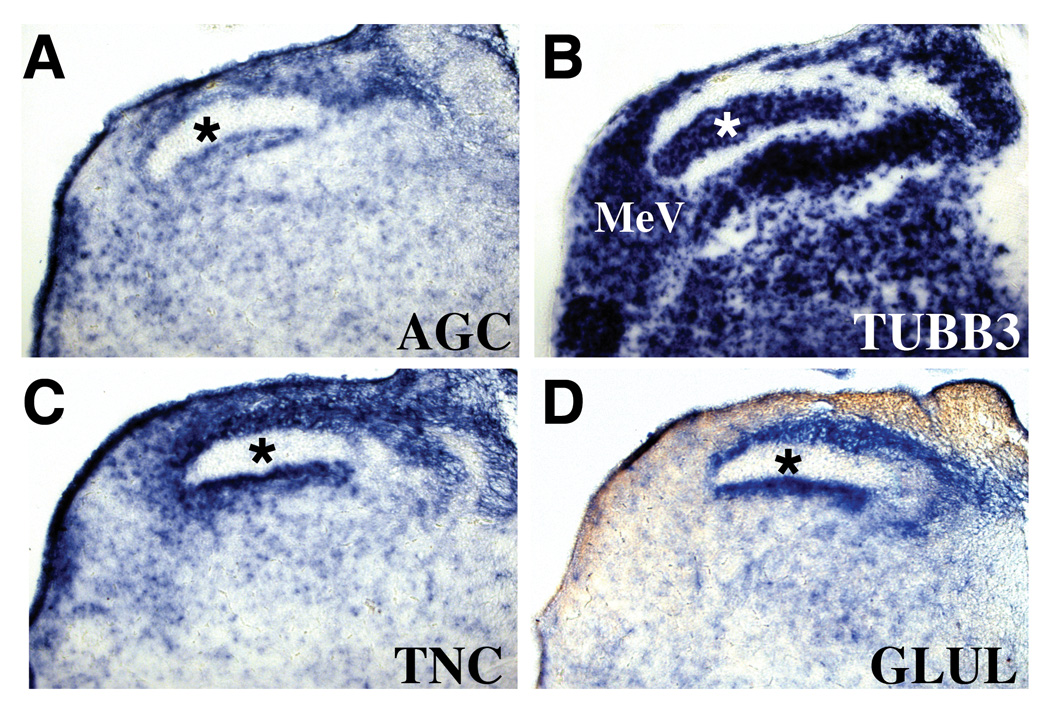
Aggrecan gene expression follows the distributions of glial markers in auditory relay nuclei of the chick E12 hindbrain. A–D, Serially adjoining cross-sections probed for the expression of aggrecan/AGC (A), the neuronal marker TUBB3 (B), and the glial markers tenascin-C/TNC (C) and glutamine synthetase/GLUL (D). Asterisk in B marks the nucleus magnocellularis, which does not express AGC and is free of the astrocyte markers TNC and GLUL. MeV, medial vestibular nucleus.
Figure 4.

Time-series histogram depiction of quantitative Northern blot data on brain aggrecan (AGC) levels compared with those of a panel of early and late astroglial markers (GLAST, GLUL, GFAP). Blots of total RNA isolated from whole brains at the indicated ages were hybridized with message-specific [32P]-labeled cDNA probes. Glial RNA levels relative to those of GAPDH were calculated with Quantity One software from Bio-Rad.
These gene expression data document a close spatiotemporal association in mid-embryogenesis between AGC-expressing cells and cells harboring the glial markers GLAST, GS, BLBP, NES and tenascin C. The observation that all these mRNA patterns differ from each other, at least in part, indicated a clear heterogeneity among these cells, with subpopulations differing in their patterns of differentiation and perhaps also in their functions. Faced with this heterogeneity we sought direct evidence that AGC-rich cells also express glial identity markers.
Aggrecan-expressing cells are in the glial lineage
Cellular co-expression experiments on E8 chick brainstem sections established that some cells expressing mRNA markers of glial identity also contain aggrecan protein. For these preparations, we employed the species-specific monoclonal antibody S103L, which recognizes an epitope in the chondroitin-sulfate-binding region of the aggrecan core protein (Krueger et al., 1990). On tissue sections, S103L immunochemistry strongly stained intracellular Golgi-associated aggrecan core protein and diffusely labeled tracts where aggrecan-expressing cells with migratory morphology are found. Strong S103L staining was found within cells expressing the glial lineage markers nestin (Fig. 5A) and GLAST (Fig. 5B), although the apparent subcellular localization of the mRNAs and the aggrecan core protein was often distinct.
Figure 5.

Aggrecan protein and message are enriched in cells expressing glial markers. A, B, E8 chick midbrain cross-sections double-labeled for aggrecan immunoreactivity (brown) and nestin/NES and GLAST mRNAs (blue). Arrows point to cells that both contain intracellular aggrecan core protein and express the glial marker transcripts. C, Confocal XY projections of E9 hindbrain cross-sections processed for fluorescent in situ hybridization (FISH) for aggrecan/AGC mRNA (green) and for immunohistological localization of the glial marker BLBP (red). D, Confocal XY projections of FISH for aggrecan/AGC mRNA (green) and immunostaining for the astrocyte marker GFAP (red) from E13 midbrain cross-sections. Confocal overlay projections identify cells expressing aggrecan and the radial glial marker or the astrocyte marker (arrowheads). ZY projections of the merged pictures through the marked axes (white lines) are shown in the left side panels.
Despite the Golgi association of the aggrecan immunoreactivity, we were concerned that the immunolabeling we saw could have been glial accumulation of extracellular aggrecan protein. We addressed this possibility in two ways. First, control sections hybridized with an AGC probe and immunostained with S103L antibody demonstrated many co-localization profiles (Schwartz and Domowicz, 2004). Second, we carried out two-color fluorescent in situ hybridization combined with confocal imaging to test for cellular co-expression. Z projections of cells doubly labeled to detect AGC and either GLAST mRNA or BLBP protein confirmed co-expression of AGC and the glial markers (Fig. 5C; Fig. S2). Finally, at later stages (E13) when GFAP is abundant, we could detect AGC-positive cells expressing GFAP protein (Fig 5D). Importantly, though, aggrecan message was not found in all GLAST- or BLBP-positive cells, confirming that it is a subpopulation of radial glial cells that expresses aggrecan.
We next developed a culture system to explore whether populations of aggrecan-expressing cells have gliogenic potential. We harvested VZ tissue from E11 chick midbrain, a source rich in AGC-expressing cells (Fig. S3). Dissociated cells plated in DMEM supplemented with EGF and bFGF, divided rapidly and established a monolayer of AGC-expressing cells (Fig. 6). This population also expressed NES and BLBP (not shown), but did not express markers of postmitotic neurons (TUBB3) or differentiated glial cells (PLP1, GFAP). After seven days in vitro, we switched the medium to a glial differentiation formula (DMEM/F12 supplemented with G5 and 1.5 µM trans-retinoic acid). Two days later, we found a marked increase in cells expressing the glial markers GS, GLAST and GFAP. We also saw a sparse population of PLP1-positive cells with morphologies consistent with those of oligodendrocytes precursors. As expected, no cells labeled with the neuronal marker TUBB3 were detected (Fig. 6). Since DNA levels increased 75% during the two days of culture in glial-differentiation media and no morphological evidence of cell death was observed (data not shown), we conclude that aggrecan-expressing cells in vitro have the ability to differentiate into glia.
Figure 6.
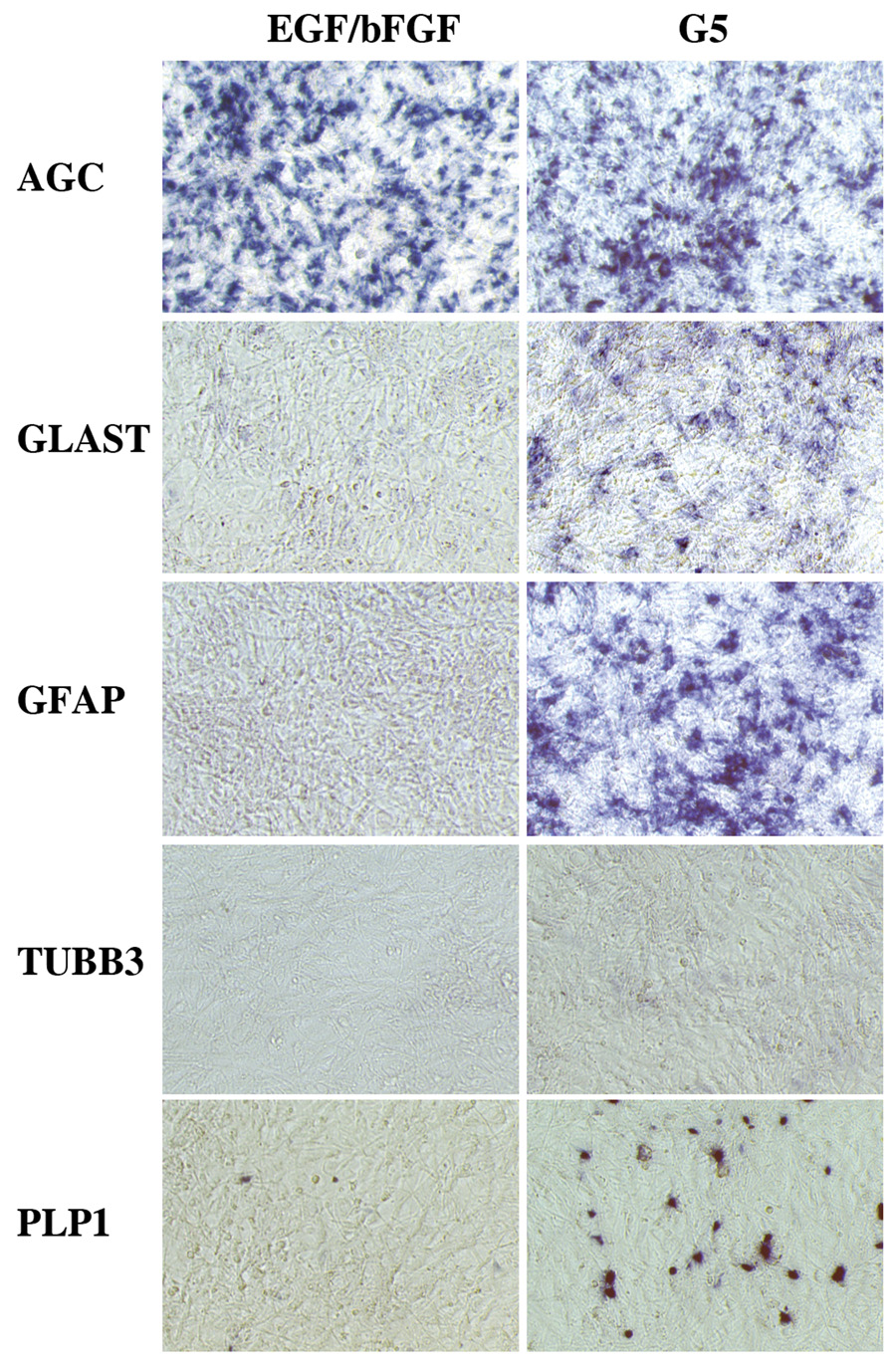
E11 midbrain VZ cultures enriched in aggrecan-expressing cells have gliogenic potential. Left panels: Gene expression profiling by in situ hybridization of VZ cell cultures harvested after nine days in vitro in DMEM/10% fetal calf serum with 40ng/ml EGF and 40ng/ml bFGF added. This preparation yielded bulk cultures of aggrecan-expressing cells (AGC) free of differentiated astrocytes (GLAST, GFAP), neurons (TUBB3) or oligodendrocytes (PLP1). Right panels: Parallel cultures shifted after seven days in EGF/bFGF medium to F12/DMEM with G5 and 1.5 µM trans-retinoic acid for two days. This shift elicited the induction of the glial markers GLAST, GFAP and PLP1 in the aggrecan-rich cultures. In situ hybridization demonstration of culture gene expression was carried out directly on the Primaria culture dishes on which the cells were grown.
An aggrecan requirement for normal glial development
Nanomelia (nm) is a mutant chicken line deficient in aggrecan. Molecular studies have identified an aggrecan gene frame-shift mutation that produces a truncated aggrecan core protein that is unable to mature through the secretory pathway (Domowicz et al., 1996; Domowicz et al., 2000; Li et al., 1993; Vertel et al., 1994). Nanomelic homozygotes do not hatch. At E17 nm brains had mild hydrocephaly of the lateral ventricles and a 10% reduction in wet brain weight compared to that of control flock mates (data not shown). We studied nanomelic brain development to test for a requirement for aggrecan function in gliogenesis.
We first assessed quantitative mRNA levels for glial markers at E20, by which time many astrocyte markers have reached their maximal levels (Fig. 4). Two-fold increases in GFAP and GS RNA and a 30% increase in GLAST RNA were found in nanomelic brain total RNA extracts relative to those prepared from flock mate controls. By contrast, no changes were recorded in the levels of the oligodendrocyte marker PLP1 or the neuronal marker TUBB3 (Fig. 7).
Figure 7.
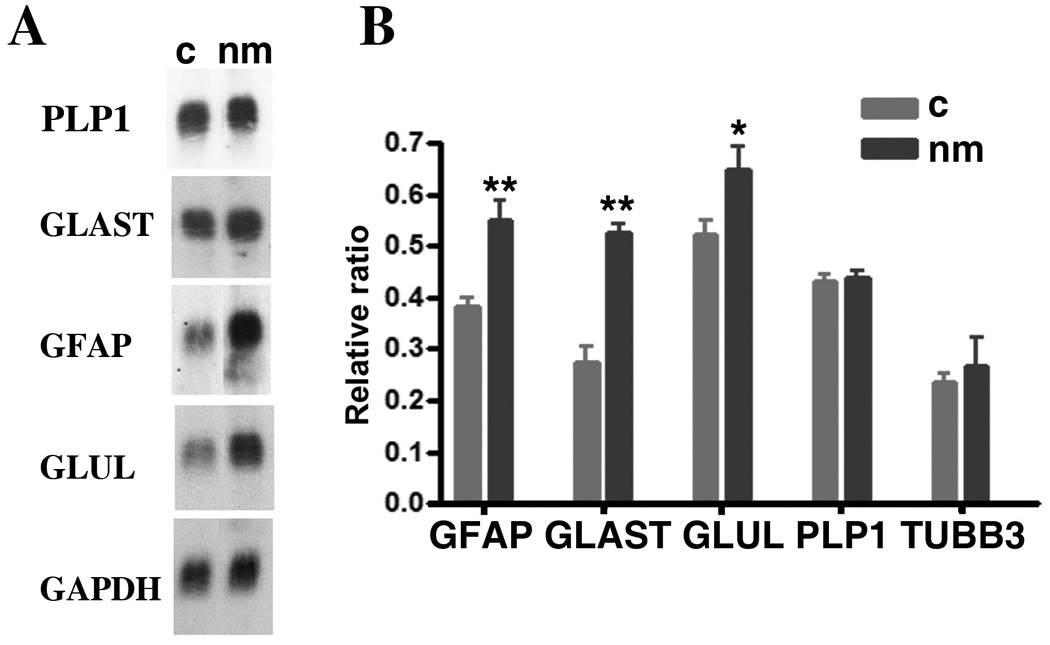
Elevation of astroglial gene expression levels in chick embryos lacking functional aggrecan. A, Blots of total brain RNA isolated from flock mate (c, left column) and nanomelic (nm, right column) E20 embryos were hybridized with the [32P]-labeled cDNA probes for GLUL, GFAP, GLAST and PLP1. B, In a separate experiment, mRNA levels relative to those of GAPDH were calculated for triplicate independent brain samples from flock mate (c) and nanomelic (nm) E18 embryos. ** p<0.01; *p<0.04 by Student’s t-test analysis.
In situ hybridization histology on E17–E21 nanomelic brains also demonstrated increased levels of the astrocyte mRNAs GFAP, GS and GLAST in all brain regions examined (Fig. 8, Fig. 9 and data not shown). That increased GFAP protein expression accompanied the GFAP message rise was shown by immunocytochemistry (Fig 9 C, F). Interestingly, the distribution pattern of GFAP was also affected. As illustrated in Fig. 9 for the E18 optic tectum, GFAP mRNA was denser in the VZ but reduced at the pial surface in nanomelic brains (Fig. 9 B, E). At earlier ages (E12–E15), however, the levels and patterns of mRNA expression for the glial markers GLAST, GS, NES and GFAP appeared unchanged in nanomelic brains (data not shown).
Figure 8.
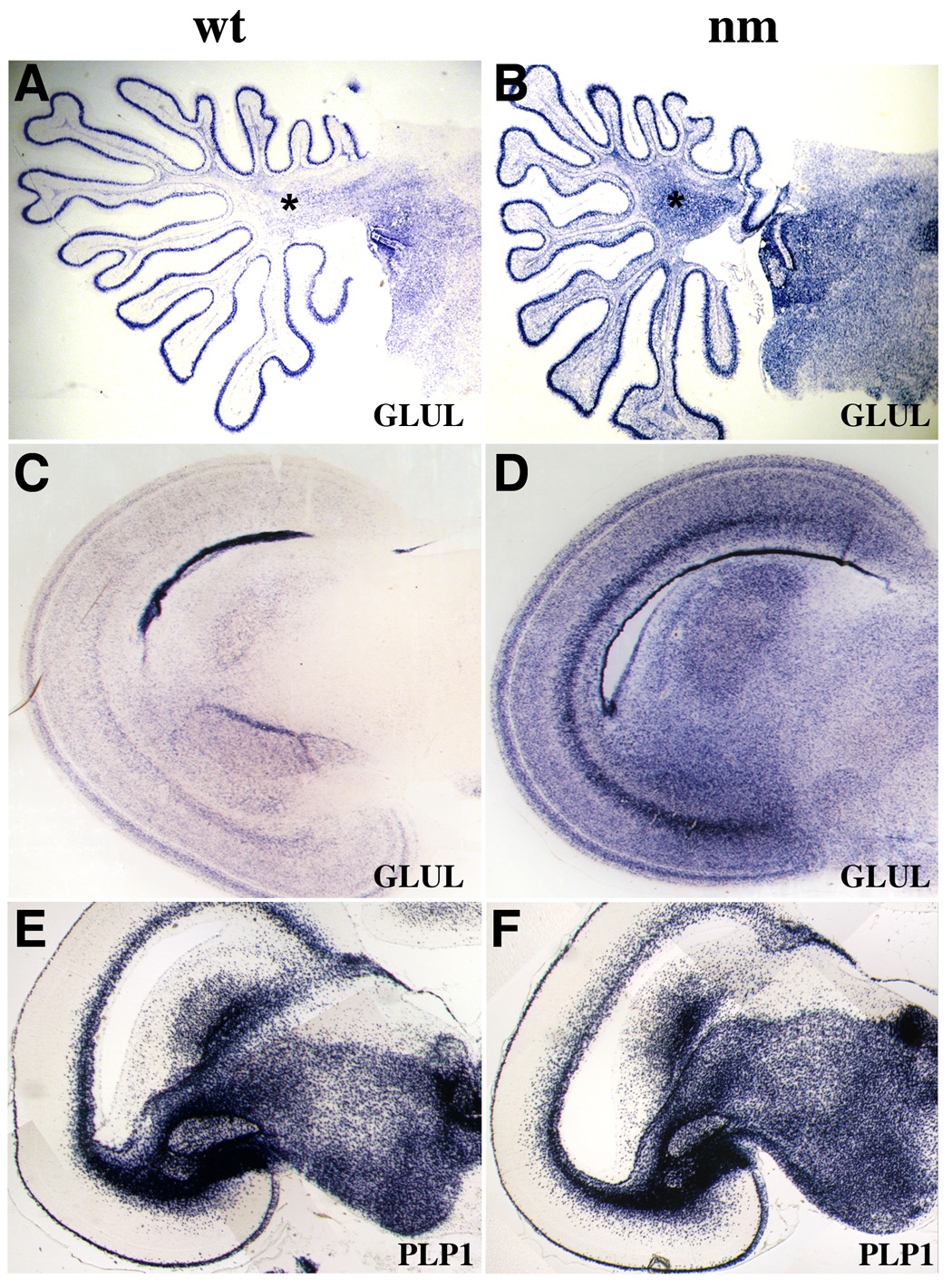
In situ hybridization experiments demonstrate clear upregulation of astrocyte-specific gene expression in nanomelic chick embryos. A–D, GLUL gene expression is increased in E20 nanomelic brains as demonstrated in sagittal sections of cerebellum (A, B) and coronal sections of midbrain (C, D). Asterisk in A marks the territory of cerebellar white matter studied in Fig. 11. E, F, No change is seen in the expression of the astrocyte marker PLP1 in cross-sections from E20 midbrain.
Figure 9.
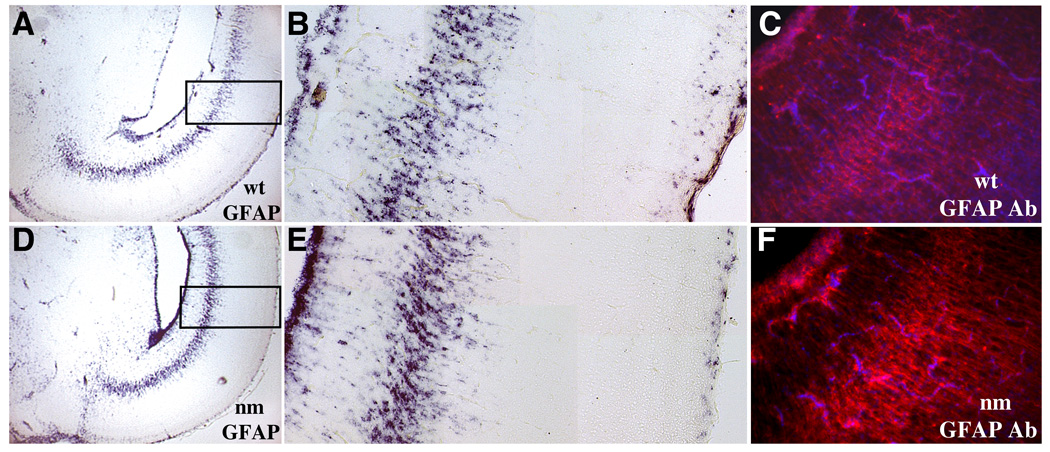
Spatial distributions of the astrocyte marker GFAP are altered in nanomelic embryonic brains. Illustrated are GFAP in situ hybridization experiments on coronal sections from wild type (A–C) and nanomelic (D–F) E18 midbrain. Boxes in A and D indicate areas of optic tectum presented at higher magnification in B and E, respectively. Note that levels of GFAP at the VZ are increased (left side, B and E) and those near and at the pial surface are decreased (right side, B and E) in the nanomelic (E) as compared with the wt control (B) tissue. Immunocytochemistry with GFAP antibody confirmed increased expression of GFAP protein in the nanomelic midbrain (F) compared with wild type (C). Sections were counter-stained with DAPI.
The late embryonic increase in astrocytic gene expression could have been due to a reactive gliosis secondary to the nm hydrocephaly or some other indirect trauma to the nanomelic brain. To address this possibility, we tested for the microglial activation that accompanies the reactive astrocytic response to injury. Northern blot analysis of nm and control brains showed that levels of the microglial marker PTGS2/COX2 in E18 nm brains were unchanged from those of controls (not shown). It was also possible that the nm embryos had cerebrovascular abnormalities that might affect astrocytes whose end-feet surround the endothelial cells forming the blood-brain barrier. We imaged the brain circulatory system in E18 chick embryos by intra-cardiac injection of 100 µl of fluorescein-albumin (2.5mg/ml) five minutes before sacrifice. Fluorescence microscopy analysis of nm brain sections showed intravascular distributions of fluorescein-albumin similar to those seen in wild type brain sections, indicating that no major disruption of the blood-brain barrier occurs in the mutant (not shown). Finally, we repeated the culturing of VZ cells from E11 nm and normal midbrains. Nanomelic cultures showed dramatic upregulation of astrocyte differentiation markers compared to the normal cultures (Fig. 10 and Fig. S4). These data argue that the astrocytic marker up-regulation seen in nanomelic brains is not a result of post-E11 systemic trauma and further supports our hypothesis that extracellular aggrecan exerts a regulatory effect on astrocyte differentiation.
Figure 10.

E11 midbrain VZ cultures from nanomelic (nm) embryos exhibited elevated astroglial gene expression with respect to flock-mate control (c) levels. Left panels: Expression of AGC and GFAP detected by in situ hybridization in VZ cell cultures harvested after nine days in vitro in DMEM/10% fetal calf serum with 40 ng/ml EGF and 40 ng/ml bFGF added. Right panels: Expression of AGC and GFAP detected by in situ hybridization in parallel VZ cultures maintained for seven days in EGF/bFGF medium then shifted to F12/DMEM with G5 and 1.5 µM trans-retinoic acid for two days.
The increase in astrocyte gene expression in nm brains could be due to an increase in astrocyte number, either from increased cell division or reduced cell death. We looked for increased proliferation by astrocyte precursor cells in two ways. First, mitotic figures in E15 and E18 cerebellar white matter were identified with anti-phosphohistone H3 immunolabeling. No differences in the densities of mitotic figures were found between nanomelic and control brains at either age (Fig 11A). Second, we found no significant increase in the densities of E18 cerebellar white matter cells labeled by BrdU retention three hours after a pulse delivery (Fig 10B). We looked for changes in apoptotic cell death by measuring the levels of caspase 3/7 activity of nanomelic and control brain regions at E12, E15 and E18. No significant differences were seen (Fig. S5). These results indicate that in the absence of native aggrecan, astrocyte gene expression levels, but probably not astrocyte numbers, are elevated in embryonic brain development.
Figure 11.
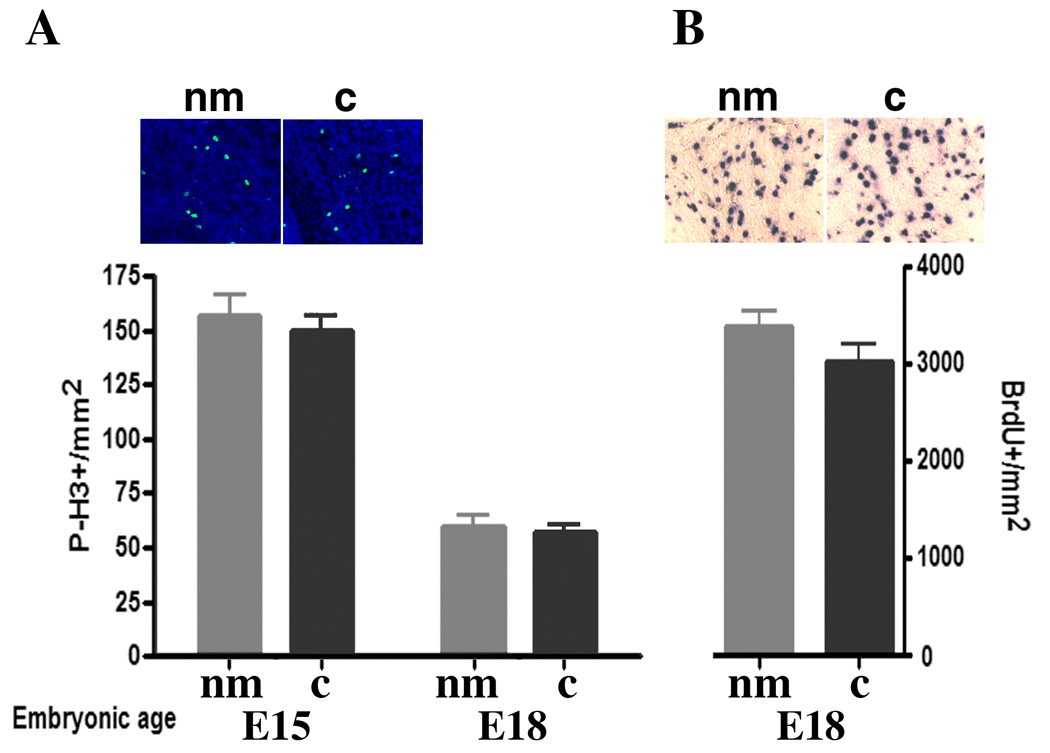
Quantitation of cell proliferation markers in cerebellar white matter suggests that increased cell division cannot account for the increase in astrocyte gene expression levels seen in nanomelia. A, Densities of phosphohistone H3-positive cells at E15 and E18 are unchanged between nanomelics (nm) and flock mate controls (c). B, Densities of BrdU-labeled cells four hours after a BrdU pulse are not significantly increased in E18 nanomelic cerebellar white matter. Insets show representative pictures of E15 phosphohistone H3-positive cells and E18 BrdU-labeled cells from nanomelics (nm) and flock mate controls (c). Student’s t-test analysis indicated no significant difference.
DISCUSSION
Although CSPGs are major components of embryonic brain ECM, their functional role in regulating cell behavior during CNS development remains unknown. We report here several lines of evidence that indicate a role for the CSPG aggrecan in avian astrocyte development: (a) the onset of aggrecan expression in the VZ coincides with the end of neurogenesis and the beginning of gliogenesis in the developing avian brain (Tsai et al., 1981a; Tsai et al., 1981b); (b) aggrecan is co-expressed with the radial glial markers BLBP, GLAST and NES, and later in development is found in GFAP-rich cells; (c) populations of VZ cells expressing aggrecan can differentiate to GFAP-positive cells when cultured in differentiation media; and (d) animals lacking functional aggrecan present marked upregulation of astrocyte gene expression levels.
For astrocytes, determination of the mechanism regulating their development has been hindered by a lack of markers to identify astrocyte precursors in vivo (Liu and Rao, 2004; Zhang, 2001). Our findings add aggrecan to the list of native astrocyte precursor markers. In addition, our observation of partially overlapping patterns of radial glia and astrocyte precursor marker expression suggest the presence of multiple subpopulations of astrocyte precursors as has been proposed for neural precursors (Gal et al., 2006). Astrocytes with radically different functions have been described in adult brain (for example, perineuronal and perivascular astrocytes) (Hartfuss et al., 2001; Haydon and Carmignoto, 2006), but distinctions in astrocyte subclass developmental origin remains an open question. Our results suggest strongly that further astrocyte expression profiling will help delineate subpopulations of functionally distinct astrocytes that may differ in their developmental origins.
The increase in astrocyte marker expression in nanomelic chick embryos could be due to a variety of causes: a rise in glial progenitor numbers, an increase in the rate or degree of astrocyte differentiation, or possibly a reactive gliosis. With respect to an increase in astrocytic marker expression due to trauma, we found that expression of a microglial marker that accompanies the macroglial response in injury (Ridet et al., 1997) does not change in nanomelic brains. Furthermore, cerebrovascular abnormalities or a massive neuronal loss that might trigger an injury response (Johnston et al., 2001; Joseph et al., 1992; Taylor et al., 1999) were not observed. The possibility that aggrecan regulates glial precursor cell division is an important one, since an increased cell-cycle rate has been observed in the nanomelic cartilage, a tissue that normally expresses high levels of aggrecan (Wong et al., 1992). However, no significant increase in cell proliferation could be documented in nanomelic brains using two independent measurements. Our findings therefore favor the conclusion that aggrecan influences astrocyte differentiation independent of astrocyte cell numbers.
Examples in the literature of genetic defects producing increased astrocyte numbers or gene expression levels are limited. FGFR3 knockout mice have increased numbers of astrocytes in the gray matter, suggesting a role for the FGF pathway in astrocyte-precursor populations (Pringle et al., 2003). CD81 null mice develop an increased number of astrocytes and microglia that in turn produce larger brains, possibly due to lack of proliferation control secondary to alterations in contact-inhibition mechanisms (Geisert et al., 2002). Double mutants in the proneural genes Neurogenin2 and Mash1 exhibit premature and excessive generation of astrocytic precursors at the expense of the pool of neuronal precursors (Nieto et al., 2001). Our findings add the CSPG aggrecan to this short list of astrocyte developmental regulators. In particular, our observations in the nanomelic mutant are supported by a recent study showing that non-specific degradation of CSPGs in the VZ alters the self-renewing capacity of radial glia, diminishes neurogenesis and increases astrogenesis (Sirko et al., 2007). These results, together with the finding of late VZ expression of the CSPG brevican (Jaworski et al., 1995), raises the possibility that CSPGs apart from aggrecan may also participate in gliogenesis.
Our findings suggest that a subpopulation of astrocyte precursors regulate astrocyte differentiation through extracellular matrix signaling mechanisms involving aggrecan. How might aggrecan act? One mechanism could be that aggrecan directly regulates the concentration of extracellular growth factors involved in glial differentiation, possibly by sequestration. Notable in this regard is the finding from in vitro studies that heavily sulfated CS chains can bind the midkine family members (midkine and pleiotrophin), and some FGF family members (FGF-1, -2, -16, -18) (Deepa et al., 2002; Umehara et al., 2004). Alternatively, aggrecan may, like other members of the aggrecan family, bind directly to other glial-derived matrix components such as tenascin-C and modulate their activity (Rauch, 2004; Yamaguchi, 2000). Perhaps by shaping the composition of the ECM, aggrecan could affect cell migratory patterns and change the signaling environment that glial precursors encounter. Independent of speculations about mechanism, however, our data provide direct genetic evidence that the composition of the embryonic brain extracellular matrix regulates glial cell differentiation.
Supplementary Material
Acknowledgments
We thank J. Henry, M. Mueller, N. Navarro, J. Olson and N. Wadlington for excellent technical assistance and E. Pirok for helpful discussion. We are grateful to W. K. Chen, J. Thomas, R. Chiquet-Ehrismann and D. Cleveland for providing us with GLUL, PLP1, TNC and TUBB3 cDNAs respectively. This work was supported by U.S. Public Health Service Grants HD009402 and NS35680.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Asher RA, Morgenstern DA, Shearer MC, Adcock KH, Pesheva P, Fawcett JW. Versican is upregulated in CNS injury and is a product of oligodendrocyte lineage cells. J Neurosci. 2002;22:2225–2236. doi: 10.1523/JNEUROSCI.22-06-02225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barritt AW, Davies M, Marchand F, Hartley R, Grist J, Yip P, McMahon SB, Bradbury EJ. Chondroitinase ABC promotes sprouting of intact and injured spinal systems after spinal cord injury. J Neurosci. 2006;26:10856–10867. doi: 10.1523/JNEUROSCI.2980-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch S, Husmann K, Schachner M, Bartsch U. The extracellular matrix molecule tenascin: expression in the developing chick retinotectal system and substrate properties for retinal ganglion cell neurites in vitro. Eur J Neurosci. 1995;7:907–916. doi: 10.1111/j.1460-9568.1995.tb01078.x. [DOI] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- Brakebusch C, Seidenbecher CI, Asztely F, Rauch U, Matthies H, Meyer H, Krug M, Bockers TM, Zhou X, Kreutz MR, Montag D, Gundelfinger ED, Fassler R. Brevican-deficient mice display impaired hippocampal CA1 long-term potentiation but show no obvious deficits in learning and memory. Mol Cell Biol. 2002;22:7417–7427. doi: 10.1128/MCB.22.21.7417-7427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittis PA, Silver J. Multiple factors govern intraretinal axon guidance: a time-lapse study. Mol Cell Neurosci. 1995;6:413–432. doi: 10.1006/mcne.1995.1031. [DOI] [PubMed] [Google Scholar]
- Cafferty WB, Yang SH, Duffy PJ, Li S, Strittmatter SM. Functional axonal regeneration through astrocytic scar genetically modified to digest chondroitin sulfate proteoglycans. J Neurosci. 2007;27:2176–2185. doi: 10.1523/JNEUROSCI.5176-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulli D, Rhodes KE, Fawcett JW. Upregulation of aggrecan, link protein 1, and hyaluronan synthases during formation of perineuronal nets in the rat cerebellum. J Comp Neurol. 2007;501:83–94. doi: 10.1002/cne.21231. [DOI] [PubMed] [Google Scholar]
- Deepa SS, Umehara Y, Higashiyama S, Itoh N, Sugahara K. Specific molecular interactions of oversulfated chondroitin sulfate E with various heparin-binding growth factors. Implications as a physiological binding partner in the brain and other tissues. J Biol Chem. 2002;277:43707–43716. doi: 10.1074/jbc.M207105200. [DOI] [PubMed] [Google Scholar]
- Dino MR, Harroch S, Hockfield S, Matthews RT. Monoclonal antibody Cat-315 detects a glycoform of receptor protein tyrosine phosphatase beta/phosphacan early in CNS development that localizes to extrasynaptic sites prior to synapse formation. Neuroscience. 2006;142:1055–1069. doi: 10.1016/j.neuroscience.2006.07.054. [DOI] [PubMed] [Google Scholar]
- Domowicz MS, Krueger RC, Li H, Mangoura D, Vertel BM, Schwartz NB. The nanomelic mutation in the aggrecan gene is expressed in chick chondrocytes and neurons. Int. J. Dev. Neurosci. 1996;14:191–201. doi: 10.1016/0736-5748(96)00007-x. [DOI] [PubMed] [Google Scholar]
- Domowicz MS, Mangoura D, Schwartz NB. Aggrecan regulates telencephalic neuronal aggregation in culture. Brain Res Dev Brain Res. 2003;143:207–216. doi: 10.1016/s0165-3806(03)00133-0. [DOI] [PubMed] [Google Scholar]
- Domowicz MS, Pirok EW, III, Novak TE, Schwartz NB. Role of the C-terminal G3 domain in sorting and secretion of aggrecan core protein and ubiquitin-mediated degradation of accumulated mutant precursors. J Biol Chem. 2000;275:35098–35105. doi: 10.1074/jbc.275.45.35098. [DOI] [PubMed] [Google Scholar]
- Faissner A, Clement A, Locheter A, Streit A, Mandl C, Schachner M. Isolation of a neural chondroitin sulfate proteoglycan with neurite outgrowth promoting properties. J. Cell Biol. 1994;126:783–799. doi: 10.1083/jcb.126.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood J, Garcion E, Dobbertin A, Heck N, Calco V, ffrench-Constant C, Faissner A. The extracellular matrix glycoprotein Tenascin-C is expressed by oligodendrocyte precursor cells and required for the regulation of maturation rate, survival and responsiveness to platelet-derived growth factor. Eur J Neurosci. 2004;20:2524–2540. doi: 10.1111/j.1460-9568.2004.03727.x. [DOI] [PubMed] [Google Scholar]
- Geisert EE, Jr, Williams RW, Geisert GR, Fan L, Asbury AM, Maecker HT, Deng J, Levy S. Increased brain size and glial cell number in CD81-null mice. J Comp Neurol. 2002;453:22–32. doi: 10.1002/cne.10364. [DOI] [PubMed] [Google Scholar]
- Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Maurer P. Proteoglycans in the nervous system--the quest for functional roles in vivo. Matrix Biol. 2001;20:23–35. doi: 10.1016/s0945-053x(00)00137-2. [DOI] [PubMed] [Google Scholar]
- Haydon PG, Carmignoto G. Astrocyte control of synaptic transmission and neurovascular coupling. Physiol Rev. 2006;86:1009–1031. doi: 10.1152/physrev.00049.2005. [DOI] [PubMed] [Google Scholar]
- Hemken PM, Bellin RM, Sernett SW, Becker B, Huiatt TW, Robson RM. Molecular characteristics of the novel intermediate filament protein paranemin. Sequence reveals EAP-300 and IFAPa-400 are highly homologous to paranemin. J Biol Chem. 1997;272:32489–32499. doi: 10.1074/jbc.272.51.32489. [DOI] [PubMed] [Google Scholar]
- Jaworski DM, Kelly GM, Hockfield S. The CNS-specific hyaluronan-binding protein BEHAB is expressed in ventricular zones coincident with gliogenesis. J Neurosci. 1995;15:1352–1362. doi: 10.1523/JNEUROSCI.15-02-01352.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AR. Contact inhibition in the failure of mammalian CNS axonal regeneration. Bioessays. 1993;15:807–813. doi: 10.1002/bies.950151206. [DOI] [PubMed] [Google Scholar]
- Johnston MV, Trescher WH, Ishida A, Nakajima W. Neurobiology of hypoxic-ischemic injury in the developing brain. Pediatr Res. 2001;49:735–741. doi: 10.1203/00006450-200106000-00003. [DOI] [PubMed] [Google Scholar]
- Joseph R, Tsering C, Grunfeld S, Welch KM. Further studies on platelet-mediated neurotoxicity. Brain Res. 1992;577:268–275. doi: 10.1016/0006-8993(92)90283-f. [DOI] [PubMed] [Google Scholar]
- Kimata K, Barrach H-J, Brown KS, Pennypacker JP. Absence of proteoglycan core protein in cartilage from cmd/cmd (cartilage matrix deficiency) mice. J. Biol. Chem. 1981;256:6961–6968. [PubMed] [Google Scholar]
- Krueger RC, Fields TA, Hildreth J, Schwartz NB. Chick cartilage chondroitin sulfate proteoglycan core protein I. Generation and characterization of peptides and specificity for glycosaminoglycan attachment. J Biol Chem. 1990;265:12075–12087. [PubMed] [Google Scholar]
- Krueger RC, Hennig AK, Schwartz NB. Two immunologically and developmentally distinct chondroitin sulfate proteoglycans in embryonic chick brain. J Biol Chem. 1992;267:12149–12161. [PubMed] [Google Scholar]
- Landauer W. Nanomelia, a lethal mutation of the fowl. J. Hered. 1965;56:131–138. doi: 10.1093/oxfordjournals.jhered.a107392. [DOI] [PubMed] [Google Scholar]
- Lebaron RG. Versican. Perspectives in Dev. Neurobiol. 1996;3:261–271. [PubMed] [Google Scholar]
- Lee JA, Cole GJ. Localization of transitin mRNA, a nestin-like intermediate filament family member, in chicken radial glia processes. J Comp Neurol. 2000;418:473–483. doi: 10.1002/(sici)1096-9861(20000320)418:4<473::aid-cne8>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Li H, Domowicz MS, Hennig A, Schwartz NB. S103L reactive chondroitin sulfate proteoglycan (aggrecan) mRNA expressed in developing chick brain and cartilage is encoded by a single gene. Mol. Brain Res. 1996;36:309–321. doi: 10.1016/0169-328x(95)00269-x. [DOI] [PubMed] [Google Scholar]
- Li H, Schwartz NB, Vertel BM. cDNA cloning of chick cartilage chondroitin sulfate (aggrecan) core protein and identification of a stop codon in the aggrecan gene associated with the chondrodystrophy, nanomelia. J Biol Chem. 1993;268:23504–23511. [PubMed] [Google Scholar]
- Liu Y, Rao MS. Glial progenitors in the CNS and possible lineage relationships among them. Biol Cell. 2004;96:279–290. doi: 10.1016/j.biolcel.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Margolis RU, Margolis RK. Chondroitin sulfate proteoglycans as mediators of axon growth and pathfinding. Cell Tissue Res. 1997;290:343–348. doi: 10.1007/s004410050939. [DOI] [PubMed] [Google Scholar]
- Matthews RT, Kelly GM, Zerillo CA, Gray G, Tiemeyer M, Hockfield S. Aggrecan glycoforms contribute to the molecular heterogeneity of perineuronal nets. J Neurosci. 2002;22:7536–7547. doi: 10.1523/JNEUROSCI.22-17-07536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae PA, Rocco MM, Kelly G, Brumberg JC, Matthews RT. Sensory deprivation alters aggrecan and perineuronal net expression in the mouse barrel cortex. J Neurosci. 2007;27:5405–5413. doi: 10.1523/JNEUROSCI.5425-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon LD, Asher RA, Fawcett JW. Limited growth of severed CNS axons after treatment of adult rat brain with hyaluronidase. J Neurosci Res. 2003;71:23–37. doi: 10.1002/jnr.10449. [DOI] [PubMed] [Google Scholar]
- Moon LD, Asher RA, Rhodes KE, Fawcett JW. Regeneration of CNS axons back to their target following treatment of adult rat brain with chondroitinase ABC. Nat Neurosci. 2001;4:465–466. doi: 10.1038/87415. [DOI] [PubMed] [Google Scholar]
- Morgenstern DA, Asher RA, Fawcett JW. Chondroitin sulphate proteoglycans in the CNS injury response. Prog Brain Res. 2002;137:313–332. doi: 10.1016/s0079-6123(02)37024-9. [DOI] [PubMed] [Google Scholar]
- Napier A, Yuan A, Cole GJ. Characterization of the chicken transitin gene reveals a strong relationship to the nestin intermediate filament class. J Mol Neurosci. 1999;12:11–22. doi: 10.1385/JMN:12:1:11. [DOI] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Popp S, Andersen JS, Maurel P, Margolis RU. Localization of aggrecan and versican in the developing rat central nervous system. Dev Dyn. 2003;227:143–149. doi: 10.1002/dvdy.10282. [DOI] [PubMed] [Google Scholar]
- Pringle NP, Yu WP, Howell M, Colvin JS, Ornitz DM, Richardson WD. Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development. 2003;130:93–102. doi: 10.1242/dev.00184. [DOI] [PubMed] [Google Scholar]
- Rauch U. Extracellular matrix components associated with remodeling processes in brain. Cell Mol Life Sci. 2004;61:2031–2045. doi: 10.1007/s00018-004-4043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch U, Zhou XH, Roos G. Extracellular matrix alterations in brains lacking four of its components. Biochem Biophys Res Commun. 2005;328:608–617. doi: 10.1016/j.bbrc.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Ridet JL, Malhotra SK, Privat A, Gage FH. Reactive astrocytes: cellular and molecular cues to biological function. Trends Neurosci. 1997;20:570–577. doi: 10.1016/s0166-2236(97)01139-9. [DOI] [PubMed] [Google Scholar]
- Rittenhouse E, Dunn LC, Cookingham J, Calo C, Spiegelman M, Dooher GB, Bennett D. Cartilage matrix deficiency (cmd): a new autosomal recessive lethal mutation in the mouse. J. Embryol. Exp. Morph. 1978;43:71–84. [PubMed] [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalfeldt M, Bandtlow CE, Dours-Zimmermann MT, Winterhalter KH, Zimmermann DR. Brain derived versican V2 is a potent inhibitor of axonal growth. J Cell Sci. 2000;113(Pt 5):807–816. doi: 10.1242/jcs.113.5.807. [DOI] [PubMed] [Google Scholar]
- Schwartz NB, Domowicz M. Proteoglycans in brain development. Glycoconj J. 2004;21:329–341. doi: 10.1023/B:GLYC.0000046278.34016.36. [DOI] [PubMed] [Google Scholar]
- Shibata T, Yamada K, Watanabe M, Ikenaka K, Wada K, Tanaka K, Inoue Y. Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J Neurosci. 1997;17:9212–9219. doi: 10.1523/JNEUROSCI.17-23-09212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirko S, von Holst A, Wizenmann A, Gotz M, Faissner A. Chondroitin sulfate glycosaminoglycans control proliferation, radial glia cell differentiation and neurogenesis in neural stem/progenitor cells. Development. 2007;134:2727–2738. doi: 10.1242/dev.02871. [DOI] [PubMed] [Google Scholar]
- Taylor DL, Edwards AD, Mehmet H. Oxidative metabolism, apoptosis and perinatal brain injury. Brain Pathol. 1999;9:93–117. doi: 10.1111/j.1750-3639.1999.tb00213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HM, Graber BB, Larramendi LMH. 3H-Thymidine autoradiographic analysis of telencephalic histogenesis in the chick embryo:I. Neuronal birthdate of telencephalic comparments in situ. J. Comp. Neurol. 1981a;198:275–292. doi: 10.1002/cne.901980207. [DOI] [PubMed] [Google Scholar]
- Tsai HM, Graber BB, Larramendi LMH. 3H-Thymidine autoradiographic analysis of telencephalic histogenesis in the chick embryo:II. Dynamics of neuronal migration, displacement and aggregation. J. Comp. Neurol. 1981b;198:293–306. doi: 10.1002/cne.901980208. [DOI] [PubMed] [Google Scholar]
- Umehara Y, Yamada S, Nishimura S, Shioi J, Robakis NK, Sugahara K. Chondroitin sulfate of appican, the proteoglycan form of amyloid precursor protein, produced by C6 glioma cells interacts with heparin-binding neuroregulatory factors. FEBS Lett. 2004;557:233–238. doi: 10.1016/s0014-5793(03)01506-0. [DOI] [PubMed] [Google Scholar]
- Vertel BM, Grier BL, Li H, Schwartz NB. The chondrodystrophy, nanomelia: biosynthesis and processing of the defective aggrecan precursor. Biochem J. 1994;301:211–216. doi: 10.1042/bj3010211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M, Lawton T, Goetinck PF, Kuhn JL, Goldstein SA, Bonadio J. Aggrecan core protein is expressed in membranous bone of the chick embryo. Molecular and biomechanical studies of normal and nanomelia embryos. J Biol Chem. 1992;267:5592–5598. [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Versican in the developing brain: lamina-specific expression in interneuronal subsets and role in presynaptic maturation. J Neurosci. 2005;25:8457–8467. doi: 10.1523/JNEUROSCI.1976-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y. Lecticans: organizers of the brain extracellular matrix. Cell Mol Life Sci. 2000;57:276–289. doi: 10.1007/PL00000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yick LW, Wu W, So KF, Yip HK, Shum DK. Chondroitinase ABC promotes axonal regeneration of Clarke's neurons after spinal cord injury. Neuroreport. 2000;11:1063–1067. doi: 10.1097/00001756-200004070-00032. [DOI] [PubMed] [Google Scholar]
- Yuasa S. Development of astrocytes in the mouse hippocampus as tracked by tenascin-C gene expression. Arch Histol Cytol. 2001;64:149–158. doi: 10.1679/aohc.64.149. [DOI] [PubMed] [Google Scholar]
- Zhang SC. Defining glial cells during CNS development. Nat Rev Neurosci. 2001;2:840–843. doi: 10.1038/35097593. [DOI] [PubMed] [Google Scholar]
- Zhou XH, Brakebusch C, Matthies H, Oohashi T, Hirsch E, Moser M, Krug M, Seidenbecher CI, Boeckers TM, Rauch U, Buettner R, Gundelfinger ED, Fassler R. Neurocan is dispensable for brain development. Mol Cell Biol. 2001;21:5970–5978. doi: 10.1128/MCB.21.17.5970-5978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


