Abstract
Background
Syphilis spirochete Treponema pallidum ssp. pallidum remains the enigmatic pathogen, since no virulence factors have been identified and the pathogenesis of the disease is poorly understood. Increasing rates of new syphilis cases per year have been observed recently.
Results
The genome of the SS14 strain was sequenced to high accuracy by an oligonucleotide array strategy requiring hybridization to only three arrays (Comparative Genome Sequencing, CGS). Gaps in the resulting sequence were filled with targeted dideoxy-terminators (DDT) sequencing and the sequence was confirmed by whole genome fingerprinting (WGF). When compared to the Nichols strain, 327 single nucleotide substitutions (224 transitions, 103 transversions), 14 deletions, and 18 insertions were found. On the proteome level, the highest frequency of amino acid-altering substitution polymorphisms was in novel genes, while the lowest was in housekeeping genes, as expected by their evolutionary conservation. Evidence was also found for hypervariable regions and multiple regions showing intrastrain heterogeneity in the T. pallidum chromosome.
Conclusion
The observed genetic changes do not have influence on the ability of Treponema pallidum to cause syphilitic infection, since both SS14 and Nichols are virulent in rabbit. However, this is the first assessment of the degree of variation between the two syphilis pathogens and paves the way for phylogenetic studies of this fascinating organism.
Background
Treponema pallidum subspecies pallidum (TPA) is the causative agent of syphilis, a sexually transmitted disease affecting more than 12 million people worldwide each year [1]. After a period of decline in the 1990s, the number of reported cases of primary and secondary syphilis has been raising annually since 2000 in the United States [2]. Sequencing of the 1.14 Mbp genome of the Nichols strain of TPA in 1998 [3] greatly stimulated study of this unculturable pathogen. One important direction not yet developed is use of the Nichols sequence for comparative studies to determine variation between different syphilis isolates, how representative Nichols is of TPA, and the genetic differences between closely related treponemes causing different diseases (e.g. syphilis, yaws, bejel, pinta). To sample strains on a sufficient scale, rapid, inexpensive, and highly accurate sequencing methods are needed. Traditional whole genome shotgun sequencing methods using dideoxy-terminators (WGS-DDT) are relatively slow and costly to be applied to numerous samples. Here we sequence a treponemal genome by Comparative Genome Sequencing (CGS) [4], which provides an alternative to WGS-DDT sequencing of closely related genomes. CGS was previously used for mutation discovery in viruses [5], in mutagenized laboratory bacterial and fungal strains [4,6-9], in clinical isolates of bacteria [10,11], and for whole genome scale comparative studies [12-14].
The TPA isolate Street Strain 14 (SS14) was isolated in 1977 in Atlanta from a patient with secondary syphilis [15] who did not respond to erythromycin therapy that was used because of a penicillin allergy [16]. In vitro testing of SS14 revealed it to be less susceptible to a variety of antibiotics when compared to Nichols [16]. Nichols strain was isolated in 1912 in Washington, D.C. from cerebrospinal fluid of the patient with neurosyphilis [17]. Previous studies (D. Šmajs, G. M. Weinstock, unpublished results) showed SS14 had all genes of the Nichols genome as judged by hybridization to a microarray containing PCR products of all annotated Nichols open reading frames (ORFs) [18]. To compare these closely related, yet phenotypically distinct strains, we sequenced the SS14 genome by CGS.
Results
Identification of heterologous regions and sequence changes between Nichols and SS14 strains
In the first mapping stage of CGS, no regions with significantly stronger labeled SS14 DNA signals were observed, indicating no increase in gene copy number in the SS14 genome. Regions giving significantly weaker SS14 signals indicated 1731 candidate regions of variation encompassing 1 or more overlapping oligonucleotide targets. The sequencing data identified 213 SNPs in the SS14 genome. An additional 17 questionable SNPs were suggested in repeated sequences of the genome but did not score well in a SNP uniqueness algorithm [4], and thus could represent false positives due to cross hybridization with the other repeats. DDT sequencing of 12 such regions revealed 5 real SNPs, 6 false positives, and one position with 2 alleles within the SS14 population (intrastrain heterogeneity). Therefore these questionable SNPs were not included in the final sequence, unless they were verified by DDT sequencing (data not shown). An additional 62 positions out of the 213 SNPs identified by CGS were DDT sequenced. 60 SNPs were confirmed (Tables 1, 2, 3 last column) and 2 false positives were found.
Table 1.
DDT sequencing of 38 hypervariable regions where SNPs could not be identified by CGS
| Region no. | ORFa | Region size (nt) | Size of sequenced region (nt) | Left coordinatea | Right coordinatea | Newly found changes in the regions suggested by CGS | Newly found changes not suggested by mapping phase of CGS | Confirmation of SNPs identified by CGS in this regionb |
| 1 | TP0012 | 37 | 390 | 12322 | 12711 | 3 nt deletion | - | - |
| 2 | TP0076 | 29 | 529 | 83788 | 84316 | - | 1 solitary SNP | - |
| 3 | TP0117 | 86 | 699 | 134808 | 135506 | 7 clustered SNPs | - | - |
| 4 | TP0117 | 86 | 3 clustered SNPs | - | - | |||
| 5 | TP0126 | 29 | 393 | 147948 | 148340 | 1 solitary SNP | - | - |
| 6 | upstream | 29 | 460 | 149103 | 149562 | 2 clustered SNPs | - | - |
| of TP0128 | 1 nt + 5 nt insertions | |||||||
| 7 | TP0131 | 421 | 723 | 150925 | 151647 | 3 clustered SNPs, 1 solitary SNP | 2 clustered SNPs | - |
| 8 | upstream of TP0136 | 29 | 404 | 156348 | 156751 | 64 nt deletion | - | - |
| 9 | TP0136 | 1087 | 1609 | 156752 | 158360 | 19 clustered SNPs | 8 clustered SNPs | 21 SNPs |
| 1 nt + 1 nt + 1 nt + 6 nt deletions | 2 solitary SNPs | |||||||
| 10 | TP0272 | 29 | 480 | 288647 | 289126 | - | - | - |
| 11 | TP0304 | 37 | 466 | 318761 | 319226 | 3 nt deletion | - | - |
| 12 | TP0326 | 79 | 452 | 345605 | 346056 | 8 clustered SNPs | - | 1 SNP |
| 13 | TP0352 | 29 | 465 | 376926 | 377390 | - | - | - |
| 14 | TP0394 | 29 | 505 | 420353 | 420857 | - | - | 1 SNP |
| 15 | TP0431 | 29 | 465 | 458973 | 459437 | - | - | 1 SNP |
| 16 | TP0457 | 29 | 465 | 487935 | 488399 | - | - | - |
| 17 | TP0484 | 29 | 468 | 514441 | 514908 | - | - | - |
| 18 | TP0486 | 29 | 494 | 517297 | 517790 | - | 1 nt insertion, 1 nt deletion | - |
| 19 | TP0493 | 29 | 478 | 529146 | 529623 | - | - | - |
| 20 | TP0515 | 44 | 506 | 555754 | 556259 | 3 clustered SNPs | - | 4 SNPs |
| 21 | TP0544 | 29 | 611 | 585940 | 586550 | 6 nt insertion | - | - |
| 22 | TP0548 | 835 | 1189 | 591557 | 592745 | 22 clustered SNPs | 2 clustered SNPs | 5 SNPs |
| 3 nt + 4 nt + 5 nt insertions | 1 solitary SNP | |||||||
| 23 | TP0577 | 37 | 405 | 628247 | 628651 | 1 solitary SNP | - | - |
| 24 | TP0598 | 29 | 550 | 648851 | 649400 | - | 4 1 nt insertions | - |
| 25 | TP0620–TP0621 | 51 | 3469 | 670958 | 674426 | - | 4 clustered SNPs | - |
| 26 | TP0668 | 37 | 462 | 730080 | 730541 | 6 nt deletion | - | - |
| 27 | TP0699 | 51 | 469 | 766143 | 766611 | 1 solitary SNP | - | - |
| 28 | TP0785 | 29 | 438 | 851631 | 852068 | - | - | - |
| 29 | TP0814 | 29 | 476 | 882990 | 883465 | - | - | - |
| 30 | TP0865 | 29 | 480 | 943847 | 944326 | 3 nt insertion | - | 1 SNP |
| 31 | TP0866 | 29 | 543 | 944677 | 945219 | - | 1 nt insertion | - |
| 32 | TP0868 | 29 | 454 | 947257 | 947710 | 7 nt deletion | - | - |
| 33 | TP0896–TP0898 | 667 | 3038 | 974053 | 977090 | 4 SNPsc and 7 variable regionsd | 1 SNP | |
| 34 | TP0898 | 27 | 416 | 978349 | 978764 | - | - | - |
| 35 | TP0933 | 29 | 164 | 1014034 | 1014197 | - | - | - |
| 36 | TP0973 | 44 | 396 | 1057660 | 1058055 | 1 solitary SNP | - | 1 SNP (igr) |
| 37 | TP1030–TP1031 | 1507 | 402 | 1123660 | 1124061 | 18 clustered SNPs | 1 nt insertion, | 16 SNPs |
| 1775 | 1124256 | 1126030 | 1 solitary SNP | |||||
| 38 | TP1036 | 29 | 550 | 1132558 | 1133107 | - | - | - |
ORF – open reading frame; nt – nucleotide; SNP – single nucleotide polymorphism; igr – intergenic region; aas described in [3]; bSNPs identified using CGS in these regions were verified by DDT sequencing; c two SNPs represent the group of 17 SNPs in non-unique sites, originally excluded from list of total changes; didentified variable regions in TP0897 were identical to the variable regions V1–V7 described previously [22–24].
Table 2.
DDT sequencing of regions selected based on pilot SS14/Nichols comparison using microarray hybridization experiments
| Region no. | ORFa | Size of sequenced region (nt) | Left coordinatea | Right coordinatea | Newly found changes | Confirmation of SNPs identified by CGSb |
| 1 | TP0017 | 848 | 18454 | 19301 | - | - |
| 2 | TP0070 | 339 | 75493 | 75831 | - | - |
| 3 | TP0094 | 1011 | 102879 | 103889 | - | - |
| 4 | TP0123 | 1083 | 143207 | 144289 | - | - |
| 5 | TP0192 | 748 | 206663 | 207410 | - | - |
| 6 | TP0200 | 264 | 210183 | 210446 | - | - |
| 7 | TP0291 | 834 | 304706 | 305539 | - | - |
| 8 | TP0319 | 1014 | 334847 | 335860 | - | 1 SNP |
| 9 | TP0321–TP0322 | 2640 | 336149 | 338788 | - | - |
| 10 | TP0323 | 851 | 338885 | 339735 | - | 1 SNP |
| 11 | TP0376 | 806 | 400903 | 401708 | - | - |
| 12 | TP0377 | 78 | 401851 | 401928 | - | - |
| 13 | TP0516 | 1533 | 556351 | 557883 | - | - |
| 14 | TP0519 | 1277 | 559215 | 560491 | - | - |
| 15 | TP0580 | 1242 | 630328 | 631569 | - | - |
| 16 | TP0587 | 183 | 639620 | 639802 | - | - |
| 17 | TP0633 | 776 | 691437 | 692212 | - | - |
| 18 | TP0683 | 1047 | 746899 | 747945 | - | - |
| 19 | TP0799–TP0800 | 2168 | 866136 | 868303 | - | - |
| 20 | TP0806 | 1397 | 875808 | 877204 | - | - |
| 21 | TP0807 | 165 | 877407 | 877571 | - | - |
| 22 | TP0808 | 187 | 877632 | 877818 | - | - |
| 23 | TP0877 | 998 | 953710 | 954707 | - | 2 SNPs |
| 24 | TP0933 | 2023 | 1013098 | 1015120 | - | - |
| 25 | TP0952 | 1438 | 1032341 | 1033778 | - | 1 SNP |
| 26 | TP0961 | 1216 | 1041973 | 1043188 | - | - |
| 27 | TP0980 | 975 | 1063047 | 1064021 | - | - |
ORF – open reading frame; nt – nucleotide; SNP – single nucleotide polymorphism; aas described in [3]; bSNPs identified using CGS in these regions were verified by DDT sequencing.
Table 3.
DDT sequencing of regions showing different whole genome fingerprint profiles in SS14 strain
| Region no. | ORFa | Difference from WGF on the gel | Size of sequenced region (nt)b | Left coordinatea | Right coordinatea | Newly found changes | Confirmation of SNPs identified by CGSc |
| 1 | TP0124–TP0134 | insertion | 3245 + 1255 insertion | 145858 | 149102 | 1255 nt insertion, 2 nt deletion | - |
| 1362 | 149563 | 150924 | - | - | |||
| 252 | 151648 | 151899 | - | - | |||
| 3465 | 152043 | 155507 | 1 nt insertion | 2 SNPs | |||
| 2 | TP0135–TP0138 | deletion | 662 | 155686 | 156347 | - | (+ 64 nt deletion as in Table 1) |
| 894 | 158391 | 159284 | 1 nt deletion | ||||
| 3 | TP0433–TP0434 | insertion | 481 + 419 insertion | 461058 | 461538 | insertion of 7 repeats of 60 nt region altogether 14 repetitions, consensus sequence of the repeat CGTGAGGTGGAGGACGYGCCGRRGGTAGTG GAGCCGGCCTCTGRGCRTGARGGAGGGGAG | - |
| 4 | TP0468–TP0471 | deletion | 3571 | 495308 | 498878 | 2 nt deletion + 1 nt insertion + deletion of seven 24 nt repetitions, consensus sequence of the repeat CTCCGCCTCCTTGCGCCGGGCTTC | 1 SNP |
nt – nucleotide; SNP – single nucleotide polymorphism; aas described in [3]; bregions previously described in Table 1 were excluded; cSNPs identified using CGS in these regions were verified by DDT sequencing.
1674 out of 1731 candidate regions were identified as SNPs in the second sequencing stage but there were 57 regions encompassing 124 oligonucleotide targets where sequence changes could not be determined. These represented possible hypervariable regions with multiple differences from Nichols in the sequencing 29 mers. DNA regions comprising these sites were grouped into 38 larger regions (29–1507 bp), amplified by PCR and DDT sequenced. In 21 of the 38 cases, mostly closely spaced SNPs and/or short insertions or deletions (indels ranging from 1 to 7 nts) were found while no changes were seen in 17 cases (Table 1, column 7), which is in agreement with data obtained by others [12]. DDT sequencing of hypervariable regions suggested by the first phase of CGS identified nucleotide changes in these regions (Table 1, column 7) and also in the vicinity of these regions, where results of the first CGS phase suggested no changes (Table 1, column 8), indicating the need for extension of DDT sequenced regions of at least 100 bp in both directions. Additional short indels were discovered during DDT sequencing of regions identified by WGF (Table 3). Altogether, 2 false positive SNPs (data not shown), 19 false negative SNPs and an additional 16 indels (Tables 1, 3) were found in these DDT sequenced regions (42,344 bp). The overall confirmation of data suggests that repeated regions of the genome are limitations for SNP discovery and almost half of possible hypervariable regions are false positive results.
The accuracy of CGS was determined by comparison to the results of DDT sequencing for 27 regions encompassing 27,141 bp (Table 2). Selection of these regions was focused on possible variable regions in SS14/Nichols hybridization experiments (D. Šmajs, G. M. Weinstock, unpublished results) using a microarray of TPA coding sequences [18] and thus was not completely random. These regions included 5 SNPs and no false positive or false negative SNPs/indels were found. These results indicate an error frequency comparable to or lower than that of high quality finished DDT sequence.
Assessment of reproducibility of CGS experiments
To test the reproducibility of the method, the genome of TPA SS14 was sequenced twice with the CGS approach, using 2 independent DNA isolations from two subsequent inoculations of rabbit testes (i.e. 4950 and 4951, respectively). When most of the variable genomic regions were excluded from the analysis (CGS cannot identify closely spaced SNPs and/or short indels), CGS discovered 198 SNPs in each DNA preparation. The experiments agreed at 192 SNPs (97%), and 12 SNPs were predicted by only one CGS experiment. Out of these 12 SNPs, 7 were found to be real, as shown by DDT sequencing (data not shown), three loci showed intrastrain heterogeneity in one of the two SS14 DNA isolations, with one allele identical to the Nichols genome sequence and a second allele identical to the base change found by CGS. Two SNPs were predicted falsely, and in both cases the false SNP was located next to a real SNP. The reproducibility of the CGS method is thus likely to be limited by the presence of SNP clusters and influenced by genetically different subpopulations in the test sample.
Physical mapping of treponemal chromosome
To verify the complete sequence of SS14 strain, to screen for possible discrepancies in cross-reacting repeat regions (tpr genes) and insertions of unique sequences, WGF was performed. This physical mapping approach showed the order of the ORFs along the chromosome is identical to Nichols genome and 4 large indel regions were identified. A 64 bp deletion upstream of TP0136 was found by both CGS and WGF methods. Three additional indels were found only by WGF, two insertions (between genes TP0126–TP0127 and within overlapping genes TP0433–TP0434) and one deletion (in TP0470) (Table 3). A deletion in TP0470 and an insertion in TP0433–TP0434 comprised tandem repeats of 24 and 60 bp, respectively. Similar analysis of the Nichols strain revealed length differences in genes TP0433–TP0434 compared to the published sequence [GenBank:AE000520] as described previously [19]. Moreover, intrastrain heterogeneity in the Nichols strain was observed in regions comprising TP0126–TP0127 and upstream of TP0136 with one allele identical to the published sequence. In the Nichols BAC library [20], similar intrastrain heterogeneity was found in the vicinity of gene TP0126. This region comprises a 1255 bp insertion between genes TP0126 and TP0127 in SS14 strain. A similar region was previously described in another syphilitic strain (Chicago, [GenBank:AY587909]) and was found to contain a sequence similar to tprK and is believed to be recipient site of the tprK conversion [21]. Altogether, three large indels were not detected by CGS. We suggest probable reasons for this fact are (1) the length of the repeats is similar to/longer than oligonucleotides used on the array and (2) sequence changes were found in Nichols DNA when compared to published complete sequence used for mapping array design [GenBank:AE000520].
Analysis of whole genome interstrain heterogeneity between Nichols and SS14
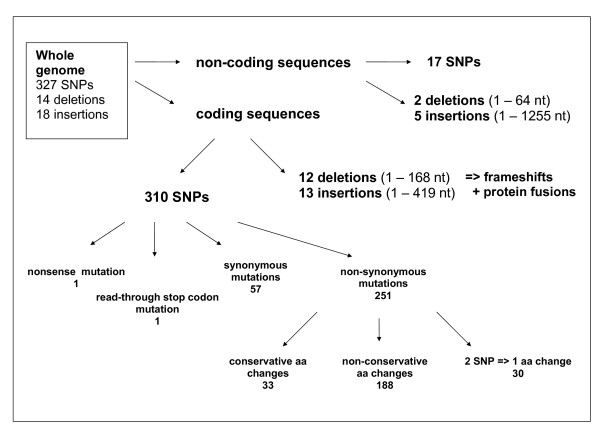
When results of CGS, WGF and DDT sequencing were combined, 327 SNPs (224 transitions and 103 transversions), 14 deletions and 18 insertions were identified (Fig. 1). Sequence changes of variable regions V1–V7 of TP0897, tprK, were not included, because sequences of these regions were found to differ greatly in both length and sequence within the SS14 population, in agreement with investigations published previously [22-24]. Obtained data have been used to compile the sequence of the SS14 genome [GenBank:CP000805]. The GenBank entry contains Ns in the positions of variable regions V1–V7 of tprK gene. All discovered sequence changes are listed in Table S1 (See Additional file 1: Supplemental data).
Figure 1.
Scheme to identify sequence changes in the SS14 genome.
Interstrain sequence heterogeneity discovered between strains Nichols and SS14 included silent mutations, amino acid alterations/indels, gene fusions, and truncations and elongations of open reading frames due to indels. Among the SNPs found by CGS was an adenine to guanine transition in both copies of 23S rDNA in SS14 strain. This sequence change was previously described in association with the SS14 erythromycin resistance [25]. Many discovered indels did not disrupt the open reading frames and represented variable number of nucleotides in homopolymeric tracts (e.g. in TP0012, TP0127), variable number of short motif repeats of 3 and 6 nucleotides (e.g. in TP0136, TP0304, TP0544, TP0668, TP0865), and variable number of longer motif repeats of 60 and 24 nts (TP0433–TP0434, TP0470).
Frameshift mutations and other changes affecting protein length are presented in Table 4. Besides 11 hypothetical proteins (including two possible surface proteins – Tp75 and p83/100), FlaB1 and Tex protein were affected. Sequence changes in four cases led to fusion of ORFs (TP0006 and TP0007 – elongation of Tp75 protein; hypothetical proteins TP0433 and TP0434, TP0597 and TP0598; conserved hypothetical proteins TP0468 and TP0469). Three of these genes (TP0006, TP0470, TP0486) were predicted to code for possible surface protein virulence factors [26]. Moreover, antigen p83/100, hypothetical gene TP0127, conserved hypothetical gene TP0470 and the fused proteins TP0433–TP0434 and TP0468–TP0469 were described to be antigenic in rabbits [27]. Two of the frameshift changes were confirmed to be present in the Nichols strain genomic DNA (TP0486 and TP0598).
Table 4.
Genes with mutations that significantly affect protein length
| ORFa | SNPs | Other changes | Result of mutation | Protein function |
| TP0006 | 1 | read-through stop codon | longer protein (+262 aa), fusion with TP0007 | Tp75 protein (possible surface protein) |
| TP0127 | 0 | 1 deletion (2 nt) | frameshift (-103 aa) | hypothetical protein |
| TP0132 | 0 | 1 insertion (1 nt) | frameshift (-44 aa) | hypothetical protein |
| TP0433–TP0434 | 1 | insertion of tandem repeats | fusion of 2 ORFs (604 aa) | hypothetical proteins (resulting fusion – arp proteinc) |
| TP0468–TP0469 | 0 | 1 insertion (2 nt) 1 deletion (1 nt) |
fusion of 2 ORFs (650 aa) | conserved hypothetical proteins |
| TP0470 | 0 | deletion of 7 tandem repeats (7 × 24 nt) | shorter protein (-56 aa) | conserved hypothetical protein |
| TP0486 | 0 | 1 deletion (1 nt)b | frameshift (+9 aa) | antigen, p83/100 (possible surface protein) |
| TP0598 | 1 | 4 insertion (4 nt)b | frameshift (+81 aa) fusion with TP0597 | hypothetical protein |
| TP0868 | 0 | 1 deletion (7 nt) | frameshift (-168aa) | flagellar filament 34.5 kDa core protein (FlaB1) |
| TP0924 | 1 | nonsense mutation | shorter protein (-250 aa) | Tex protein |
| TP1030 | 7 | 1 insertion (1 nt) | frameshift (-46 aa) | hypothetical protein |
ORF – open reading frame; SNP – single nucleotide polymorphism; aa – amino acid; aas described in [3]; bsame sequence change detected in Nichols Houston strain genomic DNA; c same sequence change described in [19].
SNPs in SS14 were found to be non-uniformly distributed with the number of SNPs per ORF varying from 0 to 49. Hypervariable regions are listed in Table 5 and include ORFs encoding 3 hypothetical proteins, Tpr proteins (TprC, TprL) and outer membrane protein TP0326. TP0326 was predicted to be a virulence factor [26] and was experimentally verified to be an antigen [27]. It is of interest that the most variable region of the genome represents TP0136 (and sequence upstream of this gene) which encodes a protein that is antigenic in both rabbit and human infections [27,28] and was found to serve as fibronectin and laminin binding protein [29].
Table 5.
ORFs with the highest number of detected SNPs (+ indels)
| ORFa | SNPs | aa changes | Other changes | Result of mutation | Protein function |
| TP0117 | 10 | 6 | Tpr protein C (TprC) | ||
| TP0136 | 49 | 38 | 4 deletions (9 nt) | 3 aa missing | hypothetical proteinb |
| TP0326 | 12 | 9 | outer membrane protein | ||
| TP0515 | 10 | 10 | conserved hypothetical protein | ||
| TP0548 | 30 | 21 | 3 insertions (12 nt) | 4 aa inserted | hypothetical protein |
| TP1031 | 31 | 23 | Tpr protein L (TprL) |
ORF – open reading frame; SNP – single nucleotide polymorphism; aa – amino acid; nt – nucleotide; aas described in [3]; bthis protein was described to be fibronectin and laminin protein [29].
The distribution of SNPs in coding and non-coding sequences of SS14 was not significantly different. ORFs represent 92.9% of total genomic sequence; 94.8% of all SNPs were in coding sequences corresponding to 310 SNPs in genes (212 transitions and 98 transversions) and 17 SNPs (5.2%) in intergenic regions (12 transitions, 5 transversions). The frequency of SNPs was different among putative protein classes (Table 6). The highest frequency of SNPs was in hypothetical genes, lowest in housekeeping genes. In addition, housekeeping genes had the lowest number of SNPs altering amino acid sequences indicating conservation of these gene products.
Table 6.
Distribution of SNPs in different gene function groups and their effects on protein sequences
| Putative gene function | whole genomea | % | affected ORFsb | % | SNPsc | % | aa changesd | % |
| Hypothetical | 316 | 30.4 | 52 | 38.2 | 199 | 64.2 | 148 | 67.0 |
| Conserved hypothetical | 177 | 17.0 | 21 | 15.4 | 34 | 11.0 | 22 | 10.0 |
| Metabolic functions | 167 | 16.1 | 19 | 14.1 | 23 | 7.4 | 19 | 8.6 |
| Housekeeping genes | 223 | 21.5 | 24 | 17.6 | 25 | 8.0 | 10 | 4.4 |
| Other function | 156 | 15.0 | 20 | 14.7 | 29 | 9.4 | 22 | 10.0 |
| Total | 1039 | 100 | 136 | 100 | 310 | 100 | 221 | 100 |
anumber of genes (ORFs) in the complete genome of TPA Nichols strain [3]; bnumber of all genes with sequence changes in the genome of SS14 strain; cnumber of SNP changes identified within ORF groups in the genome, other sequence changes were not included; damino acid changes caused by SNPs, changes in length of the protein molecule are listed in Table 4.
Identification of intrastrain variability in TPA population
Because DDT sequencing of some PCR products did not result in an unambiguous sequence, WGS-DDT sequencing of small insert libraries was performed. Analysis of libraries and PCR products revealed multiple (intrastrain) sequence variants in TP0117 (tprC), TP0402 (coding for flagellum-specific ATP synthase), TP0620 (tprI), TP0621 (tprJ), TP0971 (pathogen-specific membrane antigen) and TP1029 (hypothetical protein) and in the intergenic region between tprI and tprJ. Consensus sequences were mostly identical to the Nichols published sequence, but some positions had minor alternative sequences or vice versa. Altogether, intrastrain genetic heterogeneity comprised polymorphisms in 43 nucleotide positions and one polymorphism in a homopolymeric stretch (Table 7).
Table 7.
Genetic heterogeneity in the SS14 population isolated from rabbit testes
| ORFa | Genome positiona | [GenBank:AE000520] sequence | SS14 sequenceb | position in ORF (Nichols) a | aa change | note |
| TP0117 | 135098 | G | G or C (5/6) | 1600 | P534 => A534 | |
| 135107 | T | T or C (3/4) | 1591 | I531 => V531 | ||
| 135141 | G | G or A (5/2) | 1557 | no change | ||
| 135144 | T | T or C (3/4) | 1554 | no change | ||
| 135149 | C | C or T (5/2) | 1549 | A517 => T517 | ||
| 135220 | G | G or A (5/6) | 1478 | T493 => I493 | ||
| 135227 | G | G or A (6/6) | 1471 | P491 => S491 | ||
| 135235 | G | G or A (2/10) | 1463 | A488 => V488 | ||
| 135239 | C | C or T (2/10) | 1459 | G487 => R487 | ||
| 135251 | A | A or G (6/6) | 1447 | Y483 => H483 | ||
| TP0402 | 427435 | C | C or T (NA) | 401 | P134 => L134 | |
| 427737 | G | G or T (NA) | 703 | A235 => S234 | ||
| TP0620 | 671746 | T | T or C (9/3) | 1142 | Q381 => R381 | |
| 671751 | T | T or G (19/10) | 1137 | R379 => G379 | ||
| 671753 | T | T or C (19/10) | 1135 | R379 => G379 | ||
| 671763 | C | C or T (8/4) | 1125 | no change | ||
| 671982 | G | G or C (12/6) | 906 | S302 => R302 | ||
| 672004 | C | C or T (12/6) | 884 | S295 => N295 | ||
| 672016 | A | G or A (12/6) | 872 | L291 => P291 | ||
| 672025 | T | T or C (11/7) | 863 | N288 => C288 | ||
| 672026 | T | T or A (11/6) | 862 | N288 => C288 | ||
| 672027 | A | A or G (11/6) | 861 | G287 => D287 | ||
| 672028 | C | C or T (12/5) | 860 | G287 => D287 | ||
| 672036 | G | G or T (11/6) | 852 | no change | ||
| 672039 | A | A or G (NA) | 849 | P283 => N283 | ||
| 672040 | G | G or T (NA) | 848 | P283 => N283 | ||
| 672041 | G | G or T (12/6) | 847 | P283 => N283 | ||
| 672042 | G | G or A (NA) | 846 | D282 => S282 | ||
| 672043 | T | T or C (13/6) | 845 | D282 => S282 | ||
| 672044 | C | C or T (10/5) | 844 | D282 => S282 | ||
| 672154 | G | G or T (7/10) | 734 | T245 => K245 | ||
| 672286 | G | G or A (4/12) | 602 | T201 => M201 | ||
| Upstream of TP0620 | 672916-7 | (-) | (-) or C (6/6) | position -30 from TP0620 | homopolymeric stretch | |
| 672944 | A | A or G (14/6) | position -58 from TP0620 | |||
| TP0621 | 673088 | T | T or C (14/4) | 2134 | I712 => V712 | |
| 673119 | G | G or A (14/4) | 2103 | no change | ||
| 673425 | C | C or T (2/8) | 1797 | no change | ||
| 673428 | A | A or G (2/8) | 1794 | no change | ||
| 673511 | A | A or C (6/6) | 1711 | F571 => V571 | ||
| 673545 | C | C or T (9/4) | 1677 | no change | ||
| 673550 | A | A or G (10/6) | 1672 | F558 => L558 | ||
| 673554 | C | C or T (10/6) | 1668 | no change | ||
| TP0971 | 1054447 | T | T or C (NA) | 301 | K101 => E101 | |
| TP1029 | 1123796 | G | G or A (5/6) | 15 | no change | |
ORF – open reading frame; aa – amino acid; NA – not available; aas described in [3]; bnumbers in parentheses show sequence reads for each alternative sequence.
Discussion
Obtaining the complete genome sequence of a second syphilis spirochete (SS14) shows the utility of the CGS strategy for treponemal comparative genomics. This is the first application of this approach to sequence an entire genome. This approach can be used when highly similar genomes are investigated and one genome sequence of closely related organism is known. The CGS strategy represents a rapid (days to weeks) and scalable methodology to sequence multiple syphilitic strains and clinical isolates. In the present study there was a need to further investigate some variable regions, but the directed DDT sequencing required was much less than needed to sequence a whole genome, thus lowering the total cost of obtaining the genome sequence.
There are some of the TPA-specific limitations of this approach to whole genome sequencing. Because the CGS strategy uses genomic DNA as a probe, accuracy is affected by the presence of repeated sequences. Repeat regions hybridize to more than one oligonucleotide on a tiling array resulting in both reduced sensitivity to detect changes, as well as ambiguity in assigning locations for the variants detected. Precautions have to be taken when inspecting tpr regions and others (arp gene, TP0470) which cross-react based on sequence similarity. Such regions, together with highly variable regions, need to be analyzed by WGF and sequenced by DDT to reveal true nucleotide changes and numbers of repeated regions. Another possible restriction of this methodology arises from the character of the TPA population. Multiple sequence variants in the Nichols strain population were both described previously and identified in this work, and hybridization based sequence changes discovery in these regions is influenced by the ratio between/among different sequence variants in the population. Finally, the accuracy of the genome sequence produced by CGS is dependent on the accuracy of the reference genome sequence. As suggested by two newly revealed frameshifts in Nichols strain sequence, discovered sequence changes have to be verified in Nichols sequence to describe real sequence changes compared to Nichols genome.
The SS14 genome brings a first insight into the whole genome variability within TPA. Both Nichols and SS14 cause infection in rabbits and so are not believed to be attenuated to cause infection in man, thus it is very probable none of the differences may affect the ability of the bacteria to cause the disease. The examples of interstrain heterogeneity and multiple alleles in a population of haploid organisms are candidates for antigenic variation, contingency genes and other types of SSR (short sequence repeats) [30,31]. Changes resulting in significant differences in protein sequences (frameshifts and sequence changes causing protein length shifts) and hypervariable regions affected novel genes, membrane antigens and Tpr proteins. The Tpr protein family includes 12 Treponema pallidum repeat proteins, found uniquely in this bacterium and showing sequence similarity to major sheath protein of Treponema denticola. 8 out of 12 tpr genes (66%) were found to be affected by sequence changes representing a higher proportion than the whole genome rate (13.1%). Positions showing interstrain and intrastrain heterogeneity or both were found in tpr genes. Altogether 53 SNPs and 38 intrastrain variable nucleotide positions, with at least one allele identical to the sequence of the Nichols genome, were found in tpr genes (V1–V7 regions of tprK were excluded from this analysis). Based on the fact that tpr genes share a high degree of similarity on the DNA level, we expect differences could be underestimated due to the limitations of the hybridization method for repeated sequences. Multiple alleles of tpr genes were described among and within TPA strains [22-24,32,33] and some TPA repeated regions (tpr genes, arp gene) were used as loci for typing of clinical isolates [34-38]. Newly identified hypervariable regions (Table 5) represent candidate sequences to screen clinical isolates and have potential to be used as typing markers of strains and isolates. In addition, different strains of TPA have already been tested for association with higher risk for neuroinvasion in rabbits [39] and identification of underlying sequence changes will enable prediction of such risks. The identified variation in novel genes suggests other targets besides tpr genes could be responsible for antigenic variation in TPA, or without support of further expression and antigenicity data, these could represent pseudogenes.
Conclusion
The CGS strategy combined with WGF represents a rapid and simply scalable method to assess genome-wide genetic variability within TPA strains and subspecies, which share a very unusual degree of sequence similarity and lack genome rearrangements (as shown in this study). We expect this method to be combined with new sequencing technologies to produce high quality genome sequences to provide important data to design genotyping systems for more intensive strain sampling. Sequence variants could be readily used for molecular typing and identification of SS14 and Nichols strains and, with accumulation of additional data from other TPA genomes, for epidemiologic applications and clinical discrimination between reinfection and reactivation of syphilitic processes. Moreover, the ability to now sequence numerous TPA strains, especially those showing different degrees of virulence, will allow phenotype to be correlated with sequence. This is a significant development for an organism of important public health impact, but for which standard bacterial genetic methods are untenable.
Methods
DNA isolation
TPA strains Nichols and SS14 were maintained by rabbit inoculation and purified by Hypaque gradient centrifugation as described previously [40]. Chromosomal DNA was prepared as described previously [3].
Comparative genome sequencing
100 ng of treponemal genomic DNA was amplified to approximately 100 μg using the REPLI-g kit (Qiagen, Valencia, CA). For each array hybridization, 5 μg of amplified genomic DNA was digested with 0.005 U DNase I in 1× One-Phor-All Buffer (Amersham Pharmacia Biotech, Piscataway, NJ) for 5 min at 37°C, followed by inactivation at 95°C for 15 min. To label the digested DNA fragments, 4 μl 5× Terminal Transferase Buffer (Promega, Madison, WI), 1 nmol Biotin-N6-ddATP, and 25 U Terminal Transferase were added directly to the inactivated digestion mix and incubated at 37°C for 90 min, followed by inactivation at 95°C for 15 min.
Mutation mapping microarrays were designed to map mutations by selecting a 29 mer oligonucleotide every 7 bases for both strands of the complete TPA Nichols genome sequence [3], [GenBank:AE000520]. All 325,138 oligonucleotides were synthesized in parallel as described by others [41,42].
Arrays were hybridized to digested, labeled genomic DNA of Nichols and SS14 strains separately and processed as described in [4] with an additional step after second wash in stringent buffer consisting of staining with a solution containing Cy3-Streptavidin conjugate (Amersham Pharmacia Biotech) for 10 min, and washing again with non-stringent wash buffer. The Cy3 signal was amplified by secondary labeling of the DNA with biotinylated goat anti-streptavidin (Vector Laboratories, Burlingame, CA). The secondary antibody was washed off with non-stringent wash buffer, and arrays were re-stained with the Cy3-Streptavidin solution.
Finally, the stain solution was removed, and arrays were washed in non-stringent wash buffer followed by two 30 sec washes in 0.5 × SSC and a 15 sec wash in 70% EtOH. Arrays were spun dry in a custom centrifuge and stored until scanning.
Microarray scanning, data analysis and sequencing microarray design and procedure were described previously [4]. The second array designed to sequence SS14 strain contained 392,000 oligonucleotides, with 8 oligos per base position (4 for each strand) and 48,600 bases were sequenced in total. Because mutations are sequenced in step two, inclusion of false positives from the mapping arrays does not affect the final data set.
Dideoxy-terminator sequencing of heterologous SS14 genome regions
After the second sequencing stage of the array analysis, some regions (Table 1) of the SS14 genome showed clear differences but SNPs could not be clearly identified. These regions were sequenced by DDT sequencing. Coordinates of these regions were extended with at least 150 bp in both directions and amplified with Taq-polymerase using oligonucleotide primers designed with Primer3 software [43]. The resulting PCR products were purified using QIAquick PCR purification Kit (Qiagen) and DDT sequenced using the original amplification primers and internal primers where applicable. Due to sequence similarity between tpr (Treponema pallidum repeat) genes, 3 of the heterologous regions (comprising genes TP0620–TP0621, TP0896–TP0898, TP1029–TP1030) were XL PCR amplified using primers annealing to unique regions in the vicinity of the desired sequence. XL PCR products were purified and mechanically sheared to fragments 500 – 1000 bp in length. These fragments were cloned into the pUC18 vector resulting in small insert libraries and recombinant plasmids isolated from at least 48 colonies were DDT sequenced to multiple coverage using pUC18 primers. All sequence reads were analyzed using Lasergene software (DNASTAR, Inc., Madison, WI).
Whole genome fingerprinting
Whole genome fingerprinting was performed as described previously [44]. The chromosomal DNA was amplified in 102 Treponema pallidum interval (TPI) regions with median length of 12,204.5 bp (ranging from 1,778 to 24,758 bp) using the GeneAmp® XL PCR Kit (Applied Biosystems, Foster City, CA). The primer pairs for these amplifications are listed in Table S2 (See additional file 1: Supplemental data). Each PCR product was digested with BamH I, EcoR I and Hind III (New England Biolabs, Ipswich, MA) or their combinations. To asses the possible presence of indels in restriction fragments ≥ 4 kb, additional digestions using Acc I, Cla I, EcoR V, Kpn I, Mlu I, Nco I, Nhe I, Rsr II, Sac I, Spe I, Xba I or Xho I were performed as needed. The resulting fingerprints of TPA Nichols and SS14 strains were compared.
Nucleotide sequence accession number
The complete sequence of TPA SS14 strain was deposited in the GenBank under the accession number CP000805.
Authors' contributions
GMW designed the study. PM performed genome sequence analysis, finishing using DDT sequencing and wrote the manuscript. MS and ES performed WGF analysis. JEN, JS, TAR, MNM, TJA composed the CGS technique team and analyzed hybridization data. JFP contributed to SNP and proteome analysis. DS, TP, SJN and GMW provided critical expertise of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplemental material consists of two tables containing list of all identified sequence changes in TPA SS14 genome compared to [GenBank:AE000520] (Table S1) and list of primers used for WGF analysis (Table S2).
Acknowledgments
Acknowledgements
This work was supported by grants from the U.S. Public Health Service to G.M.W. (R01 DE12488 and R01 DE13759), S.J.N. (R01 AI49252) and T.P. (AI45842) and by the grants of the Grant Agency of the Czech Republic (310/04/0021 and 310/07/0321) and the Ministry of Health of the Czech Republic (NR/8967-4/2006) to D.S. and by the institutional support (MSM0021622415).
The authors want to thank Donna Muzny and the DNA sequencing team at the HGSC for their assistance.
Contributor Information
Petra Matějková, Email: matej@med.muni.cz.
Michal Strouhal, Email: strouhal@med.muni.cz.
David Šmajs, Email: dsmajs@med.muni.cz.
Steven J Norris, Email: steven.j.norris@uth.tmc.edu.
Timothy Palzkill, Email: timothyp@bcm.tmc.edu.
Joseph F Petrosino, Email: jpetrosi@bcm.tmc.edu.
Erica Sodergren, Email: esodergr@wustl.edu.
Jason E Norton, Email: jnorton@nimblegen.com.
Jaz Singh, Email: jaz@nimblegen.com.
Todd A Richmond, Email: todd@nimblegen.com.
Michael N Molla, Email: molla@bu.edu.
Thomas J Albert, Email: talbert@nimblegen.com.
George M Weinstock, Email: geowei@mac.com.
References
- World Health Organization . Tech Report No WHO/HIV_AIDS/20 WHO/CDS/CSR/EDC/200110. World Health Organization; 2001. Global prevalence and incidence of selected curable sexually transmitted infections: overview and estimates. [Google Scholar]
- Centers for Disease Control and Prevention . Syphilis Surveillance Report. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; 2006. Sexually Transmitted Disease Surveillance 2005 Supplement. [Google Scholar]
- Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- Albert TJ, Dailidiene D, Dailide G, Norton JE, Kalia A, Richmond TA, Molla M, Singh J, Green RD, Berg DE. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat Methods. 2005;2:951–953. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]
- Wong CW, Albert TJ, Vega VB, Norton JE, Cutler DJ, Richmond TA, Stanton LW, Liu ET, Miller LD. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Alder JD, Silverman JA. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BØ. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nature Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- Manjunatha UH, Boshoff H, Dowd CS, Zhang L, Albert TJ, Norton JE, Daniels L, Dick T, Pang SS, Barry CE., 3rd Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:431–436. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Hosaka T, Tokuyama S, Okamoto S, Ochi K. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2) J Bacteriol. 2007;189:3876–3883. doi: 10.1128/JB.01776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman SJ, Beskhlebnaya V, Chesnokova V, Chattopadhyay S, Stamm WE, Hooton TM, Sokurenko EV. Differential stability and trade-off effects of pathoadaptive mutations in the Escherichia coli FimH adhesin. Infect Immun. 2007;75:3548–3555. doi: 10.1128/IAI.01963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytia-Trees EK, Ribot EM, Fields PI, Whittam TS, Swaminathan B. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 2006;16:757–767. doi: 10.1101/gr.4759706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SR, Chakicherla AY, Chain PS, Schmidt R, Shin MW, Legler TC, Scow KM, Larimer FW, Lucas SM, Richardson PM, Hristova KR. Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibium petroleiphilum PM1. J Bacteriol. 2007;189:1931–1945. doi: 10.1128/JB.01259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rikihisa Y. Virulence potential of Ehrlichia chaffeensis strains of distinct genome sequences. Infect Immun. 2007;75:3604–3613. doi: 10.1128/IAI.02028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LV, Kerner TC, Jr, Bankaitis VA, Bassford PJ., Jr Identification and preliminary characterization of Treponema pallidum protein antigens expressed in Escherichia coli. Infect Immun. 1983;41:709–721. doi: 10.1128/iai.41.2.709-721.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LV, Stapleton JT, Bassford PJ., Jr In vitro assay to demonstrate high-level erythromycin resistance of a clinical isolate of Treponema pallidum. Antimicrob Agents Chemother. 1988;32:164–169. doi: 10.1128/aac.32.2.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols HJ, Hough WH. Demonstration of Spirochaeta pallida in the cerebrospinal fluid. JAMA-J Am Med Assoc. 1913;60:108–110. [Google Scholar]
- Smajs D, McKevitt M, Howell JK, Norris SJ, Cai WW, Palzkill T, Weinstock GM. Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J Bacteriol. 2005;187:1866–1874. doi: 10.1128/JB.187.5.1866-1874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rodes B, George R, Steiner B. Molecular characterization and analysis of a gene encoding the acidic repeat protein (Arp) of Treponema pallidum. J Med Microbiol. 2007;56:715–721. doi: 10.1099/jmm.0.46943-0. [DOI] [PubMed] [Google Scholar]
- Smajs D, McKevitt M, Wang L, Howell JK, Norris SJ, Palzkill T, Weinstock GM. BAC library of T. pallidum DNA in E. coli. Genome Res. 2002;12:515–522. doi: 10.1101/gr.207302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centurion-Lara A, LaFond RE, Hevner K, Godornes C, Molini BJ, Van Voorhis WC, Lukehart SA. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol Microbiol. 2004;52:1579–1596. doi: 10.1111/j.1365-2958.2004.04086.x. [DOI] [PubMed] [Google Scholar]
- LaFond RE, Centurion-Lara A, Godornes C, Rompalo AM, Van Voorhis WC, Lukehart SA. Sequence diversity of Treponema pallidum subsp. pallidum tprK in human syphilis lesions and rabbit-propagated isolates. J Bacteriol. 2003;185:6262–6268. doi: 10.1128/JB.185.21.6262-6268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFond RE, Centurion-Lara A, Godornes C, Van Voorhis WC, Lukehart SA. TprK sequence diversity accumulates during infection of rabbits with Treponema pallidum subsp. pallidum Nichols strain. Infect Immun. 2006;74:1896–1906. doi: 10.1128/IAI.74.3.1896-1906.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LV, Bergen HL. The sequence-variable, single-copy tprK gene of Treponema pallidum Nichols strain UNC and Street strain 14 encodes heterogeneous TprK proteins. Infect Immun. 2000;68:6482–6486. doi: 10.1128/IAI.68.11.6482-6486.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LV, Bergen HL. A point mutation associated with bacterial macrolide resistance is present in both 23S rRNA genes of an erythromycin-resistant Treponema pallidum clinical isolate. Antimicrob Agents Chemother. 2000;44:806–807. doi: 10.1128/AAC.44.3.806-807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock GM, Hardham JM, McLeod MP, Sodergren EJ, Norris SJ. The genome of Treponema pallidum: new light on the agent of syphilis. FEMS Microbiol Rev. 1998;22:323–332. doi: 10.1111/j.1574-6976.1998.tb00373.x. [DOI] [PubMed] [Google Scholar]
- McKevitt M, Brinkman MB, McLoughlin M, Perez C, Howell JK, Weinstock GM, Norris SJ, Palzkill T. Genome scale identification of Treponema pallidum antigens. Infect Immun. 2005;73:4445–4450. doi: 10.1128/IAI.73.7.4445-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman MB, McKevitt M, McLoughlin M, Perez C, Howell J, Weinstock GM, Norris SJ, Palzkill T. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J Clin Microbiol. 2006;44:888–891. doi: 10.1128/JCM.44.3.888-891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman MB, McGill MA, Petterson J, Rogers A, Matejkova P, Smajs D, Weinstock GM, Norris SJ, Palzkill T. A novel Treponema pallidum antigen, TP0136 is an outer membrane protein that binds human fibronectin. Infect Immun. 2008;76:1848–1857. doi: 10.1128/IAI.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss CD, Field D, Moxon ER. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J Clin Invest. 2001;107:657–662. doi: 10.1172/JCI12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centurion-Lara A, Godornes C, Castro C, Van Voorhis WC, Lukehart SA. The tprK gene is heterogeneous among Treponema pallidum strains and has multiple alleles. Infect Immun. 2000;68:824–831. doi: 10.1128/IAI.68.2.824-831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centurion-Lara A, Sun ES, Barrett LK, Castro C, Lukehart SA, Van Voorhis WC. Multiple alleles of Treponema pallidum repeat gene D in Treponema pallidum isolates. J Bacteriol. 2000;182:2332–2335. doi: 10.1128/JB.182.8.2332-2335.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molepo J, Pillay A, Weber B, Morse SA, Hoosen AA. Molecular typing of Treponema pallidum strains from patients with neurosyphilis in Pretoria, South Africa. Sex Transm Infect. 2007;83:189–192. doi: 10.1136/sti.2006.023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay A, Liu H, Chen CY, Holloway B, Sturm AW, Steiner B, Morse SA. Molecular subtyping of Treponema pallidum subspecies pallidum. Sex Transm Dis. 1998;25:408–414. doi: 10.1097/00007435-199809000-00004. [DOI] [PubMed] [Google Scholar]
- Pillay A, Liu H, Ebrahim S, Chen CY, Lai W, Fehler G, Ballard RC, Steiner B, Sturm AW, Morse SA. Molecular typing of Treponema pallidum in South Africa: cross-sectional studies. J Clin Microbiol. 2002;40:256–258. doi: 10.1128/JCM.40.1.256-258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope V, Fox K, Liu H, Marfin AA, Leone P, Sena AC, Chapin J, Fears MB, Markowitz L. Molecular subtyping of Treponema pallidum from North and South Carolina. J Clin Microbiol. 2005;43:3743–3746. doi: 10.1128/JCM.43.8.3743-3746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MY, Liu H, Steiner B, Pillay A, Mickey T, Finelli L, Morse S, Markowitz LE, St Louis ME. Molecular subtyping of Treponema pallidum in an Arizona County with increasing syphilis morbidity: use of specimens from ulcers and blood. J Infect Dis. 2001;183:1601–1606. doi: 10.1086/320698. [DOI] [PubMed] [Google Scholar]
- Tantalo LC, Lukehart SA, Marra CM. Treponema pallidum strain-specific differences in neuroinvasion and clinical phenotype in a rabbit model. J Infect Dis. 2005;191:75–80. doi: 10.1086/426510. [DOI] [PubMed] [Google Scholar]
- Baseman JB, Nichols JC, Rumpp JW, Hayes NS. Purification of Treponema pallidum from Infected Rabbit Tissue: Resolution into Two Treponemal Populations. Infect Immun. 1974;10:1062–1067. doi: 10.1128/iai.10.5.1062-1067.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TJ, Norton J, Ott M, Richmond T, Nuwaysir K, Nuwaysir EF, Stengele KP, Green RD. Light-directed 5'-->3' synthesis of complex oligonucleotide microarrays. Nucleic Acids Res. 2003;31:e35. doi: 10.1093/nar/gng035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir EF, Huang W, Albert TJ, Singh J, Nuwaysir K, Pitas A, Richmond T, Gorski T, Berg JP, Ballin J, McCormick M, Norton J, Pollock T, Sumwalt T, Butcher L, Porter D, Molla M, Hall C, Blattner F, Sussman MR, Wallace RL, Cerrina F, Green RD. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editor. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Weinstock GM, Norris SJ, Sodergren E, Smajs D. Identification of virulence genes in silico: infectious disease genomics. In: Brogden KA, Roth JA, Stanton TB, Bolin CA, Minion FC, Wannemuehler MJ, editor. Virulence Mechanisms of Bacterial Pathogens. 3. Washington, D.C.: ASM Press; 2000. pp. 251–261. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material consists of two tables containing list of all identified sequence changes in TPA SS14 genome compared to [GenBank:AE000520] (Table S1) and list of primers used for WGF analysis (Table S2).