Abstract
Treatment of Arabidopsis (Arabidopsis thaliana) alternative oxidase1a (aox1a) mutant plants with moderate light under drought conditions resulted in a phenotypic difference compared with ecotype Columbia (Col-0), as evidenced by a 10-fold increase in the accumulation of anthocyanins in leaves, alterations in photosynthetic efficiency, and increased superoxide radical and reduced root growth at the early stages of seedling growth. Analysis of metabolite profiles revealed significant changes upon treatment in aox1a plants typical of combined stress treatments, and these were less pronounced or absent in Col-0 plants. These changes were accompanied by alteration in the abundance of a variety of transcripts during the stress treatment, providing a molecular fingerprint for the stress-induced phenotype of aox1a plants. Transcripts encoding proteins involved in the synthesis of anthocyanins, transcription factors, chloroplastic and mitochondrial components, cell wall synthesis, and sucrose and starch metabolism changed, indicating that effects were not confined to mitochondria, where the AOX1a protein is located. Microarray and quantitative reverse transcription-polymerase chain reaction analysis revealed that transcripts typically induced upon stress treatment or involved in antioxidant defense systems, especially chloroplast-located antioxidant defense components, had altered basal levels in untreated aox1a plants, suggesting a significant change in the basal equilibrium of signaling pathways that regulate these components. Taken together, these results indicate that aox1a plants have a greatly altered stress response even when mitochondria or the mitochondrial electron transport chain are not the primary target of the stress and that AOX1a plays a broad role in determining the normal redox balance in the cell.
The alternative oxidase (AOX) is a terminal oxidase found in all plants that functions as a dimer. In contrast to the multisubunit protein complexes of the cytochrome electron transport chain, AOX function only requires the expression of one nucleus-located gene (Finnegan et al., 2004). Although it is expressed during normal growth and development, it is often dramatically induced at the transcript level by a variety of treatments, frequently referred to as stresses. The increase in transcript abundance is typically accompanied by an accumulation in AOX protein in a range of plant species (Rhoads and McIntosh, 1992; Vanlerberghe and McIntosh, 1997; McCabe et al., 1998; Thirkettle-Watts et al., 2003; Escobar et al., 2004, 2006; Clifton et al., 2006). Conditions that alter the transcript abundance of AOX include the addition of compounds that inhibit the mitochondrial electron transport chain, such as antimycin A or rotenone (Vanlerberghe and McIntosh, 1997; Finnegan et al., 2004; Clifton et al., 2005; Rhoads et al., 2006), the availability of various nutrients, such as phosphate (Parsons et al., 1999; Sieger et al., 2005), nitrate, or ammonia (Escobar et al., 2004), and genetic lesions that alter mitochondrial function (Gutierres et al., 1997; Karpova et al., 2002; Dutilleul et al., 2003).
Several studies investigating the role of AOX have been carried out in tobacco (Nicotiana tabacum), in which lines that overexpress or underexpress AOX have been analyzed extensively (Vanlerberghe and McIntosh, 1997). Studies using these materials demonstrate that AOX plays a role to reduce the production of reactive oxygen species (ROS) in cell cultures in which mitochondrial function was inhibited (Maxwell et al., 1999; Yip and Vanlerberghe, 2001). Following from this, AOX has been proposed to play a role as a survival protein, by its ability to maintain mitochondrial function, playing a central role in determining ROS equilibrium in cells and plants (Robson and Vanlerberghe, 2002; Amirsadeghi et al., 2007) and playing a role in altering growth in response to nutrient availability (Sieger et al., 2005). It has also been shown that induction of AOX in tobacco plants can lead to an enhanced stress tolerance (Dutilleul et al., 2003), postulated to occur due to its ability to alter ROS equilibrium (Noctor et al., 2007). Arabidopsis (Arabidopsis thaliana) lines with altered AOX levels have also been produced, and, in agreement with the studies in tobacco, altered levels of ROS were evident, although other changes in transcript levels for selective oxidative stress-related enzymes were not observed (Umbach et al., 2005).
The role of AOX can also be deduced from situations in which it is induced. Thus, in thermogenic plants, the induction of AOX in floral organs facilitates the production of heat to attract insects for pollination (Finnegan et al., 2004). In nonthermogenic plants, it has long been proposed that AOX acts as an energy overflow pathway (Lambers, 1982). In Arabidopsis plants compromised in cyclic electron flow around PSI, AOX is induced to oxidize excess reducing equivalents produced in chloroplasts (Yoshida et al., 2007). Another situation in which AOX activity is induced is when electron transport is restricted via mutations in the cytochrome electron transport chain (Gutierres et al., 1997; Karpova et al., 2002). In Nicotiana sylvestris cytoplasmic male sterile lines (due to a deletion in mitochondrial DNA encoding subunits of complex I), the induction of AOX demonstrates its ability to affect/interact with redox and carbohydrate metabolism in plant cells, thus providing flexibility and playing a role in interorganelle communication (Dutilleul et al., 2003, 2005). The ability of AOX to affect redox and energy metabolism as well as to play a role in interorganelle communication has led to the proposal that AOX acts in reprogramming metabolism in response to changing circumstances (Arnholdt-Schmitt et al., 2006; Clifton et al., 2006). A programming role of AOX may serve to maintain energy charge and constant growth under a variety of circumstances (Moore et al., 2002).
Although AOX is rapidly induced by a variety of stress treatments in various plants (Vanlerberghe and McIntosh, 1997; Finnegan et al., 2004; Clifton, 2006), there is little evidence that a lack of AOX has a dramatic effect even under stressful conditions. Arabidopsis plants with altered AOX expression display a 20% reduction in growth in cold conditions (Fiorani et al., 2005), and in tobacco cells reduced AOX prevents cell death under a variety of situations (Robson and Vanlerberghe, 2002). Overexpression of AOX in tobacco plants also increases sensitivity to ozone treatment (Pasqualini et al., 2007), in contrast to what was observed in tobacco cytoplasmic male sterile lines, in which AOX was induced and increased tolerance to ozone was observed (Dutilleul et al., 2003). Normally, in field conditions, plants are likely to be exposed to a combination of stress conditions (Mittler, 2006). If AOX does play a role in maintaining redox and energy metabolism under stress, it is possible that this will be more apparent under a combination of stresses, rather than with the single and often severe treatments used to induce AOX in published reports. Thus, we used moderate combined stress conditions with Arabidopsis lines lacking AOX1a in order to gain further insight into the role of AOX in plants.
RESULTS
Plants Lacking AOX1a Display a Stress Phenotype
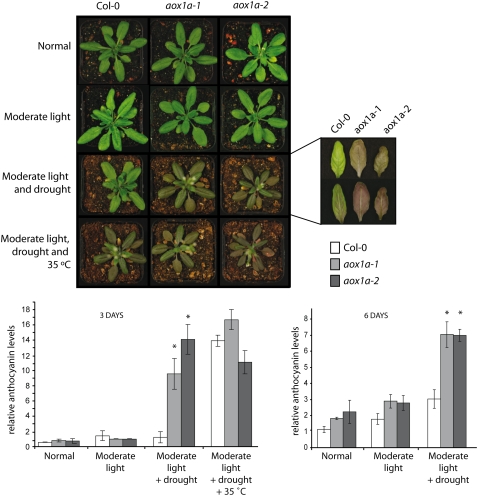
The role of AOX1a in the cellular stress response was analyzed using two independent T-DNA insertion lines inactivating AOX1a, referred to as aox1a-1 and aox1a-2. These T-DNA insertion lines were confirmed to contain a DNA insertion in exon 4 and intron 2, respectively (Supplemental Fig. S1, A and B), which resulted in the absence of an immunodetectable AOX protein with an apparent molecular mass of 34 kD (Supplemental Fig. S1C). To further investigate the role of AOX1a in a stress response, we exposed plants to drought, light, and temperature stresses, as an analysis of many microarray experiments indicated that AOX1a was highly responsive to abiotic stresses (Clifton et al., 2006). Transferring plants from a normal light regime (100 μE m−2 s−1) to a moderately elevated regime (250 μE m−2 s−1) had little effect on plants after 3 or 6 d, with no significant difference in anthocyanin content measured (Fig. 1). Likewise, withholding water for 3 to 6 d had no apparent effect on phenotype (data not shown). However, transfer to moderate light during drought stress, imposed by not watering for 4 d before transfer, produced a notable difference in leaf color between Col-0 and aox1a plants (Fig. 1). The purple pigmentation of the aox1a plants correlated with a greater than 10-fold difference in anthocyanin content between Col-0 and aox1a after both 3 and 6 d of treatment (Fig. 1). Transfer of plants to moderate light and elevated temperature (35°C) during drought resulted in both aox1a and Col-0 plants accumulating anthocyanin pigments (Fig. 1). Notably, the aox1a plants that were light and heat treated during drought were dead after 6 d, whereas Col-0 plants were still alive, and after being returned to normal growth conditions and watered, they flowered and produced viable seeds (data not shown).
Figure 1.
The response of aox1a plants to moderate light, drought, and temperature stress. Col-0 and aox1a plants were grown for 4 weeks at 22°C, 16 h of light and 8 h of dark under 100 μE m−2 s−1 light, referred to as normal conditions. Plant were transferred to 250 μE m−2 s−1 and watered as normal, referred to as moderate light; water was withheld for 3 d and then plants were transferred to 250 E m−2 s−1 with no subsequent watering, referred to as moderate light and drought; and water was withheld for 4 d and plants were transferred to 250 μE m−2 s−1 with no subsequent watering at 35°C, referred to as moderate light, drought, and 35°C. Photographs were taken at 3 d after transfer to the moderate light conditions; the inset shows the upper (top) and lower (bottom) surfaces of the leaves. The histograms indicate the relative anthocyanin content of leaves from these plants. Asterisks indicate significant differences (P > 0.05) between the anthocyanin content of Col-0 leaves and aox1a leaves. Note that for 6-d light-, drought-, and heat-treated plants, the aox1a plants were dead; thus, the amount of anthocyanin could not be determined.
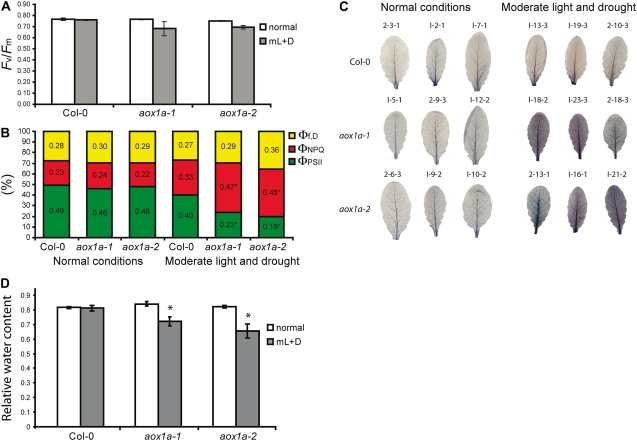
Different parameters were measured in moderate light- and drought-treated and untreated Col-0 and aox1a plants to determine the effects on photosynthesis and photoprotection. The maximum quantum efficiency of PSII photochemistry (Fv/Fm) remained unchanged for Col-0 plants grown under both conditions, and while it decreased for aox1a plants under stress, the decrease was not significant. Measurement of electron transport rates and chlorophyll fluorescence quenching for the three genotypes under the treatment conditions revealed that the absence of AOX1a mostly affects both photochemical and nonphotochemical quenching (NPQ). Both aox1a lines dissipated more excitation energy by NPQ than Col-0 under moderate light and drought treatment, as evidenced by the significantly higher light-dependent thermal dissipation component of NPQ (ΦNPQ) and lower photochemical efficiency (ΦPSII; Fig. 2B; Supplemental Table S1). Despite the differences in photosynthetic capacity, chlorophyll and carotenoid composition and the chlorophyll a/b ratio were unaltered in the aox1a lines compared with Col-0 (data not shown) and were comparable to published results (DellaPenna and Pogson, 2006). This would indicate no substantive changes in ratios of PSII to PSI and antennae to photosystems. ROS, superoxide radical (O2−), and hydrogen peroxide (H2O2) were qualitatively measured in leaves isolated from plants subjected to normal conditions or moderate light and drought treatment (Fig. 2C). O2− was assayed by visualizing the amount of precipitate formed after incubation with nitroblue tetrazolium and H2O2 using 3,3′-diaminobenzidine. There were no differences between Col-0 and aox1a lines for H2O2 in normal or moderate light and drought treatment (data not shown). However, the stress treatment did increase the levels of O2− in both aox1a lines compared with Col-0. The pattern of ROS accumulation was consistent with what is typically found in Arabidopsis plants exposed to stress (Fryer et al., 2003). The relative water content between aox1a and Col-0 leaves did not differ under normal conditions, but aox1a plants had a 10% to 20% decrease in relative water content after 4 d of stress treatment (Fig. 2D). This reduction in water content was probably not due to a difference in cuticle thickness or an osmostic shock response, as the dehydration rates of detached leaves were similar for both Col-0 and aox1a mutants (data not shown).
Figure 2.
Analysis of the stress imposed on Col-0 and aox1a plants upon moderate light/drought treatment. Plants were grown for 4 weeks at 22°C, 16 h of light and 8 h of dark under 100 μE m−2 s−1 light, referred to as normal conditions. Plants were then subjected to either continued normal growth conditions or transferred to moderate light of 250 μE m−2 s−1 and withholding water for 4 d prior to transfer and no subsequent watering, referred to as moderate light and drought conditions. Measurements were made after 4 d of treatment. A, Fv/Fm. Data shown are means and se of two different experiments with four replicates per line per treatment per experiment. B, ΦPSII, ΦNPQ, and Φf,D were determined at normal light and moderate light and drought. Mean values are indicated. Asterisks denote significant differences (P < 0.05) between aox1a plants and Col-0 using a t test (see Supplemental Table S1 for details). C, Leaves were harvested from three plants per line per treatment per experiment and assayed qualitatively for O2− by nitroblue tetrazolium. Representative images from individual plants for experiments 1 and 2 are shown and labeled as follows: 2-13-1, experiment 2, plant 13, leaf 1. D, Relative water content of Col-0 and aox1a plants grown under normal conditions or moderate light and drought conditions. Weights of whole rosettes were measured directly after excision from the plant (Fw), after submersion in water for 4 h (Tw), or after drying at 80°C overnight (Dw). Relative water content = (Fw − Dw) ÷ (Tw − Dw). Averages and sd of four plants are shown.
A number of additional parameters were analyzed to determine whether there was any physiological difference between Col-0 and aox1a plants. Analysis of root growth on vertical agar plates revealed that aox1a plants displayed an approximately 10% decrease in root length after 9 d (Table I). However, no differences were detectable with leaf growth under normal growth conditions after 3 or 4 weeks. Measurement of leaf disc respiratory rates revealed no significant differences between Col-0 and aox1a plants in total oxygen consumption, with rates of 2,000 nmol O2 min−1 g−1 dry weight (±10%) obtained under both normal and moderated light and drought treatment conditions. As expected, a clear difference in alternative oxidase capacity was measured between aox1a and Col-0; in the former, oxygen consumption in the presence of potassium cyanide was barely detectable, whereas in the latter, a capacity of almost 500 nmol O2 min−1 g−1 dry weight (±10%) was present.
Table I.
Measurements of root growth in 9-d-old Col-0 and aox1a seedlings
Seedlings were germinated on vertical Murashige and Skoog agar plates, images were taken after 9 d, and root growth was measured using the software ImageJ. Mean (millimeter) and se values are shown from three independent sets of measurements. Student's t test was performed comparing each aox1a line with Col-0. NA, Not applicable.
| Genotype | Mean | se | P |
|---|---|---|---|
| Col-0 | 52.189 | 2.003 | NA |
| aox1a-1 | 45.895 | 2.183 | 0.041 |
| aox1a-2 | 37.950 | 2.112 | 1.19E-05 |
aox1a Plants Display Altered Metabolite Profiles
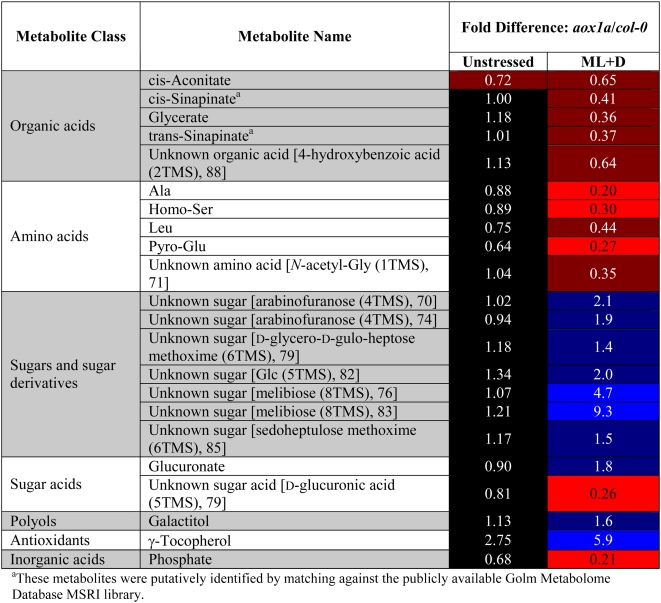
Quantitative profiles of major metabolites from leaves of Col-0 and aox1a plants before and after the light and drought treatment were generated using gas chromatography-mass spectrometry (GC-MS) analysis of derivatized compounds from methanol-soluble leaf extracts. In both Col-0 and aox1a samples, a series of significant changes were recorded in response to the treatment. These included 24 metabolites that were significantly increased in abundance in both Col-0 and aox1a plants, nine of which were significantly induced only in Col-0, nine that were significantly induced only in aox1a, and only four metabolites that were significantly decreased by the treatment in either genotype (Supplemental Table S2). A range of these metabolites could be unambiguously identified by comparison with standards libraries, while others can be placed in broad classes with best matches to related compounds found in NIST05 MS libraries. The general patterns of increases in both Col-0 and aox1a treated plants were indicative of drought stress, as exemplified by the increased abundance of disaccharides and trisaccharides (Seki et al., 2007), but the fold changes were generally larger in the aox1a plant samples.
Comparative analysis of Col-0 versus aox1a metabolite levels showed that nine compounds were more abundant and 13 were less abundant in the aox1a plants compared with Col-0 following the combined light and drought treatment. Comparative analysis of Col-0 versus aox1a also revealed almost no significant differences in these metabolites between the plants under normal growth conditions. Notably, these differences could be broadly classed as decreases in amino acids and organic acids and increases in sugars. This kind of metabolic pattern is consistent with the notion that loss of AOX1a could form a constriction of primary metabolism in aox1a plants by decreasing the capacity of the tricarboxylic acid (TCA) cycle to operate uncoupled from oxidative phosphorylation (Table II; Lambers, 1982; Vanlerberghe and McIntosh, 1997).
Table II.
Metabolite level differences between aox1a lines and Col-0 under normal growth and environmental stress conditions
Metabolite levels were compared between aox1a and Col-0 lines under normal watering and light conditions and after 3 d of moderate light and drought using GC-MS-based metabolite profiling (see “Materials and Methods”). The mean fold difference (between aox1a and Col-0) in GC-MS signal levels (across the two independent aox1a lines) are shown for metabolites that responded similarly in both aox1a lines. The fold differences under normal growth conditions are shown in the “Unstressed” column, while the fold differences under the stress conditions are given in the “ML+D” (moderate light and drought) column. Metabolite signals that were significantly higher (P < 0.05, n = 5) in both aox1a lines are highlighted in blue, while signals that were significantly lower (P < 0.05, n = 5) are highlighted in red. Signals that were not significantly different from Col-0 in either aox1a line are shown in black. Color intensities are related to metabolite response intensities, with more strongly responsive signals highlighted in brighter tones. Compounds shown in square brackets are unknown metabolites with mass spectral homology to the indicated compound, with the number after the compound name being the “simple” match score reported by AMDIS when searched against the NIST02 MS library.
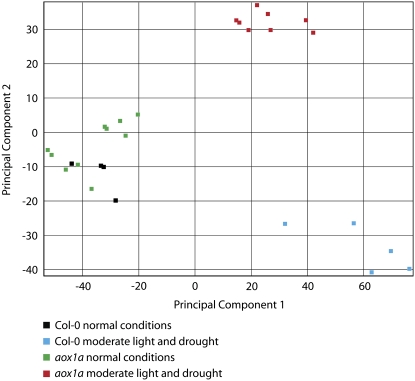
To obtain an overall picture of the differences in metabolite abundance between Col-0 and aox1a and upon treatment, the metabolite profile was subjected to principal component analysis (PCA; Fig. 3). Under normal growth conditions, there were no apparent differences between Col-0 and aox1a, but upon treatment the genotypes diverged. It was also clear that both aox1a lines diverged together, away from Col-0. Thus, in addition to the visually apparent differences between Col-0 and aox1a in the production of the secondary metabolite anthocyanin pigments (Fig. 1), there were clear differences in the primary metabolite profile upon treatment.
Figure 3.
PCA of metabolites from Col-0 and aox1a under normal conditions and after combined light and drought treatment. To obtain an overview of the differences between the genotypes and with treatment, PCA analysis was carried out on all metabolites (Supplemental Table S2).
An Altered Transcriptome during Stress Is Apparent in aox1a Plants
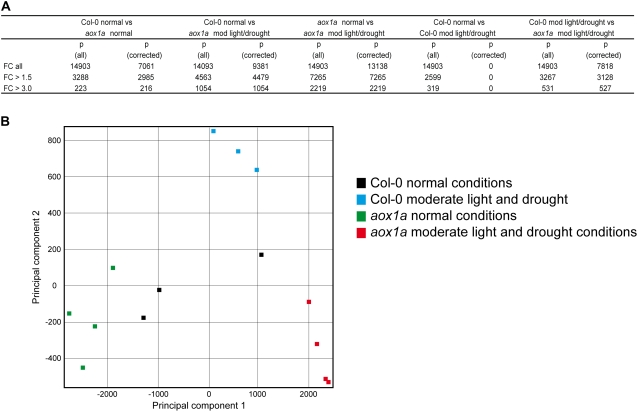
To further investigate the stressed phenotype, microarray analyses were performed. This revealed significant differences between Col-0 and aox1a plants, both under normal growth conditions and in response to the moderate light and drought conditions. Using a false discovery rate (FDR) correction factor of q = 0.1 and a fold change of >3, 216 and 1,054 transcripts were altered in aox1a plants compared with Col-0 plants under normal conditions and under moderate light and drought conditions, respectively (Fig. 4A). If aox1a plants grown under normal conditions are compared with aox1a plants subjected to combined moderate light and drought stresses, 2,219 transcripts changed in abundance by 3-fold or greater (Fig. 4A). Comparing aox1a treated with Col-0 treated plants, 527 transcripts were changed by greater than 3-fold. Overall, this indicated that there was widespread rearrangement of the aox1a transcriptome both at the basal level and, more dramatically, upon treatment. Note that no significant changes were observed between Col-0 plants grown under normal and treated conditions with FDR correction, indicating that the treatment regime was moderate for Col-0 plants. The large differences in the transcriptome between aox1a and Col-0 are supported by PCA (Schoelkopf et al., 1998; Fig. 4B). Based on principal component 1 (PC1), there is a large difference between aox1a in normal conditions versus treatment. Differences between aox1a and Col-0 under normal conditions are also evident based on PC1, and while there is some difference between Col-0 normal and treated samples based on PC2, overall this difference is smaller in terms of scale than the difference observed for aox1a.
Figure 4.
Whole transcriptome analysis of the response of Col-0 and aox1a lines to moderate light and drought treatment. A, Changes in transcript abundance between Col-0 and aox1a under moderate light and drought treatment. FC (all) refers to the number of genes called present in the array, FC > 1.5 refers to the number of genes with an expression fold change of 50% or more, and FC > 3.0 refers to the number of genes with an expression fold change greater than 200%. P (all) represents the number of genes matching the FC criteria for which a P value was calculated, while P (corrected) represents the number of genes matching the FC criteria with a P value of 0.05 or less when corrected for FDR according to a method based on Benjamini and Hochberg (1995), a q value computation. B, PCA of the above changes to obtain an overall view of the differences between genotypes and treatments.
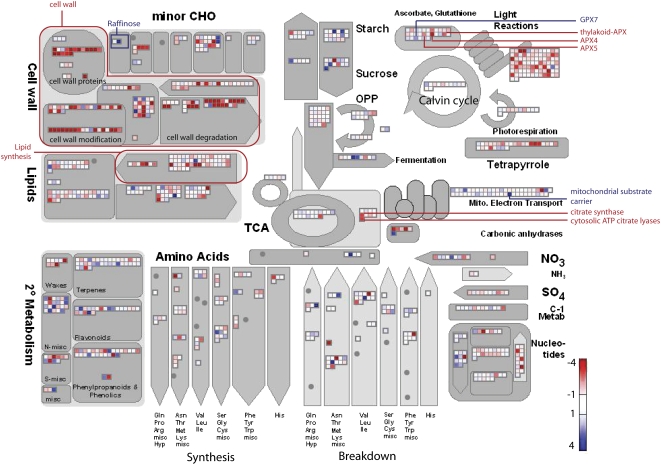
To gain an overview of these changes in the aox1a lines compared with Col-0 under normal conditions and light and drought treatment with regard to cellular metabolism, the significantly changing transcripts, after FDR correction, were displayed using MapMan software (Thimm et al., 2004; Usadel et al., 2005). Under normal conditions, there were increases in transcript abundance for genes encoding proteins involved in cell wall modification and synthesis and lipid synthesis (Supplemental Fig. S2A). Likewise, there were increases in transcripts for genes encoding proteins involved in photorespiration and some genes involved in antioxidant defense (Supplemental Fig. S2A). Upon treatment, decreases in transcript abundance for genes encoding proteins involved in photosynthesis, photorespiration, and cell wall metabolism were observed, while increases in transcripts for genes encoding components of Suc and starch metabolism and flavanoid production were observed (Fig. 5). Notably, if aox1a untreated plants were compared with aox1a treated plants, the magnitude of the changes was greater (Supplemental Fig. S2B). For instance, transcripts associated with cell wall proteins, cell wall synthesis, and modifications were drastically decreased, suggesting that these processes may be blocked at the conversion of Glc-1-P to d-GlcUA (Supplemental Fig. S2, B and C). Metabolite data also support this conclusion, as upon moderate light and drought treatment aox1a plants accumulate more than double the amount of Glc compared with Col-0 plants (Table II; Supplemental Fig. S2C), whereas the amount of d-GlcUA is approximately 5 times lower than the amount observed in Col-0 plants after treatment (Table II; Supplemental Fig. S2C). Taken together, this may indicate a shift in transcripts encoding components of central metabolism away from the growth and synthesis of complex energy stores toward the breakdown and mobilization of internal carbon stores in the aox1a plants upon treatment.
Figure 5.
Overview of changes in transcript abundance associated with primary metabolism. Transcripts associated with primary metabolism are significantly changed after FDR correction between aox1a and Col-0 under moderate light and drought treatment conditions. Changes are displayed using the MapMan software (http://gabi.rzpd.de/projects/MapMan/). The abundance ratio or fold change is shown by the color scale, with red indicating a decrease and blue indicating an increase in transcript abundance. Key genes and areas are highlighted in red and blue based on the direction of transcript abundance changes. CHO, Carbohydrate metabolism; OPP, oxidative phosphorylation pathway.
Studies have shown that impaired photorespiration can restrict photosynthesis under conditions that lead to high oxygenation of Rubisco, for example, increased temperature, high light, or stress conditions, which lead to stomatal closure, and low intracellular CO2 concentrations, such as during drought (Wingler et al., 2000). High sugar levels have also been shown to inhibit photosynthesis and can act in a variety of signaling pathways (Koch, 1996). Our metabolite data showed large accumulations in an array of sugars, for example, raffinose increased by 22 times during treatment of aox1a plants (Supplemental Table S2). Such effects may lead to a down-regulation of transcripts encoding proteins involved in photosynthesis (Supplemental Fig. S2B; Supplemental Table S2).
Some general trends are noticeable in the proportion of transcripts that changed in abundance that can be ascribed to specific component and function categories (Supplemental Fig. S3). Genes encoding ribosomal (6.29% versus 1.68% in the genome), chloroplastic and plastidic (12.75% and 6.87% versus 7.98% and 2.70% in the genome), and mitochondrial (4.99% versus 3.71% in the genome) proteins are significantly up-regulated in the aox1a lines under normal conditions (Supplemental Fig. S3). The ribosomal (5.22% versus 1.68% in the genome) and chloroplastic (11.89% versus 7.98% in the genome) and plastidic (6.10% versus 2.70% in the genome) groups are significantly down-regulated in aox1a upon treatment (Supplemental Fig. S3), whereas the mitochondrial components are significantly up-regulated upon aox1a treatment (4.75% versus 3.71% in the genome; Supplemental Fig. S3; Supplemental Table S3). Analysis of the changes that occurred in aox1a plants both under normal and treated conditions indicated that there were substantial changes in transcript abundance for genes encoding transcription factors and mitochondrial and chloroplastic components (Supplemental Table S3). The large number of changes of 2-fold or more, including almost 500 transcription factors and chloroplastic components and 250 mitochondrial components (Supplemental Table S3), indicate that a lack of AOX1a has widespread effects beyond mitochondrial function.
Analysis of transcripts for genes encoding mitochondrially located components revealed that transcript abundance for genes encoding proteins of the cytochrome electron transport chain and the TCA cycle were not dramatically changed in aox1a plants subjected to moderate light and drought treatment. This is consistent with previous findings that these components are largely unaffected by a variety of treatments (Clifton et al., 2005, 2006). However, examination of other bypasses of the respiratory chain revealed that three rotenone-insensitive NAD(P)H dehydrogenases were up-regulated, two internal (At1g07180 NDA1, +2.4-fold; At2g29990 NDA2, +3.5-fold) and one external (At4g05020 NDB2, +3.1-fold), whereas the transcript abundance of one encoding an external NAD(P)H dehydrogenase was down-regulated (At4g28220 NDB1, −3.3-fold; Fig. 5; Supplemental Table S3). Only one gene encoding an uncoupling protein was altered (At3g54110, −2.15-fold). Although some other genes encoding mitochondrial proteins known to be responsive to oxidative stress were also up-regulated (e.g. At2g41380, encoding a putative embryo-abundant protein; Tolleter et al., 2007), this was not a universal trend. Notably, At2g21640, a gene whose induction was defined as one of five hallmarks of oxidative stress (Gadjev et al., 2006) and that was shown to be induced at a protein level in mitochondria upon oxidative stress (Sweetlove et al., 2002), had a reduced transcript abundance of 4-fold in aox1a plants grown under normal conditions and upon treatment relative to Col-0 plants (Fig. 6; Supplemental Table S3). A previous analysis of a large number of microarray experiments listed 20 genes encoding mitochondrial proteins most responsive to stress treatment (Clifton et al., 2006). Fourteen of these were called present on the arrays in this study. In Col-0 plants, none of these genes changed significantly upon treatment, in contrast to aox1a plants, in which 11 changed significantly, eight were up-regulated, and three were down-regulated (Supplemental Table S3). Thus, aox1a plants were sensitive to the combined drought and light treatment, inducing transcripts normally associated with a stress response and that were not induced in Col-0 by the combined treatment.
Figure 6.
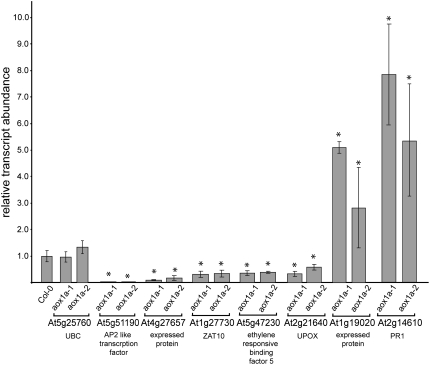
Analysis of transcript differences between Col-0 and aox1a plants. QRT-PCR analysis of transcript abundance is shown for genes considered hallmarks of oxidative stress or core environmental stress response genes between aox1a and Col-0 grown under normal conditions. Changes in gene expression are normalized to the levels in Col-0, which are set to 1. Asterisks indicate significant differences (P ≤ 0.05) between aox1a and Col-0. UBC is included as a comparison with no significant change observed. See Supplemental Table S3 for a full list of genes analyzed.
In the case of chloroplasts, many of the changes were associated with the Calvin cycle or the photosynthetic electron transport chain. The changes were dramatic: 44 genes displayed a change of 10-fold or more. These ranged across a variety of functions, from protein import (Tic20 At4g03320, +36-fold) and chloroplast nucleoid-binding protein (At1g09750, −249-fold) to starch synthase (At1g32900, +52-fold). Given the magnitude and number of changes, it appears that many chloroplast-located processes are affected. Notably, transcripts for many genes encoding proteins involved in chlorophyll synthesis were also down-regulated (Supplemental Table S3).
aox1a Plants Have an Altered Defense Equilibrium
Changes in transcript abundance between aox1a and Col-0 under normal conditions were further investigated and confirmed using quantitative reverse transcription (QRT)-PCR. We considered five genes that have been recognized as markers for oxidative stress and four core environmental stress response genes (Gadjev et al., 2006; Kilian et al., 2007). Four genes encoding mitochondrial proteins that have been reported to be responsive to stress (Clifton et al., 2006) were also included, along with PR1a, a marker for induction by salicylic acid linked to responses to biotic pathogens and oxidative stress (Green and Fluhr, 1995; Bechtold et al., 2005), and four other genes linked to stress response in a variety of studies (Rizhsky et al., 2004; Panchuk et al., 2005; Yang et al., 2005; Lee et al., 2006). Transcript abundance was analyzed for 17 genes in total, with ubiquitin (UBC) as a control (Supplemental Table S4; Czechowski et al., 2005). The basal level of transcript abundance of seven of these genes was altered significantly in aox1a plants, five were lower and two were higher (Fig. 6); notably, At2g21640, previously defined as a hallmark of oxidative stress encoding a mitochondrial protein (Sweetlove et al., 2002; Gadjev et al., 2006), was confirmed to be lower by QRT-PCR in aox1a plants. At least three of these genes encode regulators of gene expression, AP2, ZAT10, and ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR5 (Kilian et al., 2007). Along with a large increase in PR1a transcript abundance, the altered level of these marker genes suggests an altered equilibrium of signaling in aox1a plants. Despite this, the other four members of the Arabidopsis AOX multigene family do not show altered transcript expression in aox1a plants under normal or treated conditions (data not shown).
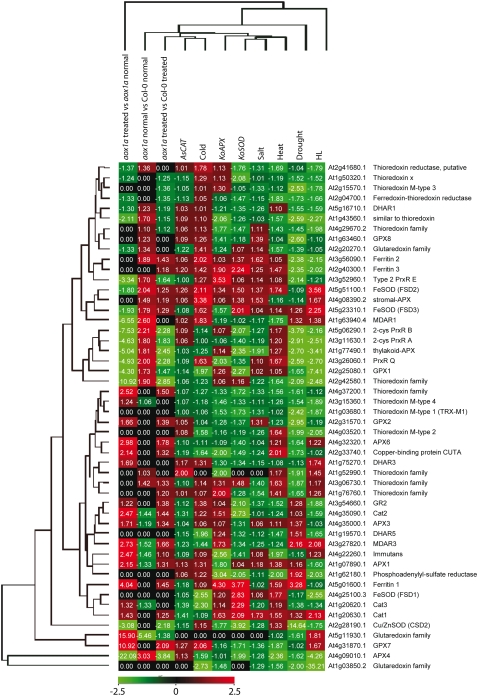
Some transcripts for these selected genes displayed altered levels in aox1a plants even under normal conditions; the known roles of these genes suggest an alteration in components that are involved in the response to oxidative stress in the aox1a plants. For example, transcript abundance for PR1a, which is induced by oxidative stress (Green and Fluhr, 1995), and ZAT10, which is responsible for the regulation of several other defense response genes (Mittler et al., 2006; Rossel et al., 2007), was significantly different in untreated aox1a plants compared with Col-0 (Fig. 6). We investigated transcript responses to alterations in all known ROS defense systems and NAD(P)H oxidases in Arabidopsis (Mittler et al., 2004) and compared these responses with those seen in the aox1a plants. Overall, the transcript abundance for genes encoding these components in the aox1a plants was not very similar to that found with any of the treatments or genotypes (Supplemental Fig. S4). In all comparisons, there were changes in the aox1a plants (i.e. compared with Col-0 under normal conditions, compared with Col-0 upon treatment, and when aox1a is grown under normal versus treated conditions). When ROS defense systems and NAD(P)H oxidases located in chloroplasts were analyzed (Fig. 7), it was apparent that the lack of AOX1a had a significant effect outside mitochondria. Thus, in comparison with Col-0 under normal conditions, transcripts for genes encoding APX4, MDAR1, and FeSOD were increased in abundance by 2-fold or more, while a glutaredoxin family member was decreased in abundance by 5-fold (Fig. 7). Overall, 13 transcripts encoding proteins involved in ROS defense and NAD(P)H oxidases changed in abundance by more than 2-fold compared with Col-0 under normal conditions, eight up-regulated and five down-regulated (Supplemental Fig. S4). Upon treatment, the differences were more dramatic: 19 versus Col-0 (16 down-regulated, three up-regulated) and 35 versus aox1a untreated (18 up-regulated, 17 down-regulated; Supplemental Fig. S4). It appeared that the magnitude of changes in aox1a was greater upon treatment; thus, when the transcript abundance in aox1a plants was compared (aox1a untreated versus aox1a treated) with Col-0 treated, the changes were more dramatic in the former. For 11 transcripts encoding chloroplast-located components, the changes were greater than 3-fold in aox1a in response to treatment (Figs. 5 and 7). Also notable was that the magnitude of individual transcript changes in the aox1a treated plants was generally greater than the changes reported in previous studies. This was especially evident for the chloroplast-located antioxidant systems; for example, an ascorbate peroxidase was down-regulated over 22-fold while GPX7 and a glutaredoxin family member were up-regulated by 10- and 15-fold, respectively. Thus, the basal and induced levels of antioxidant defense systems were altered in aox1a, and, importantly, the direction of change was altered with treatment for many (i.e. while some were up-regulated in aox1a untreated compared with Col-0, they were down-regulated in aox1a treated compared with aox1a untreated; Fig. 7).
Figure 7.
Relative transcript abundance for genes encoding antioxidant defense components located in chloroplasts. The relative expression data for antioxidant defense components for heat, drought, salt, cold, and high-light treatments, knockout superoxide dismutase, knockout catalase, and antisense ascorbate peroxidase were taken from Mittler et al. (2004). Red indicates an increase and green indicates a decrease in abundance, with numbers indicating magnitude on a linear scale; black indicates no significant change.
Analysis of transcript abundance for genes encoding proteins involved in anthocyanin biosynthesis revealed that many were up-regulated several hundred-fold (Fig. 5; Supplemental Fig. S5), consistent with the increase observed in anthocyanins (Fig. 1). Chalcone synthase (At5g13930), 9-flavonol 3-hydroxylase (At5g07990), dihydroflavonol 4-reductase (At5g42800), leucoanthocyanidin dioxygenase (At4g22870), UDP-Glc-flavonoid glycosyltransferase (At5g54060, At4g14090, and At1g22370), and glutathione S-transferase-like (At5g17220) were up-regulated from 10-fold to several hundred-fold. Also, two regulatory genes of this pathway, PRODUCTION OF ANTHOCYANIN PIGMENT1 and -2 (At1g56650 and At1g66390, respectively), were up-regulated by 80- and 327-fold, respectively. Increased anthocyanin production has been observed previously in double mutants for cytosolic and thylakoid ascorbate peroxidase and has been suggested to be a mechanism that is activated in response to increased oxidative stress (Miller et al., 2007). Thus, an increase in anthocyanins and the induction of transcripts encoding proteins of this biosynthetic pathway are consistent with altered ROS signaling in aox1a plants.
DISCUSSION
This study was undertaken to investigate the link between AOX and stress response(s) in plants. Over 20 years of study have linked AOX activity to oxidative stress (Vanlerberghe and McIntosh, 1997; Finnegan et al., 2004; Rasmusson et al., 2004; Rhoads et al., 2006), and many studies have indicated that AOX activity could play a role in allowing the TCA cycle to operate uncoupled from oxidative phosphorylation (Lambers, 1982; Vanlerberghe and McIntosh, 1997). Here, we report that a clear stress phenotype characterized by alterations in the phenotype, transcriptome, and some metabolites could be induced in aox1a plants by combining moderate light and drought. Additionally, it appears that aox1a plants under stress have altered photochemistry, as evidenced by lower levels of photochemical efficiency and higher levels of NPQ under the light regimes used. A small but significant difference was observed in root length after 9 d of growth, but no difference in shoot growth could be detected. Previously, a vegetative shoot growth phenotype was reported for plants with altered AOX grown at 12°C but not at 23°C (Fiorani et al., 2005). As seeds were cold vernalized at 4°C to synchronize germination and analysis of microarray data indicates that AOX1a is expressed in germinating seeds (Zimmermann et al., 2004), the difference may be due to the cold vernalization treatment.
One apparent effect of altering mitochondrial AOX is an alteration of the signals controlling antioxidant defense systems throughout the cell. Evidence for this comes from observations that transcript abundance for 59 genes encoding proteins involved in antioxidant defenses changed upon stress treatment in aox1a plants; in particular, transcripts for chloroplast iron and copper/zinc superoxide dismutases were down-regulated upon stress treatment (Fig. 7; Supplemental Fig. S4). Increased O2− in stress-treated aox1a plants compared with wild-type plants is consistent with patterns observed in the wild type upon severe photooxidative stress (Fryer et al., 2003; Rossel et al., 2007). This may indicate a perturbation in the balance of ROS production and amelioration, such as the 3-fold decrease of Cu/ZnSOD (CSD2) in stressed aox1a plants. In response to light stress, previous studies have observed the induction of cytosolic ROS defense systems (Karpinski et al., 1997, 1999; Rossel et al., 2002; Pnueli et al., 2003; Davletova et al., 2005). In a similar manner, a disruption in the mitochondrial electron transport chain can lead to alterations in ROS in various organelles due to the complexity and interaction of the ROS signaling and defense networks. A previous study analyzing antisense Arabidopsis plants for AOX1a also concluded that there were alterations in transcript abundance for genes encoding chloroplast proteins, including three involved in antioxidant defense (Umbach et al., 2005). Thus, it appears that inactivation of AOX1a has a greater effect outside the mitochondrion in terms of altering metabolism and antioxidant defense compared with within the mitochondrion. Although there are some apparent differences relating to the ability of AOX to reduce ROS from studies carried out in tobacco (Maxwell et al., 1999; Amirsadeghi et al., 2006) and Arabidopsis (Umbach et al., 2005), this study only detected an increase in O2− that was similar in pattern to that observed with photooxidative stress (Fryer et al., 2003). As we also observed differences in photochemistry, the O2− observed is likely to originate in chloroplasts and not as a direct result of the proposed role for AOX in reducing the production of ROS in mitochondria. The decrease in the Fv/Fm is not due to the effects of drought, as there are no marked changes in photochemistry in Arabidopsis with relative water content of 70% (N.S. Woo, M.R. Badger, and B.J. Pogson, unpublished data). This indicates that the perturbations in aox1a are not simply due to a greater susceptibility to drought and suggests a more complex interplay between photochemistry and ROS perception and detoxification in aox1a knockout plants.
When the transcriptional responses in aox1a plants under moderate light and drought treatment, distinct from Col-0 under treatment, were compared with findings from previous studies designed to induce ROS production in the chloroplast, a large overlap was observed. Comparative analysis was performed between transcripts changing in Col-0 treated versus aox1a treated plants and a study in which Arabidopsis seedlings were subjected to high light intensities (1,000 μE m−2 s−1) for 1 h (Rossel et al., 2002). Of the 66 transcripts that were significantly altered in that previous study, 27 were significantly altered after FDR correction in our study in the same direction and by a similar magnitude. Several of these transcripts encode regulatory factors, for example, HY5, a key regulator of nucleus-encoded photosynthesis-associated genes (Ruckle et al., 2007), and transcripts associated with antioxidant defense systems, for example, l-ascorbate peroxidase APX1 (At1g07890). Transcript abundance changes in Col-0 plants under treatment versus aox1a plants under treatment were also compared with those from another study in which Arabidopsis plants were treated with methyl viologen, a superoxide anion propagator in chloroplasts in the light (Scarpeci et al., 2008). Again, a substantial overlap was observed between the two studies; of the 300 transcripts changing in the previous study, 80 were altered by the same magnitude and direction in our study after FDR correction. Again, this included a number of transcripts associated with regulatory proteins, for example, a zinc finger (C3HC4-type RING finger) family protein (At1g14200) and DRE-BINDING PROTEIN2A (At5g05410). Notably, At2g21640, a transcript encoding a mitochondrial protein defined as one of the hallmarks of oxidative stress (Sweetlove et al., 2002; Gadjev et al., 2006), was also down-regulated in this previous study (Scarpeci et al., 2008) by a similar magnitude to that observed in this study. This suggests that the increase in ROS was not mitochondrial in origin. While there are a number of differences between these studies and our work, including the additional imposition of drought stress in this study and differences in sampling time (1 and 2 h, respectively, for the previous studies versus 3 d in this work), a key overlap is still observed. Taken together, these findings suggest that aox1a plants under moderate light and drought treatment display at least in part a transcriptional response to chloroplast-generated ROS.
As aox1a plants display an increase in O2− during treatment, some similar changes may theoretically occur with chemical-induced oxidative stress by menadione, which has been shown to disrupt mitochondrial metabolism (Baxter et al., 2007), or by knockdown of MnSOD, which inhibits/inactivates TCA cycle enzymes (Morgan et al., 2008). However, while there appear to be some similarities in the reprogramming of cellular metabolism with the menadione study, such as the decrease in cell wall biosynthesis and decreases in Ala, homo-Ser, and pyro-Glu, there are as many differential responses as common ones between the two studies. Treatment of Arabidopsis cells with menadione decreases the rates of synthesis of Suc, Gal, and raffinose, in contrast to an increase in many sugars observed in this study. This suggests that the oxidative stress induced in aox1a plants during light and drought treatments is distinct from the generalized oxidative assault of menadione treatment, but given the differences in the tissues and the timing of sampling in the two studies, it is not surprising that they diverge significantly. In the MnSOD study (Morgan et al., 2008), the oxidative stress is focused on the mitochondria, but this leads to steady-state increased levels of TCA cycle intermediates and oxidative inhibition of TCA cycle enzymes, while in the case of aox1a, organic acids are unchanged or depleted during stress treatment, so again the impact of AOX1a loss does not appear to be replicated by an increase in mitochondrial ROS. As AOX has been proposed to allow more rapid TCA cycle function (Lambers, 1982) and our metabolite evidence can be interpreted as a restriction in metabolism from sugars to organic acids and amino acids (Table II), some similar changes may theoretically occur with knockdown of mitochondrial isocitrate dehydrogenase (idh) mutants (Lemaitre et al., 2007). That study observed decreases in a range of free amino acid levels, including Leu and Asp, and we also found Leu and the Asp-derived metabolite homo-Ser to be decreased during the stress response (Table II). Therefore, there may be some synteny in these results. But there are a large number of steady-state differences in idh plants that are not replicated on treatment of aox1a plants, and very few steady-state differences in metabolites are observed between aox1a and Col-0 under control conditions, suggesting that this comparison also has only limited power to uncover the role of AOX1a.
So why is AOX necessary for a normal response to combined light and drought stress? The simplest explanation for the requirement of AOX for normal redox signaling is the ability of an alternative respiratory chain [i.e. an alternative NAD(P)H dehydrogenase and a terminal alternative oxidase] to oxidize excess reducing equivalents from chloroplasts (Rasmusson et al., 2004; Yoshida et al., 2006, 2007). In the absence of AOX, these excess reducing equivalents build up, resulting in an alteration in redox equivalents in various subcellular compartments. Both the QRT-PCR assays (Fig. 6) and PCA analysis (Fig. 4) of transcript abundance indicate that there are differences compared with Col-0, indicating changes in the transcriptome in aox1a plants, even under normal growth conditions (Supplemental Table S3). These changes, and the signals that triggered them, may affect the ability to respond to stress conditions; for instance, we observed a reduction in ZAT10 mRNA in the aox1a lines under normal conditions (Fig. 6). Reducing ZAT10 mRNA has been shown to impair tolerance to ROS and high light and to decrease the expression of antioxidant genes whose products are targeted to different organelles (Rossel et al., 2007).
In conclusion, the absence of AOX1a leads to a change in the transcriptome in Arabidopsis even under normal conditions. The changes in transcript levels of several genes encoding components involved in ROS defense, signaling, transcription factors, and proteins located in mitochondria and chloroplasts indicate that signaling and communication are altered. Upon combined moderate light and drought stress treatments, these alterations result in a drastically different stress response. The absence of AOX1a results in a basal ROS defense and signaling network that appears to be overwhelmed upon stress treatment.
MATERIALS AND METHODS
Plant Material Growth and Treatments
Arabidopsis (Arabidopsis thaliana) plants, ecotype Col-0 and T-DNA insertional lines for AOX1a (SALK_084897 and SAIL_030_D08), were grown at 22°C for 16 h at 100 μE m−2 s−1 light conditions and 8 h of dark. Light and drought treatments were applied as follows: 4-week-old plants grown under normal conditions with watering every 2 d were transferred to either moderate light conditions (approximately 250 μE m−2 s−1) with continued watering, moderate light conditions with no water (starting 4 d prior to transfer to ensure that soil was dry), or moderate light with no water and 35°C. For root growth measurements, Col-0 and aox1a plants were germinated on vertical Murashige and Skoog agar plates (Gamborg, 1968). Approximately 10 seeds for each genotype were placed on a single plate, and the root length of 9-d-old seedlings was measured using the software ImageJ (http://rsb.info.nih.gov/ij/). The experiments were repeated three times.
Pigment Extraction and Quantification
To determine anthocyanin levels, all leaf tissues were excised from one plant per assay, weighed, and then extracted with 99:1 (v/v) methanol:HCl at 4°C with shaking for 48 h. The optical density at 530 nm (OD530) and OD657 for each sample were measured, and the relative anthocyanin levels were determined using the following equation: OD530 − (0.25 × OD657) × extraction volume (mL) × 1 ÷ weight of tissue sample (g) = relative units of anthocyanin per gram fresh weight of tissue. For each sample condition, extractions were repeated in triplicate. Results were scaled relative to the anthocyanin levels found in Col-0 4-week-old plants grown under normal conditions. Significant changes (P < 0.05) were determined using Student's t test comparing anthocyanin levels found in aox1a lines and Col-0 under the same conditions.
Relative Water Content Analysis
Col-0 and aox1a plants were treated as described previously for normal conditions or moderate light and drought treatment. After 4 d of treatment, any bolts were removed and whole rosettes were weighed (fresh weight [Fw]). Rosettes were then completely submerged in water for 4 h before being weighed again (turgid weight [Tw]). Rosettes were then dried overnight at 80°C and weighed to obtain dry weight measurements (Dw). Four plants from each genotype for each treatment condition were measured. Relative water content was calculated as (Fw − Dw) ÷ (Tw − Dw), and Student's t test was performed to determine significance.
Measurement of Photosynthetic Parameters
Fluorescence induction kinetics at room temperature (22°C) were measured using a pulse amplitude modulation fluorometer (PAM101; H. Walz) on 33-d-old seedlings as described (Rossel et al., 2006). After a 30-min dark acclimation, the Fv/Fm was measured as (Fm − F0)/Fm, where Fm is the maximum PSII fluorescence in the dark-adapted state and F0 is the initial (minimum) PSII fluorescence in the dark-adapted state (Butler, 1978). Subsequent to illumination, the utilization of absorbed photons by the PSII antennae in photosynthetic electron transport and thermal dissipation was assessed from the quantum efficiency (Φ) of photochemical energy dissipation (ΦPSII), light-dependent (ΦNPQ) and light-independent thermal dissipation, and fluorescence energy dissipation (Φf,D), with Φf,D + ΦNPQ + ΦPSII = 1 (Hendrickson et al., 2004). Fluorescence measurements were made on four plants for each of the Col-0 and aox1a lines per experiment. The data are averages and se of two independent experiments.
ROS Determination
To visualize O2− and H2O2, the method of Forster et al. (2005) was adapted for individual leaves. Leaves were isolated from plants exposed to normal growth conditions or moderate light and drought conditions for 3 d. The detached leaves were placed in 6 mm nitroblue tetrazolium for O2− or 5 mm 3,3′-diaminobenzidine for H2O2. After 3 h, the leaves were placed into 100% boiling ethanol and incubated overnight to remove the chlorophyll. The leaves were mounted on overhead transparencies in the presence of 40% (v/v) glycerol and scanned. Three leaves per plant, three plants per genotype per experiment, were assayed for ROS. Each experiment was repeated twice, and representative images are shown from both experiments.
Leaf Respiration Rates
All oxygen consumption measurements were performed using a Clarke-type electrode, and data were collected by Oxygraph Plus version 1.01 (Hansatech Instruments). The electrode was calibrated by the addition of excess sodium dithionite to deplete all oxygen in 1 mL of deioinized water in the electrode chamber. Equal-sized leaf discs were removed from plants, scored with a scalpel while immersed in incubation medium (10 mm HEPES, 10 mm MES, and 2 mm CaCl2, pH 7.2), and incubated in the dark for 30 min. Total respiration was measured at 22°C in the dark following the addition of five leaf discs to 2 mL of incubation medium. Respiration specifically via the cytochrome or alternative oxidase pathway was measured by the addition of 500 μm n-propyl gallate or 1 mm potassium cyanide, respectively.
Extraction and Analysis of Metabolites by GC-MS Analysis
Harvesting of leaf material, 72 h after commencement of treatment and 4 h into the light period, for metabolite analysis was carried out in an identical manner to harvesting for mRNA transcript profiling. Leaves were rapidly excised by cutting at the base of the petiole, snap frozen in liquid nitrogen, and stored at −80°C until analysis. Metabolites were extracted and derivatized using a method modified from that of Roessner-Tunali et al. (2003). Derivatized metabolite samples were analyzed on an Agilent GC/MSD system (Agilent Technologies). Raw GC-MS data files in the proprietary ChemStation (.D) format were exported to generic NetCDF/AIA (.CDF) format with ChemStation GC/MSD data analysis software. The NetCDF files produced were then processed using in-house MetaMiner software to carry out all peak detection, quantification, library matching, normalization, statistical analysis, and data visualization. A detailed methodology is presented in Supplemental Data Set S1. For PCA of metabolite profiles, analyte abundance matrices were exported from MetaMiner, and chemical artifacts, internal standards, and metabolite signals that were not detected in at least 80% of the replicates for at least one genotype were removed from the data set. PCA was carried out with AVADIS Prophetic software (version 4.3; Strand Life Sciences) with mean centering and scaling of all variables to unit variance. PC1 and PC2 were plotted and together account for 88% of the variance in the data set.
Microarray Analysis
Microarray files are available from ArrayExpress, a MIAME-compliant data warehouse (Mukherjee et al., 2005), under the accession E-ATMX-32.
Analysis of the changes in transcript abundance between Col-0 and aox1a lines in 4-week-old Arabidopsis plants was performed using Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays. Col-0 tissue samples were collected in biological triplicate for each treatment and in biological duplicate for each of the aox1a lines for each treatment. For each replicate, total RNA was isolated from the leaves using the RNeasy Plant Mini Protocol (Qiagen) and quality verified using a Bioanalyzer (Agilent Technologies), and spectrophotometric analysis was carried out to determine the A260 to A280 and A260 to A230 ratios. Preparation of labeled complementary RNA from 5 μg of total RNA, target hybridization, as well as washing, staining, and scanning of the arrays were carried out exactly as described in the Affymetrix GeneChip Expression Analysis Technical Manual, using an Affymetrix GeneChip Hybridization Oven 640, an Affymetrix Fluidics Station 450, and a GeneChip Scanner 3000 7G at the appropriate steps. Data quality was assessed using GCOS 1.4 before CEL files were exported into AVADIS Prophetic (version 4.3) for further analysis.
CEL files were subjected to GC-RMA normalization, a variation of the robust multiarray average normalization algorithm. MAS5 normalization algorithms were also carried out to generate present/absent calls across the arrays. Correlation plots were examined between all chips using the scatterplot function, in all cases r ≥ 0.97 (data not shown). Probe sets identified as absent in more than half the arrays were removed. For each treatment, arrays for each of the aox1a lines were analyzed individually at first to ensure similar global fluorescence intensity values before being combined as one genotype (n = 4) for further comparative analysis with Col-0 (n = 3). Fold changes for a number of different comparisons were calculated using the Differential Expression Analysis function, and P values were calculated using an unpaired t test; P values were given as uncorrected and after correction for estimation of FDR in multiple comparisons. A method of FDR correction that is based on the Benjamini and Hochberg method (Benjamini and Hochberg, 1995; Nettleton, 2006) was used. This method utilizes an add-on in the software package R to calculate q values based on P value distributions (http://www.r-project.org; Storey, 2002). These comparisons were as follows: Col-0 untreated versus Col-0 stress treated, Col-0 untreated versus aox1a untreated, Col-0 untreated versus aox1a stress treated, aox1a untreated versus aox1a stress treated, and Col-0 stress treated versus aox1a stress treated. PCA plots were generated using the PCA tool in AVADIS Prophetic (version 4.3), reducing the 22,746 gene expression values to two dimensions with mean centering and scaling of all variables to unit variance. Principal eigenvalues associated with the principal axes were also calculated and plotted against their respective percentile contribution to determine the percentage of variance captured. Principal axes 1 and 2 were plotted as those capturing over 94% of the variance in the data set. Cluster images were generated using the hierarchical cluster function in the AVADIS software package, clustering on both rows and columns using complete linkage and Euclidean distance measures. To gain a qualitative overview of changes in transcript abundance for aox1a plants compared with Col-0 under normal conditions or moderate light and drought treatment, MapMan software was used (http://gabi.rzpd.de/projects/MapMan/; Thimm et al., 2004). Only transcripts with significant changes were displayed.
QRT-PCR Analysis of Gene Expression
QRT-PCR was performed on Arabidopsis leaf tissue. Leaf tissue was excised from 4-week-old mutant and Col-0 plants. All samples were taken in biological triplicate and snap frozen under liquid nitrogen. Total RNA isolation and cDNA synthesis were carried out as described previously (Lister et al., 2004). Transcript levels were assayed using the LightCycler 480 and the LightCycler 480 SYBR Green I Master (Roche). From each of the independent cDNA preparations, each transcript was analyzed twice. The se was calculated for every data point. Transcript abundance for Col-0 untreated samples was normalized to 1 for each gene, with all other values presented as relative transcript abundance. Significant changes were determined using Student's t test with P ≤ 0.05. QRT-PCR primers used for the genes AOX1a (At3g22370), NDB2 (At4g05020), and UCP (At3g54110) have been described previously (Clifton et al., 2005). QRT-PCR was also carried out for the following genes: UPOX (At2g21640), PR1 (At2g14610), UBC (At5g25760), At2g43510, At1g57630, At1g27730, At5g51190, At5g47230, At1g19020, At1g05340, At4g27657, At5g59820, At4g32320, At3g15210, and At4g27410. Primer sequences and annotations are listed in Supplemental Table S2.
Functional Categorization
Functional categorization using gene ontology annotations was performed on the Arabidopsis whole genome set along with gene sets defined as differentially expressed (either positively or negatively) during microarray experiments. Functional categorizations for each gene list were obtained from The Arabidopsis Information Resource using the gene ontology annotations, functional categorization function (http://www.arabidopsis.org/tools/bulk/go/index.jsp) for cellular component, and molecular function. The percentage distribution of each category for the various gene lists was compared with that of the whole genome. A χ2 test was performed for each comparison, and percentile distributions were considered to be significantly different at the 98% confidence interval.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of the insertional lines inactivating AOX1a.
Supplemental Figure S2. Overview of changes in transcript abundance associated with primary metabolism.
Supplemental Figure S3. Analysis of functional categorization of aox1a responsive transcripts.
Supplemental Figure S4. Relative transcript abundance for genes encoding antioxidant defense components located in Arabidopsis.
Supplemental Figure S5. Changes in transcript abundance for genes that encode proteins involved in anthocyanin production.
Supplemental Table S1. Photosynthetic efficiency.
Supplemental Table S2. Characteristic metabolite response profile of aox1a T-DNA insertion lines to moderate light and drought compared with Col-0 response.
Supplemental Table S3. Changes in transcript abundance between aox1a and Col-0.
Supplemental Table S4. List of 17 genes and ubiquitin, and primers used for QRT-PCR.
Supplemental Data Set S1. A detailed methodology of metabolite analysis using MetaMiner software.
Supplementary Material
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: James Whelan (seamus@cyllene.uwa.edu.au).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Amirsadeghi S, Robson CA, McDonald AE, Vanlerberghe GC (2006) Changes in plant mitochondrial electron transport alter cellular levels of reactive oxygen species and susceptibility to cell death signaling molecules. Plant Cell Physiol 47 1509–1519 [DOI] [PubMed] [Google Scholar]
- Amirsadeghi S, Robson CA, Vanlerberghe GC (2007) The role of the mitochondrion in plant responses to biotic stress. Physiol Plant 129 253–266 [Google Scholar]
- Arnholdt-Schmitt B, Costa JH, de Melo DF (2006) AOX: a functional marker for efficient cell reprogramming under stress? Trends Plant Sci 11 281–287 [DOI] [PubMed] [Google Scholar]
- Baxter CJ, Redestig H, Schauer N, Repsilber D, Patil KR, Nielsen J, Selbig J, Liu J, Fernie AR, Sweetlove LJ (2007) The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol 143 312–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Karpinski S, Mullineaux PM (2005) The influence of light environment and photosynthesis on oxidative signalling responses in plant-biotrophic pathogen interactions. Plant Cell Environ 28 1046–1055 [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodological 57 289–300 [Google Scholar]
- Butler WL (1978) Energy distribution in the photochemical apparatus of photosynthesis. Annu Rev Plant Physiol 29 345–378 [Google Scholar]
- Clifton R (2006) The alternative oxidase gene family in Arabidopsis: insights from a transcriptomic study. PhD thesis. University of Western Australia, Perth, Australia
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58 193–212 [DOI] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757 730–741 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57 711–738 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chetrit P, Foyer CH, de Paepe R (2003) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Lelarge C, Prioul JL, De Paepe R, Foyer CH, Noctor G (2005) Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol 139 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar MA, Franklin KA, Svensson AS, Salter MG, Whitelam GC, Rasmusson AG (2004) Light regulation of the Arabidopsis respiratory chain: Multiple discrete photoreceptor responses contribute to induction of type II NAD(P)H dehydrogenase genes. Plant Physiol 136 2710–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar MA, Geisler DA, Rasmusson AG (2006) Reorganization of the alternative pathways of the Arabidopsis respiratory chain by nitrogen supply: opposing effects of ammonium and nitrate. Plant J 45 775–788 [DOI] [PubMed] [Google Scholar]
- Finnegan PF, Soole KL, Umbach AL (2004) Alternative mitochondrial electron transport proteins in higher plants. In DA Day, H Millar, J Whelan, eds, Plant Mitochondria: From Genome to Function, Vol 17. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 163–230
- Fiorani F, Umbach AL, Siedow JN (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature: a study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster B, Osmond CB, Pogson BJ (2005) Improved survival of very high light and oxidative stress is conferred by spontaneous gain-of-function mutations in Chlamydomonas. Biochim Biophys Acta 1709 45–57 [DOI] [PubMed] [Google Scholar]
- Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR (2003) Control of ascorbate peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J 33 691–705 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inze D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg O (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50 151–158 [DOI] [PubMed] [Google Scholar]
- Green R, Fluhr R (1995) UV-B-induced PR-1 accumulation is mediated by active oxygen species. Plant Cell 7 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson L, Furbank RT, Chow WS (2004) A simple alternative approach to assessing the fate of absorbed light energy using chlorophyll fluorescence. Photosynth Res 82 73–81 [DOI] [PubMed] [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM (1997) Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P (1999) Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284 654–657 [DOI] [PubMed] [Google Scholar]
- Karpova OV, Kuzmin EV, Elthon TE, Newton KJ (2002) Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 347–363 [DOI] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47 509–540 [DOI] [PubMed] [Google Scholar]
- Lambers H (1982) Cyanide-resistant respiration: a non-phosphorylating electron transport pathway acting as an energy overflow. Plant Physiol 55 478–485 [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre T, Urbanczyk-Wochniak E, Flesch V, Bismuth E, Fernie AR, Hodges M (2007) NAD-dependent isocitrate dehydrogenase mutants of Arabidopsis suggest the enzyme is not limiting for nitrogen assimilation. Plant Physiol 144 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J (2004) A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe TC, Finnegan PM, Millar AH, Day DA, Whelan J (1998) Differential expression of alternative oxidase genes in soybean cotyledons during postgerminative development. Plant Physiol 118 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R (2007) Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol 144 1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11 15–19 [DOI] [PubMed] [Google Scholar]
- Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Moore AL, Albury MS, Crichton PG, Affourtit C (2002) Function of the alternative oxidase: is it still a scavenger? Trends Plant Sci 7 478–481 [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Lehmann M, Schwarzländer M, Baxter CJ, Sienkiewicz-Porzucek A, Williams TCR, Schauer N, Fernie AR, Fricker MD, Ratcliffe RG, et al (2008) Decrease in manganese superoxide dismutase leads to reduced root growth and affects TCA cycle flux and mitochondrial redox homeostasis. Plant Physiol 147 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee G, Abeygunawardena N, Parkinson H, Contrino S, Durinck S, Farne A, Holloway E, Lilja P, Moreau Y, Oezcimen A, et al (2005) Plant-based microarray data at the European Bioinformatics Institute: Introducing AtMIAMExpress, a submission tool for Arabidopsis gene expression data to ArrayExpress. Plant Physiol 139 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton D (2006) A discussion of statistical methods for design and analysis of microarray experiments for plant scientists. Plant Cell 18 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, De Paepe R, Foyer CH (2007) Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci 12 125–134 [DOI] [PubMed] [Google Scholar]
- Panchuk II, Zentgraf U, Volkov RA (2005) Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 222 926–932 [DOI] [PubMed] [Google Scholar]
- Parsons HL, Yip JY, Vanlerberghe GC (1999) Increased respiratory restriction during phosphate-limited growth in transgenic tobacco cells lacking alternative oxidase. Plant Physiol 121 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini S, Paolocci F, Borgogni A, Morettini R, Ederli L (2007) The overexpression of an alternative oxidase gene triggers ozone sensitivity in tobacco plants. Plant Cell Environ 30 1545–1556 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34 187–203 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE (2004) Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol 55 23–39 [DOI] [PubMed] [Google Scholar]
- Rhoads DM, McIntosh L (1992) Salicylic acid regulation of respiration in higher plants: alternative oxidase expression. Plant Cell 4 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Subbaiah CC, Siedow JN (2006) Mitochondrial reactive oxygen species: contribution to oxidative stress and interorganellar signaling. Plant Physiol 141 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279 11736–11743 [DOI] [PubMed] [Google Scholar]
- Robson CA, Vanlerberghe GC (2002) Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and -independent pathways of programmed cell death. Plant Physiol 129 1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel JB, Walter PB, Hendrickson L, Chow WS, Poole A, Mullineaux PM, Pogson BJ (2006) A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ 29 269–281 [DOI] [PubMed] [Google Scholar]
- Rossel JB, Wilson IW, Pogson BJ (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol 130 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel JB, Wilson PB, Hussain D, Woo NS, Gordon MJ, Mewett OP, Howell KA, Whelan J, Kazan K, Pogson BJ (2007) Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpeci TE, Zanor MI, Carrillo N, Mueller-Roeber B, Valle EM (2008) Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol Biol 66 361–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoelkopf B, Smola A, Mueller KR (1998) Non-linear component analysis as a kernel eigenvalue problem. J Neural Comp 10 1299–1319 [Google Scholar]
- Seki M, Umezawa T, Urano K, Shinozaki K (2007) Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10 296–302 [DOI] [PubMed] [Google Scholar]
- Sieger SM, Kristensen BK, Robson CA, Amirsadeghi S, Eng EW, Abdel-Mesih A, Moller IM, Vanlerberghe GC (2005) The role of alternative oxidase in modulating carbon use efficiency and growth during macronutrient stress in tobacco cells. J Exp Bot 56 1499–1515 [DOI] [PubMed] [Google Scholar]
- Storey JD (2002) A direct approach to false discovery rates. J Roy Statist Soc Ser B Methodological 64 479–498 [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32 891–904 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller L, Rhee SYMS (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Thirkettle-Watts D, McCabe TC, Clifton R, Moore C, Finnegan PM, Day DA, Whelan J (2003) Analysis of the alternative oxidase promoters from soybean. Plant Physiol 133 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Jaquinod M, Mangavel C, Passirani C, Saulnier P, Manon S, Teyssier E, Payet N, Avelange-Macherel MH, Macherel D (2007) Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell 19 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) ALTERNATIVE OXIDASE: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48 703–734 [DOI] [PubMed] [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC (2000) Photorespiration: metabolic pathways and their role in stress protection. Philos Trans R Soc Lond B Biol Sci 355 1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58 585–596 [DOI] [PubMed] [Google Scholar]
- Yip JY, Vanlerberghe GC (2001) Mitochondrial alternative oxidase acts to dampen the generation of active oxygen species during a period of rapid respiration induced to support a high rate of nutrient uptake. Physiol Plant 112 327–333 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Terashima I, Noguchi K (2006) Distinct roles of the cytochrome pathway and alternative oxidase in leaf photosynthesis. Plant Cell Physiol 47 22–31 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Terashima I, Noguchi K (2007) Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol 48 606–614 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.