Abstract
Plant cells contain different O-acetylserine(thiol)lyase (OASTL) enzymes involved in cysteine (Cys) biosynthesis and located in different subcellular compartments. These enzymes are made up of a complex variety of isoforms resulting in different subcellular Cys pools. To unravel the contribution of cytosolic Cys to plant metabolism, we characterized the knockout oas-a1.1 and osa-a1.2 mutants, deficient in the most abundant cytosolic OASTL isoform in Arabidopsis (Arabidopsis thaliana). Total intracellular Cys and glutathione concentrations were reduced, and the glutathione redox state was shifted in favor of its oxidized form. Interestingly, the capability of the mutants to chelate heavy metals did not differ from that of the wild type, but the mutants have an enhanced sensitivity to cadmium. With the aim of establishing the metabolic network most influenced by the cytosolic Cys pool, we used the ATH1 GeneChip for evaluation of differentially expressed genes in the oas-a1.1 mutant grown under nonstress conditions. The transcriptomic footprints of mutant plants had predicted functions associated with various physiological responses that are dependent on reactive oxygen species and suggested that the mutant was oxidatively stressed. Evidences that the mutation caused a perturbation in H2O2 homeostasis are that, in the knockout, H2O2 production was localized in shoots and roots; spontaneous cell death lesions occurred in the leaves; and lignification and guaiacol peroxidase activity were significantly increased. All these findings indicate that a deficiency of OAS-A1 in the cytosol promotes a perturbation in H2O2 homeostasis and that Cys is an important determinant of the antioxidative capacity of the cytosol in Arabidopsis.
Sulfur is an essential macronutrient for plant growth and development. It is found in the amino acids, Cys and Met, and in many other cellular constituents, such as reduced glutathione (GSH). In recent years, the functions of GSH have attracted considerable attention because it is the predominant nonprotein thiol and one of the major determinants of cellular redox homeostasis in plant tissues. Its roles include acting as a mobile pool of reduced sulfur, involvement in the detoxification of xenobiotics, protection against heavy metal toxicity, acting as a source of reductant in enzymatic reactions, effects on growth and development, regulation of gene expression, resistance to pathogen infection, and tolerance to environmental perturbations that promote oxidative stress (Meyer and Hell, 2005; Mullineaux and Rausch, 2005). The sulfur moiety of the majority of sulfur-containing compounds in plants, including GSH, is derived from Cys, which is the final product of the primary sulfate assimilation pathway. Therefore, the biosynthetic pathways of most sulfur-containing compounds in plants are intimately linked to Cys biosynthesis.
The Cys biosynthetic pathway involves two sequential reactions catalyzed by Ser acetyltransferase (SAT), which synthesizes the intermediary product, O-acetyl-Ser (OAS), from acetyl-CoA and Ser, and O-acetyl-Ser(thiol)lyase (OASTL), which incorporates the sulfide to OAS producing Cys. Together, both enzymes form the hetero-oligomeric Cys synthase complex that was described for the first time in bacteria and has subsequently been extensively studied in plants (Wirtz and Hell, 2006). Plant cells contain different SAT and OASTL enzymes that are localized in the cytosol, plastids, and mitochondria. This results in a complex variety of isoforms and in different subcellular Cys pools. In Arabidopsis (Arabidopsis thaliana) five SAT (Howarth et al., 2003) and nine OASTL genes (Wirtz et al., 2004) have been identified. The existence of multiple SAT and OASTL complementary DNAs (cDNAs) in plant genome databases suggests that this pattern of gene organization is similar in other plant species. Because the assimilatory reduction of sulfate takes place in the chloroplasts, it has been suggested that the chloroplastic SAT and OASTL isoforms probably participate in the primary sulfate assimilation pathway. The precise metabolic and signaling functions of the cytosolic and mitochondrial SAT and OASTL isoforms remain elusive. Therefore, it would be of particular interest to know whether each subcellular Cys pool is linked to a specific plant cell response or a particular metabolic network.
The most abundant cytosolic OASTL isoform, OAS-A1, is known to be involved in the defense responses of Arabidopsis against abiotic stresses, such as salinity and the presence of heavy metals (Barroso et al., 1999; Dominguez-Solis et al., 2001). Overexpression of the OAS-A1 gene in Arabidopsis increases the plant's tolerance to severe heavy metal stress. This increased tolerance suggests that the availability of cytosolic Cys is a limiting step for the synthesis of GSH, phytochelatins (PCs), or both (Dominguez-Solis et al., 2004). PCs are Cys-rich polypeptides that are rapidly synthesized from GSH in response to the presence of heavy metals. These peptides bind heavy metals with high affinity throughout the thiol moiety of Cys and the PC-metal complexes are then transported into the vacuoles (Clemens, 2006).
To further unravel the function of the major cytosolic OASTL OAS-A1, we report our findings on the characterization of the T-DNA tagged oas-a1.1 and oas-a1.2 mutants of Arabidopsis. Although the knockout plants have reduced Cys and GSH concentrations, they display cadmium (Cd) sensitivity but the biosynthetic pathway of PCs remained unaffected when compared to wild-type plants. To identify which metabolic network was most likely to have been influenced by the cytosolic Cys pool, we conducted a comprehensive evaluation of which genes are differentially expressed in oas-a1.1 mutant plants under nonstressed conditions using the ATH1 GeneChip. Our results indicate that the mutant has a constitutively reduced capacity to eliminate reactive oxygen species (ROS) under nonstressed conditions and this has been confirmed by a range of approaches.
RESULTS
Identification and Characterization of Arabidopsis oas-a1 Mutants
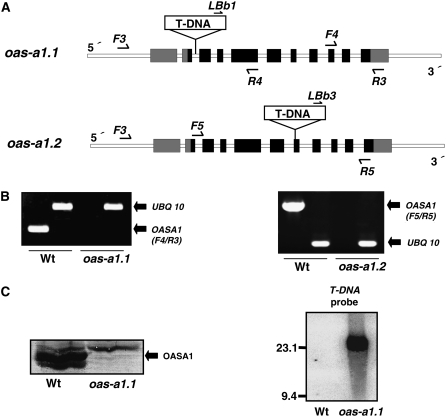
Our previous studies had showed that the overexpression of the OAS-A1 gene in Arabidopsis resulted in increased Cd accumulation, suggesting that the cytosolic OASTL isoform plays a role in the heavy metal tolerance of plants (Dominguez-Solis et al., 2001, 2004). To further elucidate the specific function(s) of the OAS-A1 isoform in the adaptation response to abiotic stress, different T-DNA insertion mutants from the SALK and SAIL collections were screened (http://signal.salk.edu/cgi-bin/tdnaexpress). Based on this screening, the SALK_072213 and the SAIL_94_E12 mutants, designated, respectively, as oas-a1.1 and oas-a1.2, were selected for further analysis. The T-DNAs are located in the second intron, downstream of the ATG initiation codon, and in the seventh exon of the OAS-A1 gene, in the oas-a1.1 and the oas-a1.2 mutants, respectively (Fig. 1A). Reverse transcription (RT)-PCR analysis of the homozygous mutant plants using specific primers revealed no detectable OAS-A1 transcript. This suggested that both oas-a1.1 and oas-a1.2 are knockout plants (Fig. 1B). The complete loss of expression in the oas-a1.1 mutant plant was further confirmed by western-blot analysis of leaf proteins using antibodies against recombinant OAS-A1 (Fig. 1C). Southern-blot analysis was also performed to determine the number of T-DNA insertions. A unique insertion site of the T-DNA in the oas-a1.1 genome was confirmed by the appearance of a single hybridization band of about 24 kb (Fig. 1C).
Figure 1.
Molecular characterization of the Arabidopsis oas-a1.1 and oas-a1.2 mutants. A, Intron-exon organization of the OAS-A1 gene (At4g14880) in the oas-a1.1 and oas-a1.2 T-DNA tagged mutants. LBb1, LBb3, F3, F4, F5, R3, R4, and R5 show the locations and directions of primers used in the screening for mutant plants. The OAS-A1 gene contains 11 exons (black squares with the untranslated regions shown in gray) and 10 introns (white squares). The gene structure and T-DNA region are not drawn to scale. B, RT-PCR analysis of the mutant plants. For RT-PCR, mRNA was prepared from the leaves of 2-week-old plants, and primers specific for OAS-A1 (F4/R3 in oas-a1.1 and F5/R5 in oas-a1.2) and UBQ10 transcripts were used. C, Western-blot (left) and Southern-blot (right) analyses of the oas-a1.1 mutant plant. For western blots, 15 μg of total protein from the leaves was electrophoresed and the OASTL isoforms were detected using antibodies raised against OAS-A1. For Southern blots, genomic DNA was digested with SpeI and hybridized with a T-DNA probe.
We also screened the oas-a1.1 and oas-a1.2 mutant plants for potential growth defects. Compared to the wild-type plants, no significant phenotypic differences were observed under nonstress growth conditions, namely, when grown either on solid medium or in hydroponic culture (data not shown). A slight increase in the size of the rosette of mutant plants, compared with wild-type plants, was found at the seedling stage but this difference was no longer evident at maturity.
Biochemical characterization of both mutant plants, oas-a1.1 and oas-a1.2, revealed that the OASTL activity was markedly reduced in leaves and that the reduction was even more marked in root tissues (Table I). This finding suggests that the cytosolic OAS-A1 isoform is the major contributor to the total OASTL activity of the plant. The SAT activity of the oas-a1.1 mutant was also measured and found to be increased relative to the wild type (Table I). Comparison of the total concentration of Cys and glutathione, as determined by HPLC in the oas-a1.1, oas-a1.2, and wild-type plants, revealed that the two thiols in the mutant were significantly decreased (Table II). In general, the amounts of Cys in plants grown in soil were slightly higher than those found in hydroponically grown plants, probably reflecting the younger developmental stage of the soil-grown plants. In addition, quantification of the glutathione also revealed an important feature of the mutants. Not only were total glutathione concentrations about 20% to 30% lower, but also the degree of oxidation of this thiol in the mutant plants was greater than in the wild-type plants (Table II). The redox potential of glutathione, which is an important determinant of cellular redox status, is a function of both the total glutathione concentration and the glutathione degree of oxidation (Mullineaux and Rausch, 2005). Thus, our results suggest a change in the cellular redox status in the oas-a1.1 and oas-a1.2 mutants.
Table I.
OASTL and SAT enzyme activities
Enzyme activities were determined in soluble extracts prepared from leaves or roots of wild-type and oas-a1.1 and oas-a1.2 mutant plants grown on soil or in hydroponic culture. Values are means ± sd of five independent experiments. nd, Not determined. **, P < 0.01.
| Plant Line | OASTL Activity
|
SAT Activity
|
||
|---|---|---|---|---|
| Soil-Grown | Hydroponics | Soil-Grown | Hydroponics | |
| U mg−1 | mU mg−1 | |||
| Leaves | ||||
| Wild type | 1.73 ± 0.01 | 2.04 ± 0.02 | 3.1 ± 0.7 | 6.2 ± 0.9 |
| oas-a1.1 | 0.48 ± 0.07 (28%)** | 0.57 ± 0.04 (28%)** | 4.1 ± 0.7 (132%)** | 7.8 ± 0.1 (126%)** |
| oas-a1.2 | 0.35 ± 0.05 (20%)** | nd | nd | nd |
| Roots | ||||
| Wild type | 3.54 ± 0.02 | 3.30 ± 0.2 | nd | nd |
| oas-a1.1 | 0.60 ± 0.03 (17%)** | 0.46 ± 0.02 (14%)** | nd | nd |
| oas-a1.2 | 0.43 ± 0.03 (12%)** | nd | nd | nd |
Table II.
Cys and glutathione contents in leaves
Thiols were determined by HPLC in leaves of wild-type and oas-a1.1 and oas-a1.2 mutant plants grown on soil or in hydroponic culture. Values are means ± sd of four independent experiments. The glutathione oxidation state was calculated according to Meyer and Hell (2005): degree of oxidation (%) = (2[GSSG]/([GSH] + 2[GSSG])) × 100. **, P < 0.01; *, P < 0.05.
| Plant Line | Total Cys
|
Total Glutathione
|
|||
|---|---|---|---|---|---|
| Soil-Grown | Hydroponics | Soil-Grown | Hydroponics | Degree of Oxidation (%) | |
| nmol g−1 fresh wt | nmol g−1 fresh wt | ||||
| Wild type | 21.7 ± 2.1 | 20.1 ± 2.0 | 333.4 ± 6 | 362.3 ± 15 | 15 |
| oas-a1.1 | 14.4 ± 1.5 * (66%) | 15.5 ± 1.2 * (77%) | 240.4 ± 10 ** (72%) | 282.7 ± 11 * (78%) | 20* |
| oas-a1.2 | 13.9 ± 1.4 * (64%) | 14.7 ± 1.3 * (73%) | 230.2 ± 8 * (69%) | 271.9 ± 14 ** (75%) | 22* |
The Mutant Plants Deficient in OAS-A1 Retain Cd Sensitivity But PC Synthesis Is Not Affected
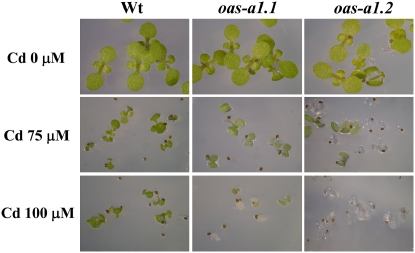
Although the oas-a1.1 and oas-a1.2 mutants did not exhibit any apparent phenotypic differences when grown under nonstress growth conditions, they showed marked sensitivity to the presence of Cd. We performed germination tests in the presence of increasing concentrations of Cd. When these were as low as 75 to 100 μm, the mutant seeds germinated but were unable to produce true leaves and their cotyledons became pale. This was in sharp contrast with the development of healthy green leaves following germination of the wild-type seeds (Fig. 2).
Figure 2.
Sensitivity of the oas-a1.1 and oas-a1.2 mutant plants to Cd stress. Wild-type and mutant seeds were germinated on solid MS containing varying concentrations of CdCl2 as stated and photographs were taken after 15 d of growth.
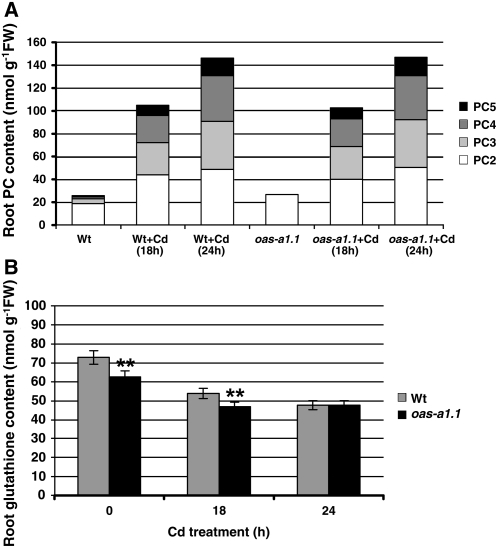
To determine whether the Cd hypersensitivity of the mutants is a consequence of a reduced rate of PC synthesis, we determined the PC contents of roots and shoots of oas-a1.1 plants grown in hydroponics. In the absence of Cd, some PC2 and PC3 were detected in the roots of both the wild-type and oas-1.1 plants, probably as a result of the copper, zinc (Zn), and other metals present in the hydroponic media. There were no detectable PCs in the shoots (data not shown). Plants were treated with a Cd concentration that did not cause visible toxicity symptoms up to 24 h, the longest period of treatment. Following exposure of plants to Cd, the PC polypeptides were detected in root extracts, with a net 6-fold accumulation of total PCs after 24 h of treatment relative to root extracts from plants grown without Cd (Fig. 3A). In shoots, only small amounts of PC2 were detected in the wild-type and mutant plants after 24 h of Cd stress. This observation indicates that the roots are mainly sensing and responding to heavy metal stress when grown under these experimental conditions. Interestingly, the responses of the wild-type and oas-a1.1 mutant plants to Cd stress were virtually identical because no differences were found in the amounts of each PC type at the two time points of analysis (Fig. 3A).
Figure 3.
A and B, Synthesis of PCs (A) and glutathione (B) in Arabidopsis roots in response to Cd stress. Wild-type and oas-a1.1 mutant plants were grown hydroponically in the absence or presence of 50 μm CdCl2 for 18 or 24 h. Values are means ± sd from at least four independent experiments. In A, sd < 10% are omitted for clarity. **, P < 0.01.
We also assessed glutathione levels in the roots of Cd-stressed and hydroponically grown wild-type and oas-a1.1 mutant plants. Similarly to the results that we obtained in the leaves, in the absence of Cd, the amount of glutathione in the roots of mutant plants was lower than that found in the wild-type plants. Cd stress caused a decrease in glutathione levels and this Cd-induced depletion was more conspicuous in the wild-type plant than in the oas-a1.1 mutant. After 24 h of Cd treatment, the wild-type and mutant plants showed the same glutathione levels (Fig. 3B). Furthermore, we found no differences in Cd concentration in the leaves of the oas-a1.1 mutant plants compared to the wild-type plants (data not shown). Therefore, the responses of the oas-a1.1 mutant plant to Cd stress in terms of Cd uptake and chelation do not appear to differ substantially from those of the wild type. In light of these findings, we suggest that the Cd hypersensitivity of the mutants deficient in OAS-A1 may be related to an alteration of a more generalized plant response to stress.
Transcriptomic Analyses
To gain further insights into the altered responses in the oas-a1.1 mutant plant, ATH1 microarrays were used. For these experiments, the biological and experimental conditions were designed according to the results of the previous experiments. Wild-type and oas-a1.1 mutant plants were grown hydroponically and, after a 2-week acclimation period, the roots and shoots were harvested separately. Total RNA was then prepared and analyzed using the Affymetrix-Arabidopsis ATH1GeneChip array. Three biological replicates were performed for each sample. We made two different comparisons to classify the differently expressed genes in the mutant plant: oas-a1.1 roots versus wild-type roots (Supplemental Table S1) and oas-a1.1 shoots versus wild-type shoots (Supplemental Table S2). Restricting the analysis to the genes whose expression was changed at least 2-fold, we identified 205 genes that exhibited alterations in their transcription level in the roots and 286 genes in the shoots. Using the 2-fold increase in expression as a threshold and a significance level of P < 0.05, we found that 44 and 110 genes were up-regulated in the roots and shoots, respectively, in the oas-a1.1 mutant plant compared to those of the wild-type plant. Similarly, using the threshold criterion of a 0.5-fold decrease in expression and P <0.05, we found that 87 and 102 genes were down-regulated in roots and shoots, respectively, of the oas-a1.1 mutant plant when compared to the wild-type plant.
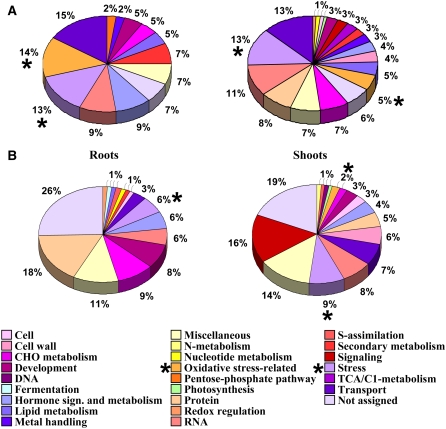
To detect physiologically relevant patterns, the genes with altered expression were assigned to functional categories based on the Arabidopsis Information Resource (TAIR), the Map-Man program, and the GENEVESTIGATOR Arabidopsis microarray database. The resulting group lists revealed that those genes with the highest expression levels in the oas-a1.1 mutant were associated with the plant's responses to stress, and, in particular, to oxidative stress. In roots, the three genes with the highest expression were At2g36690, which encodes an oxidoreductase; At3g24500, which is a putative ethylene-responsive transcriptional coactivator MBF1; and At3g30720, which encodes a highly expressed protein in senescent leaves and mature pollen (Supplemental Table S3). In shoots, the three genes with the highest expression were At5g24770 and At5g24780, which encode the vegetative storage protein 2 and 1 precursors, respectively, and At5g37260, which is probably a member of the Myb family of transcription factors (Supplemental Table S4). The results presented in Figure 4A show that a high proportion of the most up-regulated transcripts belong to functional categories related to stress adaptation.
Figure 4.
A and B, Functional distribution of induced (A) and repressed (B) genes in roots and shoots of the oas-a1.1 mutant compared with wild-type plants. An asterisk highlights the functional categories related to stress and, in particular, to oxidative stress.
The comparison of transcriptomic profiles also showed that the most down-regulated genes in the oas-a1.1 mutant belong to functional groups that were unrelated to general stress responses. In roots, the two genes with lowest expression were identified as belonging to the development process group: At4g35770 encodes a SEN1 (senescence-associated protein) and At1g15040 encodes a Gln amidotransferase-related protein. The third gene with the lowest expression, At4g33070, was grouped in the fermentation functional category and encodes a putative pyruvate decarboxylase (Supplemental Table S5). In shoots, the majority of the repressed genes were not assigned to the functional category of stress. The genes with the lowest expression were two transport-related genes, At4g36670 and At3g23550, encoding a putative mannitol transporter and a MATE efflux family protein, respectively. In addition, a third gene, At5g45890, which is described as encoding the senescence-specific SAG12 protein, was also identified (Supplemental Table S6). A summary of the distribution of the down-regulated genes in the various functional categories is presented in Figure 4B.
We performed real-time quantitative reverse transcription PCR (qRT-PCR) on a subset of genes to validate the expression data of microarray analysis. Different genes with altered expression, including up- and down-regulated in roots and in shoots, were selected. The qRT-PCR analysis was performed in both mutant plants, oas-a1.1 and oas-a1.2. Overall, there was good qualitative agreement between the results of qRT-PCR and microarray analysis, and both mutants showed the same correlation in gene expression compared to the wild type (Fig. 5).
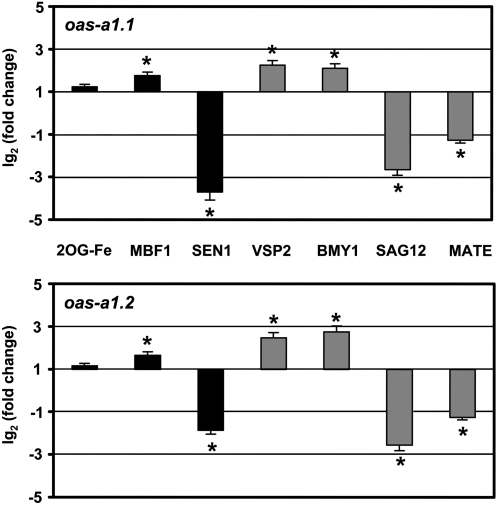
Figure 5.
Relative expression levels of selected genes in the oas-a1.1 and oas-a1.2 mutant plants. Real-time RT-PCR analysis of expression of 2OG-Fe(II) (AT2G36690), MBF1 (AT3G24500), and SEN1 (AT4G35770) genes in roots (black bars); and VSP2 (AT5G24770), BMY1 (AT4G15210), SAG12 (AT5G45890), and MATE (AT3G23550) genes in shoots (gray bars) of hydroponically grown oas-a1.1 and oas-a1.2 mutant plants. The transcript levels were normalized using the constitutive UBQ10 gene as an internal control. Data shown are mean values ± sd of three independent analyses. *, P < 0.05.
oas-a1.1 Mutant Plants Show Increased Production of ROS
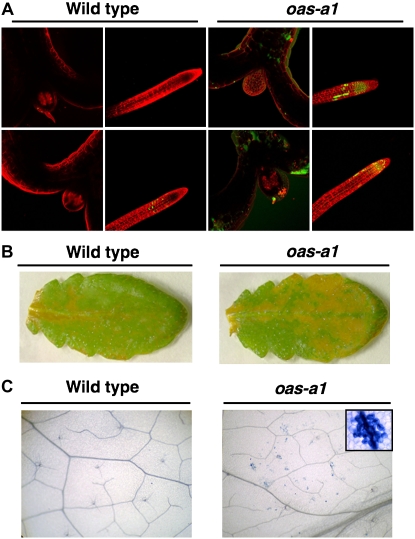
The results of our transcriptomic analyses show that under nonstress growth conditions, the oas-a1.1 mutant plant has genes that are induced as part of the plant's responses to stress, and, in particular, to oxidative stress. These results suggest that the ability for scavenging ROS in the mutant plant is modified and this affects its ROS homeostasis. To further support the transcriptomic data, we determined the redox status of the oas-a1.1 mutant plants grown under nonstress conditions using different approaches. The production of H2O2 in both roots and shoots of the mutant plants was monitored using nonfluorescent dichlorofluorescein (H2DCFDA), which yields a highly fluorescent product upon oxidation by H2O2. The oas-a1.1 mutant plants appeared to be oxidatively stressed under control growth conditions because increased fluorescence emission was observed in shoot and root tissues. This probably reflects an increased localized H2O2 production (Fig. 6A). The histochemical diaminobenizidine (DAB) method was used to examine the production of ROS in mature leaves of wild-type and osa-a1.1 mutant plants. The accumulation of H2O2 was clearly distinguishable in the mutant plant as a brown staining, which was homogeneously distributed, and more abundantly, in the mutant leaf cells compared to the wild type, but the staining was not confined to a particular tissue (Fig. 6B). H2O2 is a signaling intermediate molecule in programmed cell death (Lamb and Dixon, 1997). We consistently observed lesions characteristic of spontaneous cell death in the leaves of the oas-a1.1 mutant after staining them with trypan blue, which were not observed in the wild-type leaves (Fig. 6C). Patches of dead cells, which were visible in the leaves of the mutant plant, resembled those seen in a lesion-mimic phenotype.
Figure 6.
Accumulation of H2O2 and leaf lesion formation in the oas-a1.1 mutant plant. A, Fluorescence microscopy imaging of H2O2 in shoot and root tissues. One-week-old wild-type and oas-a1.1 mutant plants were loaded with H2DCFDA for 5 min in the presence of propidium iodide to visualize cell walls (pseudocolored in red) by confocal microscopy. H2O2 is visualized as pseudocolored in green. The experiments were repeated at least five times with similar results. B, Histochemical detection of H2O2 in the mature leaf. Detached leaves from 4-week-old wild-type and oas-a1.1 mutant plants were infiltrated with DAB and visualized. The presence of H2O2 is shown as brown staining. The experiments were repeated at least five times with similar results. C, Cell death staining of leaf lesions. Detached leaves from 4-week-old wild-type and oas-a1.1 mutant plants were stained with trypan blue for detection of dead cells. The insert contains a magnified photo of a lesion in the leaf from an oas-a1.1 mutant plant.
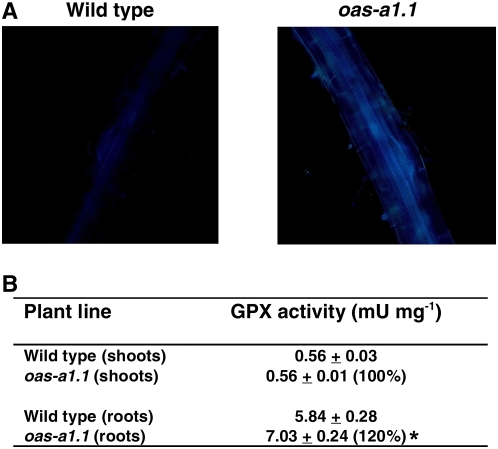
In plant tissues, many phenolic compounds, including lignin precursors, are potential ROS scavengers and the induction of phenylpropanoid metabolism and lignification is a feature of oxidative stress (Grace and Logan, 2000). We observed a significant increase in lignification in the roots of the oas-a1.1 mutant plants when compared to wild-type plants. This increased lignification was shown as enhanced blue UV autofluorescence of the vascular tissue cell walls (Fig. 7A). We also found that the increased lignin synthesis in the roots of the oas-a1.1 mutant plants correlated with an enhanced guaiacol peroxidase (GPX) activity, which was determined in identical plant tissues (Fig. 7B). Recently it was reported that an intermediary metabolite in phenylpropanoid metabolism (chlorogenic acid), in addition to being an antioxidant, acts as a reducing substrate for GPX (Grace and Logan, 2000).
Figure 7.
Deficiency of OAS-A1 causes root lignification and enhanced GPX activity. A, UV autofluorescence of lignin deposition. Roots of wild-type and oas-a1.1 mutant plants were observed using a fluorescence microscope. Bright-field images are shown in Supplemental Fig S1. B, GPX activity determined in crude extracts prepared from the shoots and roots of wild-type and oas-a1.1 mutant plants grown hydroponically. Values are means ± sd from five independent experiments. *, P < 0.05.
DISCUSSION
Plant cells contain several SAT and OASTL enzymes that are involved in the biosynthesis of Cys and whose subcellular localizations are different. This results in a complex variety of isoforms and in diverse subcellular Cys pools. At present, the reason(s) underlying the existence of so many redundant isoforms and the contributions of each subcellular Cys pool to plant cell metabolism are unknown. Much evidence coming from our gain-of-function studies suggested that the cytosolic Cys pool might be channeled to the specific plant responses against heavy metal exposure. OAS-A1 is the most abundant cytosolic OASTL isoform in Arabidopsis and was shown to be involved in the plant's responses to heavy metal stress. Overexpression of its gene in Arabidopsis enables this transgenic plant to survive under severe heavy metal stress (Dominguez-Solis et al., 2001, 2004). Our results suggested that the availability of cytosolic Cys is a limiting step for the synthesis of GSH and PC in Cd-stressed plants. However, the data presented in this investigation question the contribution of cytosolic Cys in redox signaling and ROS detoxification as part of the overall metabolic response of plants to stress.
Biochemical characterization of the T-DNA tagged mutant oas-a1.1 and oas-a1.2 plants revealed that the total OASTL activity and the total intracellular Cys concentration are reduced in these plants. As a consequence, in the mutant plants the total glutathione concentration is lowered and, more importantly, an oxidative shift in the glutathione redox state is produced, as can be inferred by comparison of the glutathione concentration and glutathione redox state in the wild-type plant. Thus, the mutant plants deficient in OAS-A1 seem to become oxidatively stressed as a result of an imbalance between the generation and removal of ROS. The transcriptomic footprints, which were determined in the oas-a1.1 mutant plants when grown under nonstressed conditions, and further validated by qRT-PCR, support this suggestion. A substantial proportion of the transcripts whose expression is altered in the oas-a1.1 mutant plant has predicted functions in different ROS-dependent physiological responses, including stress protection, pathogen responses, hormone signaling, plant growth and development, and regulation of transcription and translation (Mittler, 2002; Buchanan and Balmer, 2005). A broad survey of ROS-induced transcript expression in Arabidopsis by compiling data from publicly available microarray experiments was conducted (Gadjev et al., 2006). Several transcripts identified in that study as hallmarks of the general oxidative stress response match with the up-regulated genes that were detected in our transcriptomic analysis of the oas-a1.1 mutant. Thus, diverse transcripts encoding Toll-Interleukin-1 (TIR)-class disease resistance proteins, cytochromes P450, glutathione S-transferases, various oxidoreductases, and peroxidases are all induced in root and shoot tissues. Furthermore, the analysis of the transcriptomic data, which were generated from the different microarray experiments, revealed that a high proportion of transcription factors are up- and down-regulated by ROS (Gadjev et al., 2006). In agreement with these findings, our comparative transcriptomic analysis established that many transcription factors are specifically induced or repressed in oas-a1.1 mutant plants and highlighted the marked effect of the mutation on plant cell metabolism. A transcription factor with enhanced expression is the putative ethylene-responsive transcriptional coactivator MBF1, one of the genes that also show enhanced expression level in the oas-a1.2 mutant investigated by real-time RT-PCR.
The specific cellular responses to H2O2 have also been analyzed, using microarray technology, in Arabidopsis cultures following their exposure to exogenous H2O2 (Desikan et al., 2001). Many of the genes, including several heat shock proteins, a calcium-binding protein, transcription factors, or the Cys proteinase, which were identified as H2O2 responsive, were also revealed in our transcriptomic profile of the oas-a1.1 mutant. Furthermore, genes known to be activated by paraquat (op den Camp et al., 2003) are also induced in both the shoot and root tissues of the aos-a1.1 mutant plant.
Significantly, many transcripts with enhanced expression in the oas-a1.1 mutant plants, such as protein phosphatase 2C, calcium-binding protein, protein kinases, Zn-finger proteins, metal binding proteins, peroxidases, glutaredoxin, glutathione S-transferase, cytochromes P450, and heat shock proteins, are identical to those with enhanced expression in knockout plants deficient in cytosolic ascorbate peroxidase 1 (Davletova et al., 2005). In addition, the knockout plants that are deficient in these two cytosolic enzymes, OAS-A1 and APX1, also show similar levels and patterns of H2O2 accumulation in their leaves. This level and pattern of H2O2 accumulation cannot be compared with that observed in plants treated with exogenous H2O2 or paraquat because, in the knockout plants, H2O2 excess is produced in situ in the cytosol as a result of a defective ROS scavenging system. Interestingly, the mutant regulator of APX2 1-1, named rax1-1, which has been identified as an allele of GSH1 (γ-glutamyl-Cys synthetase), has lower glutathione levels in the leaves than the mutant plants deficient in OAS-A1. However, the foliar levels of H2O2 in the rax1-1 mutant were not affected when the plant was grown under nonstress conditions (Ball et al., 2004). In contrast to the oas-a1.1 and oas-a1.2 mutant plants, the glutathione redox state and the Cys levels in leaves were similar in rax1-1 and wild-type plants at all stages of their vegetative growth. This discrepancy could explain the different impact of the two mutations on cellular ROS levels. Furthermore, this result highlights the fact that levels of Cys, specifically in the cytosol, profoundly affect the stress defenses of the OAS-A1 knockout mutant plants through a perturbation of H2O2 homeostasis.
The redox environment of an organelle, cell, or tissue is the result of many redox couples acting simultaneously, and in particular of the most abundant ones, such as the GSH/GSSG redox couple (Schafer and Buettner, 2001). However, there is an increasing number of reports from animal cells showing that the plasma GSH/GSSG ratio is not in equilibrium with the plasma Cys/cystine pool. As a result, the balance between prooxidants and antioxidants cannot be defined by a single thiol/disulfide system (Jones, 2006). For instance, the Cys availability and the Cys/cystine redox couple regulate the p44/p42 mitogen-activated protein kinase (MAPK) pathway and cell proliferation in intestinal cells and this regulation occurs without altering the intracellular glutathione redox potential (Nkabyo et al., 2005). Furthermore, the Cys/cystine redox couple also influences cell differentiation in HT29 cells and this process is independent of the GSH/GSSG couple (Jones et al., 2004). The discrepancy between the redox status of the rax1-1 and the oas-a1.1 mutants and their respective contents in Cys and glutathione suggests that cytosolic Cys may serve as an independent node for redox signaling and control in plants. However, we cannot exclude the possibility that the cytosolic Cys supply may predominantly influence the level of cytosolic glutathione and, therefore, causes an imbalance in the steady-state redox equilibrium between the chloroplast and cytosolic compartments and/or the mitochondrial and cytosolic compartments.
Evidence reinforcing the suggestion that the OAS-A1 mutation causes a perturbation in H2O2 homeostasis is the H2O2-signaled induction of clustered cell death recently described (Dat et al., 2003). This particular pattern of cell death, in which groups of dead cells are found primarily in close proximity to the veins, can be triggered by perturbing H2O2 homeostasis in planta (Dat et al., 2003). Other evidence has come from the demonstration of increased synthesis of the polymer, lignin, which is often induced by oxidative stress and has a specific protective role as a structural component of the plant cell wall (Grace and Logan, 2000). Not only did we observe discernible differences in the extent of lignification between the oas-a1.1 mutant and wild-type plant, but also the transcriptomic analysis showed that genes encoding proteins involved in lignin biosynthesis, such as mannitol dehydrogenase and chitinase in the shoots and roots of the oas-a1.1 mutant, respectively, were up-regulated (van de Mortel et al., 2006). Furthermore, genes encoding cell wall proteins involved in strengthening the cell wall structure were also induced in the oas-a1.1 mutant, whereas genes encoding proteins involved in cell expansion, such as expansin and pectinesterase, were repressed in the shoots of the oas-a1.1 mutant.
In conclusion, our results suggest that Cys is a determinant of the antioxidative capacity of the cytosol in Arabidopsis, because a deficiency of the major OASTL isoform in the cytosol, OAS-A1, causes imbalance of H2O2 homeostasis. This conclusion is reinforced by the very recent finding that OAS-A1 is located to the peroxisomes, which are actively involved in ROS detoxification, in Arabidopsis leaves (Reumann et al., 2007). Furthermore, our results show that the enhanced sensitivity to Cd of the mutants defective in OAS-A1 is not due to a reduced supply of Cys for the synthesis of PCs required for metal chelation. Rather, the viability of the mutant plants exposed to Cd is compromised by a synergistic heavy metal-generated oxidative stress and not by PC deficiency.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Stress Treatments
Arabidopsis (Arabidopsis thaliana; accession Col-0) and the SALK_072213 and SAIL_94_E12 mutants were used in this work (Sessions et al., 2002; Alonso et al., 2003; Yamada et al., 2003). The plants were grown either in soil or perlite supplemented with Hoagland medium, in solid Murashige and Skoog (MS) medium in petri dishes, or in hydroponic culture. Plants were grown under a photoperiod of 16 h of white light (120 μE m−2 s−1) at 20°C/8-h dark at 18°C. For hydroponic culture, seeds were grown on petri dishes on MS medium containing 2.5 g L−1 Phytagel (Sigma-Aldrich) and 1% (w/v) Suc for 2 weeks and then seedlings were transferred to hydroponic culture. The nutrient solution contained 2 mm Ca(NO3)2, 0.5 mm KH2PO4, 0.75 mm MgSO4, 10 mm KNO3, 1.5 μm CuSO4, 2 μm ZnSO4, 10 μm MnSO4, 50 μm H3BO3, 0.1 μm MoO3, 50 μm KCl, and 50 μm Fe-Na-EDTA, and was replaced once a week.
Cd stress was imposed on plants growing on solid media by the addition of CdCl2 to the medium at the indicated concentrations. The Cd treatment of hydroponically grown plants was performed by adding 50 μm of CdCl2 to the above-described nutrient solution after a 2-week period of acclimation to the hydroponic conditions.
Mutant Isolation by PCR Screening
Several lines containing a T-DNA insertion in OAS-A1 (At4g14880) were selected from the SALK T-DNA and the SAIL T-DNA collections (http://signal.salk.edu/cgi-bin/tdnaexpress). Of the mutants studied in which the T-DNA insertions were located in the promoter region, namely, SALK_002902, SALK_074242, SALK_074243, SALK_126022, SALK_126023, SALK_142320, SALK_141592, SALK_123896, none showed the presence of the insertion or exhibited a significantly lower gene expression than that of the wild type, based on PCR analyses. We also screened four other mutants with the T-DNA insertions in the coding region: SALK_072212, SALK_072213, SAIL_193_D01, and SAIL_94_E12. The insertion was PCR undetectable in the SALK_072212 and the SAIL_193_D01 mutants and only the SALK_072213 and the SAIL_94_E12 mutants contained the T-DNA at the predicted sites.
To identify individuals that were homozygous for the T-DNA insertion, genomic DNA was extracted from kanamycin-resistant (30 mg mL−1) seedlings of SALK mutants or from DL-phosphinothricin resistant (25 mg mL−1) seedlings of SAIL mutants, and subjected to PCR genotyping using the following primers: F3, 5′-GAGTCGGGAGAGGAAGAGTCGCC-3′; R4, 5′-CGCCAAAATCTCTTCCGCCTT-3′; and LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′ for the SALK mutants, and F3, R5, 5′-AGCCTCGAAGGTCATGGCTTCC-3′; and LBb3, 5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′ for the SAIL mutants. PCR conditions were as follows: a denaturation cycle of 2 min at 94°C, 35 amplification cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, and an extension cycle of 5 min at 72°C.
RNA Isolation and RT-PCR Analysis
Total RNA was extracted from Arabidopsis leaves using the RNeasy plant mini-kit (QIAGEN) and retrotranscribed using oligo(dT) primer and the Superscript first-strand synthesis system for RT-PCR (Invitrogen). An aliquot of the cDNA was amplified in subsequent PCR reactions using the following primers: F4, 5′-GGCAAGGCAGCTTGCTCTTAAAGAAG-3′; R3, 5′-GGAAGCAAAGACGCAATGTAACTAAAGATG-3′; F5, 5′-CACCATGGCCTCGAGAATTGCTA-3′;and R5, 5′-AGCCTCGAAGGTCATGGCTTCC-3′, for the OAS-A1 gene; and UBQ10F, 5′-GATCTTTGCCGGAAAACAATTGGAGGATGGT-3′ and UBQ10R, 5′-CGACTTGTCATTAGAAAGAAAGAGATAACAGG-3′, for the constitutive UBQ10 gene used as a control. PCR conditions were as follows: a denaturation cycle of 2 min at 94°C, 35 amplification cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, and an extension cycle of 5 min at 72°C.
DNA Isolation and Southern-Blot Analysis
Total DNA was isolated from Arabidopsis leaves using the method described by Dellaporta et al. (1983). DNA (30 μg) was first digested with SpeI, a restriction enzyme that does not cut inside the OAS-A1 gene, and then electrophoresed, blotted onto a Z-PROBE blotting membrane (Bio-Rad) by capillary transfer, and fixed by backing at 80°C for 1 h. The membrane was prehybridized for 2 h at 65°C in 0.5 m sodium phosphate buffer (pH 7.2) containing 1 mm EDTA and 7% dodecyl sulfate (SDS). A T-DNA left border labeled probe was added and hybridization continued overnight. The membrane was washed twice in 40 mm sodium phosphate buffer (pH 7.2) containing 1 mm EDTA and 4% SDS at 65°C for 15 min each, plus two washes in 40 mm sodium phosphate buffer containing 1 mm EDTA and 1% SDS at 65°C for 15 min each. The filter was placed in front of a Fujifilm imaging plate BAS-MS 2040 for 24 h at room temperature and signals were detected and analyzed in a Cyclone storage phosphor system (Perkin-Elmer).
Western-Blot Analysis
Plant material was ground in 50 mm potassium phosphate buffer (pH 7.5), 1 mm EDTA, and 0.5 mm phenylmethylsulphonyl fluoride using a mortar and pestle with liquid nitrogen. After the crude extract had been centrifuged at 15,000g for 15 min at 4°C, the total amount of protein in the resulting supernatant was determined by the method of Bradford (1976). Protein aliquots (15 μg) of leaf extracts were electrophoresed on 12% acrylamide gels before transfer to polyvinylidene fluoride membranes using the NovaBlot system (Pharmacia), according to the manufacturer's instructions. Western blots were performed using polyclonal antibodies raised against the recombinant OAS-A1, which were generously provided by R. Hell (Heidelberg Institute of Plant Sciences, Germany). The secondary antibody was conjugated alkaline phosphatase (Sigma-Aldrich).
Assays of SAT, OASTL, and GPX Activities
SAT activity was determined in crude extracts by following the consumption of acetyl-CoA at 412 nm due to the accumulation of reduced 5,5′-dithiobis(2-nitrobenzoic acid), according to the method of Yamagata (1987). OASTL activity was measured using the method previously described by Barroso et al. (1995). GPX activity was assayed colorimetrically with guaiacol as a substrate. The reaction mixture contained 50 mm potassium phosphate buffer (pH 6.1), 0.25 mm H2O2, 6.25 mm guaiacol, and enzyme extract. The linear increase in absorption at 470 nm due to formation of tetraguaiacol was followed for 2 min (extinction coefficient of 26.6 mm−1 cm−1).
Quantification of Thiol Compounds
To quantify the total Cys and glutathione contents, thiols were extracted, reduced with NaBH4, and quantified by reverse-phase HPLC after derivatization with monobromobimane (Molecular Probes), as previously described (Dominguez-Solis et al., 2001). To determine the GSH content, the reduction step was omitted. The GSSG content was calculated as the difference between the contents of total glutathione and GSH.
The PC polypeptides were extracted and quantified by reverse-phase HPLC after postcolumn derivatization with 5,5′-dithiobis(2-nitrobenzoic acid), as described (Ramos et al., 2007). Standards of Cys and GSH (Sigma-Aldrich) and PCs (chemically synthesized by Biosyntan) were used for peak identification.
Quantification of Cd in Leaves
Hydroponically grown plants were cultured in the absence and presence of 50 μm CdCl2 for 18 and 24 h. The leaves were collected, dried, and analyzed by inductively coupled plasma atomic emission spectrometry as described (Dominguez-Solis et al., 2001).
RNA Extraction and Microarray Hybridization
Total RNA was initially isolated using the RNeasy plant mini-kit (QIAGEN) and used to synthesize biotinylated complementary RNA (cRNA) for hybridization to Arabidopsis ATH1 arrays (Affymetrix), using the 3′ amplification one-cycle target labeling kit. Briefly, 4 μg of RNA was reverse transcribed to produce first-strand cDNA using a (dT)24 primer with a T7 RNA polymerase promoter site added to the 3′ end. After second-strand synthesis, in vitro transcription was performed using T7 RNA polymerase and biotinyated nucleotides to produce biotin-labeled cRNA. The cRNA preparations (15 μg) were fragmented into 35- to 200-bp fragments at 95°C for 35 min. The fragmented cRNAs were hybridized to the Arabidopsis ATH1 microarrays at 45°C for 16 h. Each microarray was washed and stained in the Affymetrix Fluidics Station 400 following standard protocols. Microarrays were scanned using an Affymetrix GeneChip Scanner 3000.
Microarray Data Analysis
Microarray analysis was performed using the affylmaGUI R package (Wettenhall et al., 2006). The robust multiarray analysis (RMA) algorithm was used for background correction, normalization, and for summarizing expression levels (Irizarry et al., 2003). Differential expression analysis was performed using Bayes t statistics using the linear models for microarray data (Limma), which is included in the affylmGUI package. P values were corrected for multiple testing using the Benjamini-Hochberg's method (false discovery rate; Benjamini and Hochberg, 1995; Reiner et al., 2003). A cutoff value of a 2-fold change and P value of less than 0.05 were adopted to discriminate expression of genes that were differentially altered. Gene classification into functional groups was obtained from TAIR (http://www.arabidopsis.org), Map-Man (http://gabi.rzpd.de/projects/Map-Man/), and GENEVESTIGATOR (https://www.genevestigator.ethz.ch/at/).
Real-Time RT-PCR
qRT-PCR was used to validate microarray data. First-strand cDNA was synthesized as described above. Gene-specific primers were designed using the Vector NTI Advance 10 software (Invitrogen). Different genes with altered expression chosen from the microarray results and their primer sequences are as follows: The up-regulated genes in roots: AT2G36690 coding for an oxidoreductase, 2OG-Fe(II) oxygenase family protein (forward 5′-GAAGTCAGGTGGTGGTGGTG-3′; reverse 5′-AGAAGCGTCAAGAAGCCG-3′), and AT3G24500 coding for a putative ethylene-responsive transcriptional coactivator MBF1 (forward 5′-ATCGTGTGAAAGCAGAGG-3′; reverse 5′-CTGTTTCGCCAAATCCG-3′). The up-regulated genes in shoots: AT5G24770 encoding the vegetative storage protein 2 precursor VSP2 (forward 5′-GATCACTTCCAAACAGTACC-3′; reverse 5′-GTTCTTTAGGGCAAGTCC-3′), and AT4G15210 encoding the β-amylase BMY1 (forward 5′-GAAGCCCTAAGTGCTCCTC-3′; reverse 5′-CGTTCTCACCCGCAACT-3′). The down-regulated gene in roots: AT4G35770 encoding a senescence-associated protein SEN1 (forward 5′-CATAATCGGTTGTGAGAGC-3′; reverse 5′-GCAATGTCTGTGATCGCC-3′). The down-regulated genes in shoots: AT5G45890 coding for the senescence-specific SAG12 protein (forward 5′-GTTAATGATGAGCAAGCACTG-3′; reverse 5′-GGAAATCAAAACCACCTCC-3′) and AT3G23550 coding for a MATE efflux family protein (forward 5′-GCAAAGAAGGCGACTT-3′; reverse 5′-AAACCAACCCAGGCATC-3′). The real-time PCR reaction was performed using iQ SYBR Green Supermix (Bio-Rad), and the signals were detected on an iCYCLER (Bio-Rad), according to the manufacturer's instructions. The cycling profile consisted of 95°C for 10 min, 45 cycles of 95°C for 15 s, and 60°C for 1 min. A melt curve from 60°C to 90°C was run following the PCR cycling. The expression level of the genes of interest was normalized to that of the constitutive UBQ10 gene by subtracting the cycle threshold (CT) value of UBQ10 from the CT value of the gene (ΔCT). Fold change was calculated as 2−(ΔCT mutant − ΔCT wild type). The results shown are mean values ± sd of at least three independent RNA samples.
Detection of H2O2
For the fluorimetric detection of H2O2, 1-week seedlings were incubated for 5 min with 10 mm H2DCFDA (Molecular Probes) in the presence of 10 mm propidium iodide to visualize cell walls. The samples were observed using a LEICA TCS SP2 spectral confocal microscope (Leica Microsystems) with the following settings: excitation, 488 nm; emission, 500- to 550-nm range for fluorescein detection and 600- to 650-nm range for propidium detection. For the histochemical detection of H2O2, mature leaves were immersed in 1 mg mL−1 DAB (Sigma-Aldrich), fixed with a solution of 3:1:1 (v/v/v) ethanol:lactic acid:glycerol, and photographed.
Cell Death Detection
Trypan blue staining for dead cells in leaves was performed by incubating the leaves in a lactic acid-phenol-trypan blue solution (2.5 mg mL−1 trypan blue, 25% [w/v] lactic acid, 23% phenol, and 25% glycerol), then heated over boiling water for 1 min, and finally destained using a 2.5 g mL−1 chloral hydrate solution before photographing the leaves.
Microscopic Analysis of Lignification
Arabidopsis seeds were germinated and plants grown for 2 weeks on solid MS medium in petri dishes. Roots were collected and hand sections made by chopping roots on a microscope slide using a scalpel. These sections were analyzed with an Olympus BX50 microscope (Olympus) equipped with epifluorescence illumination (excitation filter WU 330–385 nm). Images were taken using a Leica DFC300 FX camera.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Bright-field images of roots shown in Figure 7.
Supplemental Table S1. Comparative transcriptomic analysis of the oas-a1.1 mutant roots versus wild-type roots.
Supplemental Table S2. Comparative transcriptomic analysis of the oas-a1.1 mutant shoots versus wild-type shoots.
Supplemental Table S3. Groups of functionally related genes up-regulated in roots of the oas-a1.1 mutant.
Supplemental Table S4. Groups of functionally related genes up-regulated in shoots of the oas-a1.1 mutant.
Supplemental Table S5. Groups of functionally related genes down-regulated in roots of the oas-a1.1 mutant.
Supplemental Table S6. Groups of functionally related genes down-regulated in shoots of the oas-a1.1 mutant.
Supplementary Material
Acknowledgments
We thank Dr. Rüdiger Hell, Heidelberg Institute of Plant Sciences (Germany) for kindly sharing the OASTL antibodies. We also thank Dr. Roberto Solano, Unidad de Genómica, Centro Nacional de Biotecnología (Spain), for help with the transcriptomic analysis.
This work was supported by Ministerio de Educación y Ciencia (grant nos. BIO2004–00784, BIO2007–62770, and AGL2005–01404) and Junta de Andalucía (grant no. CVI–273), Spain.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Cecilia Gotor (gotor@ibvf.csic.es).
The online version of this article contains Web-only data.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Ball L, Accotto GP, Bechtold U, Creissen G, Funck D, Jimenez A, Kular B, Leyland N, Mejia-Carranza J, Reynolds H, et al (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso C, Romero LC, Cejudo FJ, Vega JM, Gotor C (1999) Salt-specific regulation of the cytosolic O-acetylserine(thiol)lyase gene from Arabidopsis thaliana is dependent on abscisic acid. Plant Mol Biol 40 729–736 [DOI] [PubMed] [Google Scholar]
- Barroso C, Vega JM, Gotor C (1995) A new member of the cytosolic O-acetylserine(thiol)lyase gene family in Arabidopsis thaliana. FEBS Lett 363 1–5 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodological 57 289–300 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56 187–220 [DOI] [PubMed] [Google Scholar]
- Clemens S (2006) Evolution and function of phytochelatin synthases. J Plant Physiol 163 319–332 [DOI] [PubMed] [Google Scholar]
- Dat JF, Pellinen R, Beeckman T, Van De Cotte B, Langebartels C, Kangasjarvi J, Inze D, Van Breusegem F (2003) Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J 33 621–632 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1 19–21 [Google Scholar]
- Desikan R, Mackerness SAH, Hancock JT, Neill SJ (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Solis JR, Gutierrez-Alcala G, Vega JM, Romero LC, Gotor C (2001) The cytosolic O-acetylserine(thiol)lyase gene is regulated by heavy metals and can function in cadmium tolerance. J Biol Chem 276 9297–9302 [DOI] [PubMed] [Google Scholar]
- Dominguez-Solis JR, Lopez-Martin MC, Ager FJ, Ynsa MD, Romero LC, Gotor C (2004) Increased cysteine availability is essential for cadmium tolerance and accumulation in Arabidopsis thaliana. Plant Biotechnol J 2 469–476 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inze D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace SC, Logan BA (2000) Energy dissipation and radical scavenging by the plant phenylpropanoid pathway. Philos Trans R Soc Lond B Biol Sci 355 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth JR, Dominguez-Solis JR, Gutierrez-Alcala G, Wray JL, Romero LC, Gotor C (2003) The serine acetyltransferase gene family in Arabidopsis thaliana and the regulation of its expression by cadmium. Plant Mol Biol 51 589–598 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264 [DOI] [PubMed] [Google Scholar]
- Jones DP (2006) Redefining oxidative stress. Antioxid Redox Signal 8 1865–1879 [DOI] [PubMed] [Google Scholar]
- Jones DP, Go YM, Anderson CL, Ziegler TR, Kinkade JM, Kirlin WG (2004) Cysteine/cystine couple is a newly recognized node in the circuitry for biologic redox signaling and control. FASEB J 18 1246. [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48 251–275 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Hell R (2005) Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res 86 435–457 [DOI] [PubMed] [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7 405–410 [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Rausch T (2005) Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth Res 86 459–474 [DOI] [PubMed] [Google Scholar]
- Nkabyo YS, Go YM, Ziegler TR, Jones DP (2005) Extracellular cysteine/cystine redox regulates the p44/p42 MAPK pathway by metalloproteinase-dependent epidermal growth factor receptor signaling. Am J Physiol Gastrointest Liver Physiol 289 G70–G78 [DOI] [PubMed] [Google Scholar]
- op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, et al (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J, Clemente MR, Naya L, Loscos J, Perez-Rontome C, Sato S, Tabata S, Becana M (2007) Phytochelatin synthases of the model legume Lotus japonicus. A small multigene family with differential response to cadmium and alternatively spliced variants. Plant Physiol 143 1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19 368–375 [DOI] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Luder F, Weckwerth W, Jahn O (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer FQ, Buettner GR (2001) Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med 30 1191–1212 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel JE, Villanueva LA, Schat H, Kwekkeboom J, Coughlan S, Moerland PD, van Themaat EVL, Koornneef M, Aarts MGM (2006) Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol 142 1127–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK (2006) affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22 897–899 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Droux M, Hell R (2004) O-acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot 55 1785–1798 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R (2006) Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. J Plant Physiol 163 273–286 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846 [DOI] [PubMed] [Google Scholar]
- Yamagata S (1987) Partial purification and some properties of homoserine O-acetyltransferase of a methionine auxotroph of Saccharomyces cerevisiae. J Bacteriol 169 3458–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.