Abstract
High-grade ovarian serous papillary cancer (OSPC) and uterine serous papillary carcinoma (USPC) represent two histologically similar malignancies characterised by markedly different biological behavior and response to chemotherapy. Understanding the molecular basis of these differences may significantly refine differential diagnosis and management, and may lead to the development of novel, more specific and more effective treatment modalities for OSPC and USPC. We used an oligonucleotide microarray with probe sets complementary to >10 000 human genes to determine whether patterns of gene expression may differentiate OSPC from USPC. Hierarchical cluster analysis of gene expression in OSPC and USPC identified 116 genes that exhibited >two-fold differences (P<0.05) and that readily distinguished OSPC from USPC. Plasminogen activator inhibitor (PAI-2) was the most highly overexpressed gene in OSPC when compared to USPC, while c-erbB2 was the most strikingly overexpressed gene in USPC when compared to OSPC. Overexpression of the c-erbB2 gene and its expression product (i.e., HER-2/neu receptor) was validated by quantitative RT–PCR as well as by flow cytometry on primary USPC and OSPC, respectively. Immunohistochemical staining of serous tumour samples from which primary OSPC and USPC cultures were derived as well as from an independent set of 20 clinical tissue samples (i.e., 10 OSPC and 10 USPC) further confirmed HER-2/neu as a novel molecular diagnostic and therapeutic marker for USPC. Gene expression fingerprints have the potential to predict the anatomical site of tumour origin and readily identify the biologically more aggressive USPC from OSPC. A therapeutic strategy targeting HER-2/neu may be beneficial in patients harbouring chemotherapy-resistant USPC.
Keywords: serous papillary uterine cancer, serous papillary ovarian cancer, gene expression profiling, Her-2/neu, herceptin
Ovarian serous papillary cancer (OSPC) represents the most common histological type of ovarian carcinoma, the fourth leading cause of cancer-related death in women in the United States (Jemal et al, 2003). Endometrial cancer is the most frequent cancer of the female genital tract with endometrioid (type 1) and serous papillary (type 2) being the most common cell types (Deligdisch and Holinka, 1987; Jemal et al, 2003). Histologically indistinguishable to high-grade serous ovarian carcinoma (Carcangiu and Chambers, 1992; Sherman et al, 1992), uterine serous papillary cancer (USPC) has a propensity for early intraabdominal, lymphatic and distant metastatic spread even at presentation (Carcangiu and Chambers, 1992; Goff et al, 1994; Nicklin and Copeland, 1996) and is characterised by a highly aggressive biological behavior (Deligdisch and Holinka, 1987; Carcangiu and Chambers, 1992; Sherman et al, 1992; Goff et al, 1994; Nicklin and Copeland, 1996). Unlike OSPC, however, which is responsive to first-line combined cisplatinum-based chemotherapy in 70–80% of the cases (Kalil and McGuire, 2002), USPC is a chemotherapy-resistant disease from outset, with responses to cytostatic agents in the order of 20% and of short duration (Levenback et al, 1992; Carcangiu and Chambers, 1995; Nicklin and Copeland, 1996).
Gene expression fingerprints representing large numbers of genes have the potential to allow precise and accurate grouping of tumours endowed with similar phenotype (Giordano et al, 2001; Sorlie et al, 2001; Rosenwald et al, 2002; Zhan et al, 2002). Gene microarrays may identify cancers endowed with a more aggressive biologic behaviour (i.e., rapidly metastatic tumours) that are unresponsive to standard adjuvant therapies and may thus allow improved prediction of response and clinical outcome. Consistent with this view, in large B-cell lymphomas and breast carcinomas, gene expression profiles have been shown to identify patients who are unlikely to be cured by conventional therapy (Sorlie et al, 2001; Rosenwald et al, 2002). In ovarian carcinoma, cDNA microarray technology has recently been used to identify numerous genes differentially expressed in normal and tumour-derived ovarian epithelial cells (Ismail et al, 2000; Hough et al, 2001; Welsh et al, 2001; Schwartz et al, 2002). Interestingly, several of the most upregulated genes encode surface or secreted proteins, such as Kop, SLPI and claudin-3, making these products attractive candidate biomarkers (Ismail et al, 2000; Hough et al, 2001; Welsh et al, 2001; Schwartz et al, 2002). In contrast, very little is known about the possible genetic diversity between OSPC and USPC, two histologically similar serous carcinomas characterised by a dramatically different biological behavior and response to chemotherapy.
In this study, oligonucleotide microarrays were used to profile and compare gene expression patterns in 11 primary cultures of OSPC and USPC. We report that mRNA fingerprints readily distinguish the more biologically aggressive and chemotherapy resistant USPC from OSPC. Of interest, OSPC2, a primary OSPC with mixed clear cell features (a variant of ovarian cancer also characterised with a particularly unfavourable prognosis), clustered with USPC. Plasminogen activator inhibitor (PAI-2) was the gene most highly upregulated in OSPC relative to USPC, while the c-erbB2 gene product (HER-2/neu) was strikingly overexpressed in USPC relative to OSPC and may therefore represent a novel diagnostic and therapeutic marker for this highly aggressive subset of endometrial tumours.
MATERIALS AND METHODS
Establishment of OSPC and USPC primary cell lines
In all, 11 primary serous papillary cell lines (six OSPC and five USPC) were established after sterile processing of the tumour samples from surgical biopsies as described for ovarian and uterine carcinoma specimens (Santin et al, 2000, 2002a, 2002b). All tumour samples were obtained with appropriate consent according to IRB guidelines. Tumours were staged according to the FIGO operative staging system. Total abdominal hysterectomy and regional lymph node sampling for invasive USPC were performed in all cases. Radical tumour debulking, including a total abdominal hysterectomy and omentectomy, was performed in all ovarian carcinoma patients. No patient received chemotherapy before surgical therapy. The patient characteristics are described in Table 1. The epithelial nature and the purity of USPC and OSPC cultures was verified by immunohistochemical staining and flow cytometric analysis with antibodies against cytokeratin as described (Ismail et al, 2000; Santin et al, 2000, 2002a, 2002b). Only primary cultures which had at least 90% viability and contained >99% tumour cells were used for total RNA extraction.
Table 1. Characteristics of the patients.
| Patient | Age | Race | Stage | Chemotherapy regimen |
|---|---|---|---|---|
| USPC 1 | 66 | Afro-American | IV B | TAX+CARB |
| USPC 2 | 77 | White | III C | TAX+CARB |
| USPC 3 | 61 | Afro-American | III C | TAX+CARB |
| USPC 4 | 62 | Afro-American | III C | TAX+CARB |
| USPC 5 | 63 | Afro-American | III C | TAX+CARB |
| OSPC 1 | 42 | White | III C | TAX+CIS |
| OSPC 2 | 43 | White | III C | TAX+CARB |
| OSPC 3 | 34 | White | III C | TAX+CARB |
| OSPC 4 | 51 | White | III C | TAX+CARB |
| OSPC 5 | 59 | Afro-American | III B | TAX+CARB |
| OSPC 6 | 52 | White | III C | TAX+CARB |
RNA purification, microarray hybridisation and analysis
RNA purification, cDNA synthesis, cRNA preparation and hybridisation to the Affymetrix Human U95Av2 GeneChip microarray were performed according to the manufacturer's protocols and as reported (Zhan et al, 2002).
Data processing
All data used in our analyses were derived from Affymetrix 5.0 software. GeneChip 5.0 output files are given as a signal that represents the difference between the intensities of the sequence-specific perfect match probe set and the mismatch probe set, or as a detection of present, marginal, or absent signals as determined by the GeneChip 5.0 algorithm. Gene arrays were scaled to an average signal of 1500 and then analysed independently. Signal calls were transformed by the log base 2 and each sample was normalised to give a mean of 0 and variance of 1.
Gene expression data analysis
Statistical analyses of the data were performed with the software packages SPSS10.0. (SPSS, Chicago, IL, USA) and the significance analysis of microarrays (SAM) method (Tusher et al, 2001). Genes were selected for analysis based on detection and fold change. In each comparison, genes having ‘present’ detection calls in more than half of the samples in the overexpressed gene group were retained for statistical analysis if they showed >two-fold change between groups. Retained genes were subjected to SAM to establish a false discovery rate (FDR), then further filtered via the Wilcoxon rank-sum (WRS) test at α=0.05. The FDR obtained from the initial SAM analysis was assumed to characterise genes found significant via WRS.
Gene cluster/treeview
The hierarchical clustering of average-linkage method with the centred correlation metric was used (Eisen et al, 1998). The dendrogram was constructed with a subset of genes from 12 588 probe sets present on the microarray, whose expression levels vary the most among the 11 samples, and thus most informative. For the hierarchical clustering shown in Figures 1 and 2, only genes significantly expressed and whose average change in expression level was at least two-fold were chosen. The expression value of each selected gene was re-normalized to have a mean of zero.
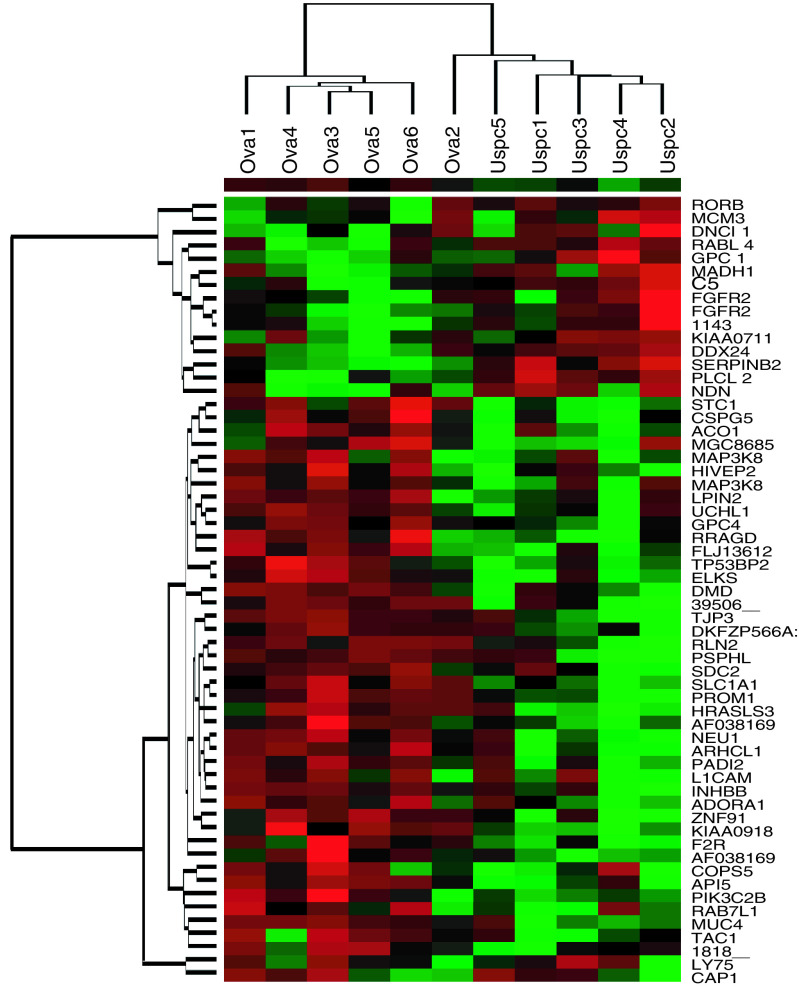
Figure 1.
Molecular profile of 11 primary OSPC and USPC cell lines. Hierarchical clustering of 59 genes with differential expression between six OSPC and five USPC groups (P<0.05) using a two-fold threshold. The cluster is colour coded using red for upregulation, green for downregulation and black for median expression. Agglomerative clustering of genes was illustrated with dendrograms. The symbol for each gene corresponding to the oligonucleotide spotted on the array is shown.
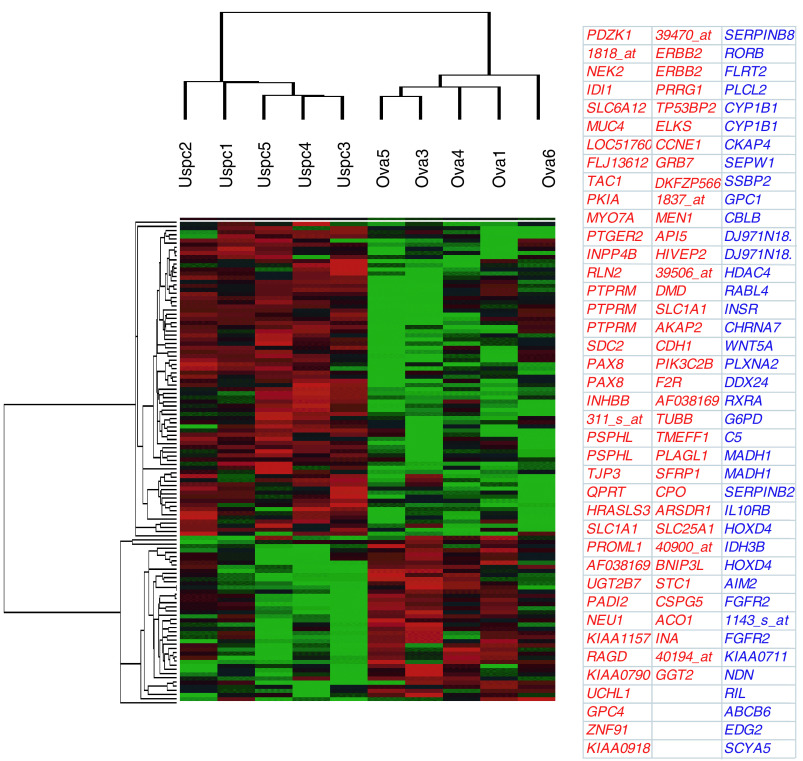
Figure 2.
Molecular profile of primary OSPC and USPC cell lines. Hierarchical clustering of 116 genes with differential expression between five OSPC and five USPC groups (P<0.05) using a two-fold threshold. The cluster is colour coded using red for upregulation, green for downregulation and black for median expression. Agglomerative clustering of genes was illustrated with dendrograms. The symbol for each gene corresponding to the oligonucleotide spotted on the array is shown. USPC upregulated genes are shown in red ink while OSPC upregulated genes are shown in blue ink.
Quantitative real-time PCR
q-RT – PCR was performed with an ABI Prism 7000 Sequence Analyzer using the manufacturer's recommended protocol (Applied Biosystems, Foster City, CA, USA) to validate differential expression of selected genes in samples from six representative primary tumour cell lines (three OSPC and three USPC). Each reaction was run in triplicate. The comparative threshold cycle (CT) method was used for the calculation of amplification fold as specified by the manufacturer. Briefly, 5 μg of total RNA from each sample was reverse transcribed using SuperScript II Rnase H Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). A value of 10 μl of reverse-transcribed RNA samples (from 500 μl of total volume) was amplified by using the TaqMan Universal PCR Master Mix (Applied Biosystems) to produce PCR products specific for PAI-2 and c-erbB2. Primers specific for 18s ribosomal RNA and empirically determined ratios of 18 s competimers (Applied Biosystems) were used to control for the amounts of cDNA generated from each sample. Sequences for primers and probes are available on request. Differences among OSPC and USPC in the q-RT – PCR expression data were tested using the Kruskal–Wallis nonparametric test. Pearson's product – moment correlations were used to estimate the degree of association between the microarray and q-RT – PCR data.
Flow cytometry
To validate microarray data on primary OSPC and USPC cell lines at the protein level, HER-2/neu receptor expression was evaluated by flow cytometry. The HER-2/neu MAb Herceptin (Genentech, San Francisco, CA, USA) was used as the primary antibody. FITC-conjugated goat anti-human F(ab)2 immunoglobulin was used as a secondary reagent (BioSource International, Camarillo, CA, USA). Analysis was conducted with a FACScan, utilising Cell Quest software (Becton Dickinson).
HER2/neu immunostaining of formalin-fixed tumour tissues
To evaluate whether the differential HER2/Neu receptor expression detected by flow cytometry on primary OSPC and USPC cell lines was comparable to the expression of HER-2/neu receptor of uncultured OSPC and USPC from which the primary cell lines were derived, protein expression was evaluated by immunohistochemical staining on formalin-fixed tumour tissue. In addition, to further confirm transcriptional profiling results, the HER2/neu marker was also evaluated by immunohistochemistry in a second independent set of 20 clinical tissue samples (i.e., 10 OSPC and 10 USPC) obtained from patients harbouring advanced stage disease (i.e., stages III and IV). Study blocks were selected after histopathologic review by a surgical pathologist. The intensity of staining was graded as 0 (staining not greater than negative control), 1+ (light staining), 2+ (moderate staining) or 3+ (heavy staining).
RESULTS
Gene expression profiles distinguish OSPC from USPC and identify differentially expressed genes
Flash frozen biopsies from ovarian and uterine tumour tissue are known to contain significant numbers of contaminant stromal cells as well as a variety of host-derived immune cells (e. g., monocytes, dendritic cells, lymphocytes). Short-term primary OSPC and USPC cell cultures, minimising the risk of a selection bias inherent in any long-term in vitro growth, provide an opportunity to study differential gene expression between relatively pure populations of tumour cells. Comprehensive gene expression profiles of six primary OSPC and five primary USPC cell lines were generated using high-density oligonucleotide arrays with 12 588 probe sets, which in total interrogated some 10 000 genes. In total, 165 genes were differentially expressed between OSPC and USPC (WRS test, P<0.05). Figure 1 shows the cluster analysis performed on hybridisation intensity values for 59 gene segments whose average difference in expression level was at least two-fold. Two major branches on the dendrogram were identified. All five USPC were grouped together in the rightmost columns. Similarly, in the leftmost columns five pure OSPC were found to cluster tightly together. Of interest, OSPC2, a serous papillary tumour with mixed clear cell features (i.e., a biologically aggressive variant of ovarian cancer characterised by a poor prognosis) clustered on a sub-branch with USPC (Figure 1). Figure 2 shows the cluster analysis on hybridisation intensity values for each gene in 10 primary cultures of OSPC and USPC showing a single type of differentiation. There were 484 genes showing >two-fold change along with ‘present’ detection calls in more than half the samples in the overexpressed group. Of these, 316 were found significant by SAM, with a median FDR of 17.4% and a 90th percentile FDR of 22.7%. Of the 484 aforementioned genes, 116 yielded P<0.05 via WRS, and all 116 were among the genes found significant by SAM. Thus, we can say with 90% confidence that the FDR among genes found significant via WRS is no higher than 22.7%. The new dendrogram shown in Figure 2 depicts a marked separation in the expression profiles of the two groups of serous papillary tumours. The tight clustering of pure OSPC from USPC was driven by two distinct profiles of gene expression. The first was represented by a group of 40 genes that were highly expressed in OSPC and underexpressed in USPC (Table 2). Many genes shown previously to be involved in ovarian carcinogenesis are present on these lists, providing a degree of validity to our array analysis. Included in this group of genes are plasminogen activator inhibitor-2 (PAI-2), fibroblast growth factor receptor-2 (FGFR2), glypican 1 (GPC1), lysophosphatidic acid receptor (EDG2), phospholipase C (PLCL2), glucose-6-phosphate dehydrogenase (G6PD) and insulin receptor (IGF1) (Table 2). The second profile was represented by 76 genes that were highly expressed in USPC and underexpressed in OSPC (Table 3). Included in this group of genes are epidermal growth factor type 2 receptor (c-erbB2), inhibin (INHBB), multiple endocrine neoplasia I (MEN1), growth factor receptor-bound protein 7 (GRB7), BCL2, E-cadherin (CDH1) and syndecan (SDC2) (Table 3). Importantly, c-erbB2 gene was the most highly differentially expressed gene in USPC when compared to OSPC (Table 3, Table 4, Figure 2). OSPC2, the only serous tumour with mixed clear cell histology evaluated in our series, was also found to highly overexpress c-erbB2 (data not shown).
Table 2. Upregulated genes expressed at least two fold higher in OSPC compared with USPC.
| Probe set name | Gene symbol | Map location | p of WRS | Ratio Ov/Ut |
|---|---|---|---|---|
| 37185_at | SERPINB2 | 18q21.3 | 0.00902344 | 21.2101742 |
| 40478_at | DJ971N18.2 | 20p12 | 0.0162936 | 7.391447995 |
| 38837_at | DJ971N18.2 | 20p12 | 0.047201768 | 6.933671714 |
| 34439_at | AIM2 | 1q22 | 0.00902344 | 6.689727463 |
| 36073_at | NDN | 15q11.2-q12 | 0.028280124 | 6.460327167 |
| 859_at | CYP1B1 | 2p21 | 0.047201768 | 4.642443935 |
| 40387_at | EDG2 | 9q32 | 0.047201768 | 4.612620508 |
| 1669_at | WNT5A | 3p21-p14 | 0.028280124 | 4.35214472 |
| 1363_at | FGFR2 | 10q26 | 0.0162936 | 3.958060853 |
| 1143_s_at | 0.028280124 | 3.948020982 | ||
| 37816_at | C5 | 9q32-q34 | 0.0162936 | 3.945622621 |
| 40071_at | CYP1B1 | 2p21 | 0.047201768 | 3.826875845 |
| 38294_at | HOXD4 | 2q31-q37 | 0.028280124 | 3.804399853 |
| 33162_at | INSR | 19p13.3-p13.2 | 0.047201768 | 3.772 |
| 34853_at | FLRT2 | 14q24-q32 | 0.047201768 | 3.471204819 |
| 40395_at | PLXNA2 | 1q32.1 | 0.028280124 | 3.371729137 |
| 39805_at | ABCB6 | 2q36 | 0.047201768 | 3.369062784 |
| 41796_at | PLCL2 | 3p24.3 | 0.00902344 | 3.280007364 |
| 1403_s_at | SCYA5 | 17q11.2-q12 | 0.047201768 | 3.158368265 |
| 33929_at | GPC1 | 2q35-q37 | 0.028280124 | 3.15594993 |
| 39566_at | CHRNA7 | 15q14 | 0.047201768 | 3.14079953 |
| 34354_at | FGFR2 | 10q26 | 0.047201768 | 2.928346342 |
| 444_g_at | HOXD4 | 2q31-q37 | 0.047201768 | 2.892672123 |
| 38042_at | G6PD | Xq28 | 0.047201768 | 2.813117012 |
| 36077_at | RABL4 | 22q13.1 | 0.028280124 | 2.720984156 |
| 36453_at | KIAA0711 | 8p23.3 | 0.047201768 | 2.688792044 |
| 32668_at | SSBP2 | 5q14.1 | 0.047201768 | 2.663148439 |
| 32610_at | RIL | 5q31.1 | 0.047201768 | 2.55031145 |
| 514_at | CBLB | 3q13.12 | 0.028280124 | 2.511893491 |
| 40112_at | IDH3B | 20p13 | 0.028280124 | 2.294973901 |
| 38271_at | HDAC4 | 2q37.2 | 0.028280124 | 2.245891142 |
| 1325_at | MADH1 | 4q28 | 0.047201768 | 2.228503651 |
| 32381_at | RORB | 9q22 | 0.028280124 | 2.205852674 |
| 32800_at | RXRA | 9q34.3 | 0.047201768 | 2.168594631 |
| 36312_at | SERPINB8 | 18q21.3 | 0.047201768 | 2.110497544 |
| 40142_at | DDX24 | 14q32 | 0.0162936 | 2.109997452 |
| 33227_at | IL10RB | 21q22.11 | 0.047201768 | 2.082986437 |
| 32529_at | CKAP4 | 12q23.3 | 0.047201768 | 2.04858844 |
| 37280_at | MADH1 | 4q28 | 0.028280124 | 2.044781456 |
| 39709_at | SEPW1 | 19q13.3 | 0.028280124 | 2.017195806 |
Table 3. Upregulated genes expressed at least two-fold higher in USPC compared with OSPC.
| Probe set name | Gene symbol | Map location | p of WRS | Ratio Ut/Ov |
|---|---|---|---|---|
| 1802_s_at | ERBB2 | 17q11.2-q12 | 0.028280124 | 17.39166248 |
| 39470_at | 0.00902344 | 14.13960749 | ||
| 41470_at | PROML1 | 4p15.33 | 0.00902344 | 11.00274366 |
| 32521_at | SFRP1 | 8p12-p11.1 | 0.047201768 | 10.49619245 |
| 33218_at | ERBB2 | 17q11.2-q12 | 0.0162936 | 9.009761458 |
| 41354_at | STC1 | 8p21-p11.2 | 0.0162936 | 7.780569927 |
| 41700_at | F2R | 5q13 | 0.028280124 | 7.299013748 |
| 38207_at | MEN1 | 11q13 | 0.028280124 | 6.578419265 |
| 36254_at | TAC1 | 7q21-q22 | 0.047201768 | 6.292979547 |
| 38268_at | SLC1A1 | 9p24 | 0.0162936 | 5.506571087 |
| 33576_at | KIAA0918 | 13q31.1 | 0.0162936 | 5.478319783 |
| 37883_i_at | AF038169 | 2q22.1 | 0.0162936 | 5.06566416 |
| 35704_at | HRASLS3 | 11q13.1 | 0.028280124 | 4.596441783 |
| 38267_at | SLC1A1 | 9p24 | 0.028280124 | 4.488128886 |
| 41376_i_at | UGT2B7 | 4q13 | 0.047201768 | 4.418941048 |
| 828_at | PTGER2 | 14q22 | 0.028280124 | 4.338041431 |
| 39506_at | 0.028280124 | 4.313685637 | ||
| 1680_at | GRB7 | 17q12 | 0.047201768 | 4.262623744 |
| 38545_at | INHBB | 2cen-q13 | 0.028280124 | 4.198823428 |
| 40679_at | SLC6A12 | 12p13 | 0.047201768 | 3.956969879 |
| 35912_at | MUC4 | 3q29 | 0.028280124 | 3.94095027 |
| 39966_at | CSPG5 | 3p21.3 | 0.047201768 | 3.918103678 |
| 32027_at | PDZK1 | 1q21 | 0.047201768 | 3.91484375 |
| 31732_at | RLN2 | 9p24.1 | 0.0162936 | 3.913095715 |
| 36202_at | PKIA | 8q21.11 | 0.047201768 | 3.89984472 |
| 37978_at | QPRT | 16q13 | 0.0162936 | 3.845374532 |
| 994_at | PTPRM | 18p11.2 | 0.047201768 | 3.812843137 |
| 37208_at | PSPHL | 7q11.2 | 0.028280124 | 3.654717567 |
| 37884_f_at | AF038169 | 2q22.1 | 0.028280124 | 3.593346825 |
| 995_g_at | PTPRM | 18p11.2 | 0.028280124 | 3.555706062 |
| 35985_at | AKAP2 | 9q31-q33 | 0.028280124 | 3.319448607 |
| 32963_s_at | RAGD | 6q15-q16 | 0.00902344 | 3.280777993 |
| 33358_at | KIAA1157 | 12q13.13 | 0.0162936 | 3.250881457 |
| 311_s_at | 0.0162936 | 3.138465417 | ||
| 35674_at | PADI2 | 1p35.2-p35.1 | 0.047201768 | 3.100307522 |
| 2021_s_at | CCNE1 | 19q12 | 0.028280124 | 3.081090355 |
| 32893_s_at | GGT2 | 22q11.23 | 0.047201768 | 3.055014721 |
| 36869_at | PAX8 | 2q12-q14 | 0.047201768 | 3.050015496 |
| 36508_at | GPC4 | Xq26.1 | 0.0162936 | 2.887073572 |
| 39901_at | MYO7A | 11q13.5 | 0.028280124 | 2.885983264 |
| 35148_at | TJP3 | 19p13.3 | 0.028280124 | 2.879832572 |
| 31892_at | PTPRM | 18p11.2 | 0.047201768 | 2.844557651 |
| 36990_at | UCHL1 | 4p14 | 0.0162936 | 2.833524684 |
| 37209_g_at | PSPHL | 7q11.2 | 0.047201768 | 2.780479031 |
| 38168_at | INPP4B | 4q31.1 | 0.00902344 | 2.645321215 |
| 36943_r_at | PLAGL1 | 6q24-q25 | 0.0162936 | 2.57527834 |
| 37258_at | TMEFF1 | 9q31 | 0.047201768 | 2.55946924 |
| 36985_at | IDI1 | 10p15.3 | 0.047201768 | 2.538587569 |
| 39075_at | NEU1 | 6p21.3 | 0.0162936 | 2.521110072 |
| 40488_at | DMD | Xp21.2 | 0.00902344 | 2.507697552 |
| 39332_at | TUBB | 6p21.3 | 0.047201768 | 2.504487188 |
| 39757_at | SDC2 | 8q22-q23 | 0.047201768 | 2.452025072 |
| 933_f_at | ZNF91 | 19p13.1-p12 | 0.028280124 | 2.445525292 |
| 37210_at | INA | 10q25.1 | 0.047201768 | 2.387532735 |
| 1860_at | TP53BP2 | 1q42.1 | 0.0162936 | 2.356857655 |
| 37869_at | ELKS | 12p13.3 | 0.028280124 | 2.356300578 |
| 33878_at | FLJ13612 | 2q36.1 | 0.0162936 | 2.319659881 |
| 35143_at | DKFZP566A1524 | 0.047201768 | 2.312331476 | |
| 38997_at | SLC25A1 | 22q11.21 | 0.00902344 | 2.304275318 |
| 40077_at | ACO1 | 9p22-p13 | 0.028280124 | 2.297124855 |
| 36261_at | LOC51760 | 16p13.13 | 0.028280124 | 2.252602915 |
| 39436_at | BNIP3L | 8p21 | 0.047201768 | 2.236567978 |
| 977_s_at | CDH1 | 16q22.1 | 0.00902344 | 2.212331718 |
| 36175_s_at | HIVEP2 | 6q23-q24 | 0.047201768 | 2.206300362 |
| 41269_r_at | API5 | 11p12-q12 | 0.0162936 | 2.189353711 |
| 1837_at | 0.047201768 | 2.180124558 | ||
| 1818_at | 0.047201768 | 2.177494716 | ||
| 366_s_at | NEK2 | 1q32.2-q41 | 0.047201768 | 2.157771457 |
| 40900_at | 0.028280124 | 2.151464435 | ||
| 40194_at | 0.028280124 | 2.133081444 | ||
| 41172_at | ARSDR1 | 14q23.3 | 0.0162936 | 2.113388456 |
| 37999_at | CPO | 3q12 | 0.028280124 | 2.100322069 |
| 35978_at | PRRG1 | Xp21.1 | 0.028280124 | 2.05552932 |
| 121_at | PAX8 | 2q12-q14 | 0.028280124 | 2.028946437 |
| 41715_at | PIK3C2B | 1q32 | 0.00902344 | 2.024856688 |
| 41644_at | KIAA0790 | 6q24.3 | 0.047201768 | 2.004743183 |
Table 4. Differentially expressed genes in USPC and OSPC ranked by significance analysis of microarrays (SAM).
| Order | Probeset | Gene ID | Score (d) | Numerator (r) | Denominator (s+s0) | Fold change | q-value (%) |
|---|---|---|---|---|---|---|---|
| 1 | 39470_at | 39470_at | 2.5193455 | 3.596293949 | 1.427471523 | 13.48750 | 5.9170776 |
| 2 | 41470_at | PROM1 | 2.1986583 | 3.825972532 | 1.740139698 | 10.60748 | 5.9170776 |
| 3 | 41354_at | STC1 | 2.0805893 | 2.751729734 | 1.322572262 | 7.74726 | 5.9170776 |
| 4 | 38207_at | 38207_at | 2.0726086 | 3.311991516 | 1.597982165 | 6.51440 | 5.9170776 |
| 5 | 1802_s_at | ERBB2 | 1.9669529 | 3.492165691 | 1.775419064 | 17.65875 | 5.9170776 |
| 6 | 39506_at | 39506_at | 1.9191495 | 3.359977858 | 1.750763964 | 4.15946 | 5.9170776 |
| 7 | 33218_at | ERBB2 | 1.8831385 | 2.592838393 | 1.376870795 | 9.12831 | 5.9170776 |
| 8 | 37978_at | QPRT | 1.8508847 | 2.471411028 | 1.335259314 | 3.80801 | 5.9170776 |
| 9 | 35912_at | MUC4 | 1.8095618 | 2.831603133 | 1.564800451 | 3.85299 | 5.9170776 |
| 10 | 41376_i_at | UGT2B7 | 1.8010151 | 3.20113721 | 1.777407186 | 4.20987 | 5.9170776 |
| 11 | 37208_at | PSPHL | 1.784549 | 3.621295275 | 2.029249576 | 3.44056 | 5.9170776 |
| 12 | 38545_at | INHBB | 1.7806599 | 3.920162398 | 2.201522201 | 4.16889 | 5.9170776 |
| 13 | 33576_at | KIAA0918 | 1.7555623 | 2.180726438 | 1.242181175 | 5.31405 | 5.9170776 |
| 14 | 32521_at | SFRP1 | 1.7432391 | 2.824069509 | 1.620012708 | 10.60682 | 5.9170776 |
| 15 | 35704_at | HRASLS3 | 1.7261492 | 2.548006268 | 1.476121705 | 4.49637 | 5.9170776 |
| 16 | 33358_at | ARHCL1 | 1.6665422 | 1.95881684 | 1.175377897 | 3.16918 | 5.9170776 |
| 17 | 31732_at | RLN2 | 1.6347725 | 2.459163154 | 1.50428461 | 3.67860 | 5.9170776 |
| 18 | 38267_at | SLC1A1 | 1.6306308 | 2.219928654 | 1.361392538 | 4.39416 | 5.9170776 |
| 19 | 37883_i_at | AF038169 | 1.6147363 | 2.219700503 | 1.374652015 | 4.98971 | 5.9170776 |
| 20 | 32963_s_at | RRAGD | 1.6121619 | 1.626523265 | 1.008908118 | 3.26944 | 5.9170776 |
| 21 | 994_at | PTPRM | 1.6097261 | 2.784507191 | 1.729801847 | 3.63111 | 5.9170776 |
| 22 | 995_g_at | PTPRM | 1.6088285 | 2.897830612 | 1.801205444 | 3.40453 | 5.9170776 |
| 23 | 311_s_at | 311_s_at | 1.5938406 | 4.139143468 | 2.596961926 | 3.01704 | 5.9170776 |
| 24 | 38268_at | SLC1A1 | 1.5621501 | 2.110626126 | 1.351103322 | 5.35166 | 5.9170776 |
| 25 | 31892_at | PTPRM | 1.5119824 | 3.112003216 | 2.058227135 | 2.74704 | 5.9170776 |
| 26 | 35148_at | TJP3 | 1.510845 | 1.998741079 | 1.322929233 | 2.73928 | 5.9170776 |
| 27 | 41700_at | F2R | 1.5014465 | 2.195953257 | 1.462558397 | 7.18646 | 5.9170776 |
| 28 | 35674_at | PADI2 | 1.479696 | 2.264084328 | 1.53010105 | 3.01842 | 5.9170776 |
| 29 | 1680_at | GRB7 | 1.4698173 | 2.027019267 | 1.379096066 | 4.25537 | 5.9170776 |
| 30 | 37209_g_at | PSPHL | 1.4664685 | 1.930835879 | 1.316656891 | 2.66282 | 5.9170776 |
| 31 | 39966_at | CSPG5 | 1.4455697 | 1.818363955 | 1.257887393 | 3.93459 | 5.9170776 |
| 32 | 36869_at | PAX8 | 1.441286 | 2.813586886 | 1.95213647 | 2.99423 | 5.9170776 |
| 33 | 36202_at | PKIA | 1.4322379 | 2.002737974 | 1.398327765 | 3.74274 | 5.9170776 |
| 34 | 828_at | PTGER2 | 1.4285229 | 1.890660776 | 1.32350749 | 4.10149 | 5.9170776 |
| 35 | 39075_at | NEU1 | 1.4246097 | 1.46915449 | 1.031268071 | 2.50012 | 5.9170776 |
| 36 | 36990_at | UCHL1 | 1.4234749 | 1.966718653 | 1.381632101 | 2.81907 | 5.9170776 |
| 37 | 36943_r_at | PLAGL1 | 1.4016117 | 1.503433722 | 1.072646422 | 2.57234 | 5.9170776 |
| 38 | 40488_at | DMD | 1.3944472 | 1.630344124 | 1.16916874 | 2.42639 | 5.9170776 |
| 39 | 35985_at | PALM2 | 1.3805351 | 2.00686626 | 1.4536872 | 3.22031 | 5.9170776 |
| 40 | 36254_at | TAC1 | 1.3634058 | 2.750616522 | 2.017459875 | 6.36232 | 5.9170776 |
| 41 | 37869_at | ELKS | 1.3454118 | 1.248133462 | 0.927696233 | 2.33294 | 5.9170776 |
| 42 | 2021_s_at | CCNE1 | 1.3294844 | 1.454719587 | 1.094198324 | 3.06396 | 5.9170776 |
| 43 | 33878_at | FLJ13612 | 1.3274937 | 1.237854188 | 0.932474615 | 2.29949 | 5.9170776 |
| 44 | 39757_at | SDC2 | 1.3043638 | 2.072350471 | 1.588782604 | 2.35737 | 5.9170776 |
| 45 | 36508_at | GPC4 | 1.2991039 | 1.843342415 | 1.41893382 | 2.86605 | 5.9170776 |
| 46 | 933_f_at | ZNF91 | 1.2741536 | 1.376374281 | 1.080226336 | 2.37962 | 5.9170776 |
| 47 | 41269_r_at | API5 | 1.2739465 | 1.209800113 | 0.949647539 | 2.16666 | 5.9170776 |
| 48 | 40679_at | SLC6A12 | 1.2660176 | 2.282007655 | 1.802508675 | 3.78353 | 5.9170776 |
| 49 | 38168_at | INPP4B | 1.2480709 | 1.359180414 | 1.089025041 | 2.54847 | 5.9170776 |
| 50 | 1860_at | TP53BP2 | 1.2409357 | 1.215816901 | 0.979758188 | 2.35894 | 5.9170776 |
| 51 | 38997_at | SLC25A1 | 1.2115515 | 1.434841782 | 1.184301064 | 2.27730 | 5.9170776 |
| 52 | 36261_at | LOC51760 | 1.2106872 | 1.239580344 | 1.023865052 | 2.21646 | 5.9170776 |
| 53 | 37258_at | TMEFF1 | 1.1973963 | 1.515221127 | 1.265429912 | 2.66426 | 5.9170776 |
| 54 | 37884_f_at | AF038169 | 1.1701381 | 1.429952351 | 1.222037264 | 3.55995 | 5.9170776 |
| 55 | 39332_at | MGC8685 | 1.1504101 | 1.560522625 | 1.356492456 | 2.45317 | 5.9170776 |
| 56 | 40077_at | ACO1 | 1.1452896 | 1.324530845 | 1.15650301 | 2.29132 | 5.9170776 |
| 57 | 40194_at | GTF2H2 | 1.1312711 | 1.17700447 | 1.040426546 | 2.11918 | 5.9170776 |
| 58 | 36985_at | IDI1 | 1.1292005 | 1.378736024 | 1.220984217 | 2.55491 | 5.9170776 |
| 59 | 121_at | PAX8 | 1.1201623 | 1.208458366 | 1.078824359 | 2.03114 | 5.9170776 |
| 60 | 977_s_at | CDH1 | 1.1193705 | 2.699730529 | 2.411829333 | 2.20046 | 5.9170776 |
| 61 | 35143_at | DKFZP566A1524 | 1.1183384 | 1.620344382 | 1.44888559 | 2.22206 | 5.9170776 |
| 62 | 37999_at | CPO | 1.1064994 | 1.06722326 | 0.964504126 | 2.06986 | 5.9170776 |
| 63 | 37210_at | INA | 1.1044125 | 1.281004592 | 1.159896882 | 2.40281 | 5.9170776 |
| 64 | 36175_s_at | HIVEP2 | 1.0992289 | 1.174149577 | 1.068157485 | 2.19097 | 5.9170776 |
| 65 | 366_s_at | NEK2 | 1.0763152 | 1.152375048 | 1.070666856 | 2.16698 | 5.9170776 |
| 66 | 32893_s_at | GGT2 | 1.0751984 | 1.434104513 | 1.33380457 | 2.99540 | 5.9170776 |
| 67 | 41172_at | RDH11 | 1.0679325 | 1.000763289 | 0.937103522 | 2.07342 | 5.9170776 |
| 68 | 35978_at | PRRG1 | 1.0611643 | 1.103335239 | 1.039740243 | 2.04432 | 5.9170776 |
| 69 | 41715_at | PIK3C2B | 1.0489539 | 0.921860026 | 0.878837519 | 2.00231 | 5.9170776 |
| 70 | 39901_at | EDIL3 | 1.0487582 | 1.199399171 | 1.143637468 | 2.81306 | 5.9170776 |
| 71 | 40900_at | MYH10 | 1.033065 | 0.971734516 | 0.940632521 | 2.13908 | 6.1516929 |
| 72 | 39436_at | BNIP3L | 1.0248207 | 1.021584326 | 0.996841974 | 2.23030 | 6.1516929 |
| 73 | 32027_at | PDZK1 | 1.0234011 | 1.294822852 | 1.265215452 | 3.74641 | 6.1516929 |
| 74 | 1818_at | 1818_at | 1.0226221 | 1.159537838 | 1.13388698 | 2.17253 | 6.1516929 |
| 75 | 41644_at | SASH1 | 1.0075712 | 0.944905271 | 0.937804973 | 1.98700 | 6.2373918 |
| 76 | 1837_at | 1837_at | 0.8980795 | 1.048441576 | 1.167426238 | 2.15603 | 7.6204787 |
| 1 | 37185_at | SERPINB2 | −2.712078 | −4.21769782 | 1.555153568 | 0.04432 | 5.9170776 |
| 2 | 34439_at | AIM2 | −1.887056 | −2.3302995 | 1.234886275 | 0.14663 | 17.36191 |
| 3 | 33162_at | INSR | −1.7303429 | −2.66873302 | 1.542314531 | 0.26385 | 17.36191 |
| 4 | 40478_at | DJ971N18.2 | −1.7156271 | −2.71817086 | 1.584359902 | 0.13539 | 17.36191 |
| 5 | 41796_at | PLCL2 | −1.6656048 | −1.85109255 | 1.111363586 | 0.30311 | 17.36191 |
| 6 | 37816_at | C5 | −1.5809083 | −2.31648603 | 1.465288041 | 0.24600 | 17.36191 |
| 7 | 859_at | CYP1B1 | −1.5772674 | −3.24419453 | 2.056844985 | 0.20802 | 17.36191 |
| 8 | 40071_at | CYP1B1 | −1.5585486 | −3.21119542 | 2.060375537 | 0.25050 | 17.36191 |
| 9 | 39566_at | CHRNA7 | −1.5536961 | −2.40918109 | 1.550612809 | 0.31177 | 17.36191 |
| 10 | 36073_at | NDN | −1.4829481 | −3.50636378 | 2.364454837 | 0.14949 | 17.36191 |
| 11 | 38837_at | DJ971N18.2 | −1.4796673 | −2.14671946 | 1.450812223 | 0.14290 | 17.36191 |
| 12 | 36077_at | RABL4 | −1.4523961 | −1.96257543 | 1.351267383 | 0.36574 | 17.36191 |
| 13 | 40395_at | PLXNA2 | −1.4373891 | −1.79826591 | 1.251064131 | 0.29518 | 17.36191 |
| 14 | 1669_at | WNT5A | −1.4102572 | −2.6444399 | 1.875147235 | 0.22963 | 17.36191 |
| 15 | 40387_at | EDG2 | −1.3772565 | −2.25220008 | 1.635280028 | 0.22311 | 17.36191 |
| 16 | 39805_at | ABCB6 | −1.3495984 | −2.0539729 | 1.521914154 | 0.30094 | 17.36191 |
| 17 | 32668_at | SSBP2 | −1.305583 | −1.39323342 | 1.067135083 | 0.38204 | 17.36191 |
| 18 | 38294_at | HOXD4 | −1.3021769 | −1.80385782 | 1.385263285 | 0.26382 | 17.36191 |
| 19 | 33929_at | GPC1 | −1.2952699 | −1.49713601 | 1.155848702 | 0.32097 | 17.36191 |
| 20 | 444_g_at | 444_g_at | −1.2845736 | −1.70421346 | 1.326676407 | 0.34439 | 17.36191 |
| 21 | 1363_at | FGFR2 | −1.2806909 | −1.57525259 | 1.230002183 | 0.24473 | 17.36191 |
| 22 | 40142_at | DDX24 | −1.2701961 | −1.27558267 | 1.004240756 | 0.46178 | 17.36191 |
| 23 | 1143_s_at | 1143_s_at | −1.2593487 | −1.61800179 | 1.284792503 | 0.24502 | 17.36191 |
| 24 | 36453_at | KIAA0711 | −1.229961 | −1.94610499 | 1.582249319 | 0.36118 | 17.36191 |
| 25 | 514_at | CBLB | −1.2010635 | −1.19235629 | 0.992750384 | 0.39810 | 17.36191 |
| 26 | 34853_at | FLRT2 | −1.2008015 | −1.40750914 | 1.172141399 | 0.29102 | 17.36191 |
| 27 | 40112_at | IDH3B | −1.1932078 | −1.31001149 | 1.097890455 | 0.43852 | 17.36191 |
| 28 | 38271_at | MGC16025 | −1.1909435 | −1.56930168 | 1.31769611 | 0.44320 | 17.36191 |
| 29 | 32381_at | RORB | −1.1900513 | −1.68115724 | 1.412676329 | 0.44795 | 17.36191 |
| 30 | 39709_at | SEPW1 | −1.1672019 | −1.17314959 | 1.005095698 | 0.49329 | 17.36191 |
| 31 | 38042_at | G6PD | −1.1666541 | −1.3074933 | 1.12072066 | 0.35407 | 17.36191 |
| 32 | 32610_at | RIL | −1.1500988 | −1.87581032 | 1.630999328 | 0.38759 | 17.36191 |
| 33 | 1325_at | MADH1 | −1.1063114 | −1.23201185 | 1.113621238 | 0.44054 | 17.36191 |
| 34 | 32800_at | RXRA | −1.0647524 | −1.19695561 | 1.124163351 | 0.46285 | 17.36191 |
| 35 | 34354_at | FGFR2 | −1.0353338 | −1.34091 | 1.295147453 | 0.33456 | 17.36191 |
| 36 | 37280_at | MADH1 | −1.03448 | −1.06866713 | 1.033047673 | 0.48063 | 17.36191 |
| 37 | 33227_at | IL10RB | −1.017976 | −1.10172876 | 1.08227379 | 0.47487 | 17.36191 |
| 38 | 32529_at | CKAP4 | −0.9990762 | −1.12343355 | 1.124472307 | 0.48333 | 17.36191 |
| 39 | 36312_at | SERPINB8 | −0.9653125 | −1.26934824 | 1.31496097 | 0.47187 | 17.36191 |
| 40 | 1403_s_at | CCL5 | −0.9613302 | −1.420866 | 1.478020843 | 0.30353 | 17.36191 |
Validation of the microarray data
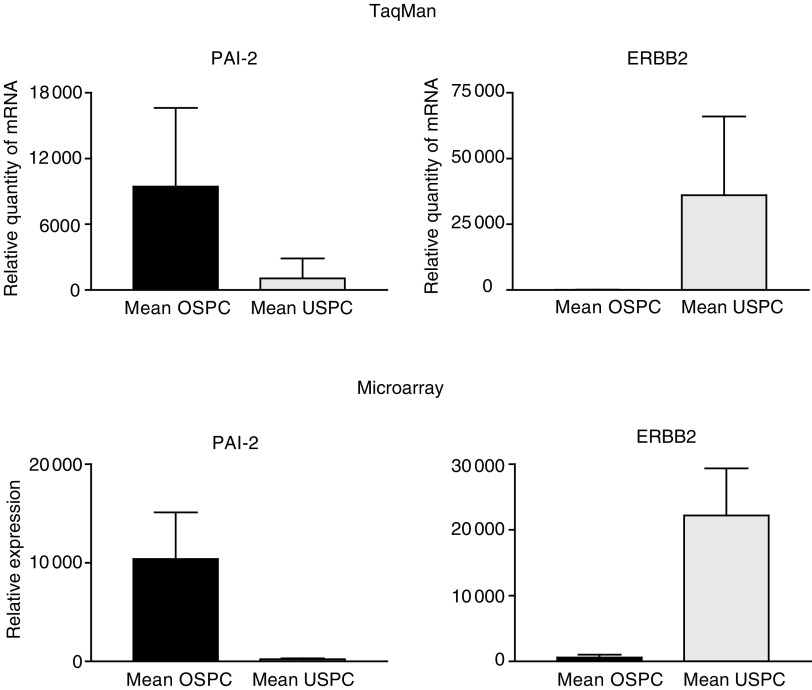
We used q-RT – PCR assays to validate the microarray data. The two most highly differentially expressed genes between OSPC and USPC (i.e., PAI-2 and c-erbB2) were selected for q-RT – PCR analysis. A comparison of the microarray and q-RT – PCR data for these genes is shown in Figure 3. Expression differences between tumour types for PAI-2 (P=0.009) and c-erbB2 (P=0.02), were readily apparent (Tables 2 and 3). Moreover, for both genes tested, the q-RT – PCR data were highly correlated to the microarray data (P<0.001) (r=0.91 and 0.71, respectively), as estimated from the 6 samples (i.e., three OSPC and three USPC) included in both the q-RT – PCR and microarray experiments. The q-RT – PCR data mirror the microarray data, both qualitatively and quantitatively, and suggest that most array probe sets are likely to accurately measure the levels of the intended transcript within a complex mixture of transcripts.
Figure 3.
Quantitative RT – PCR and microarray expression analysis of PAI-2 (SERPINB2) and c-erbB2 (ERBB2) selected genes differentially expressed between OSPC and USPC.
HER-2/neu expression
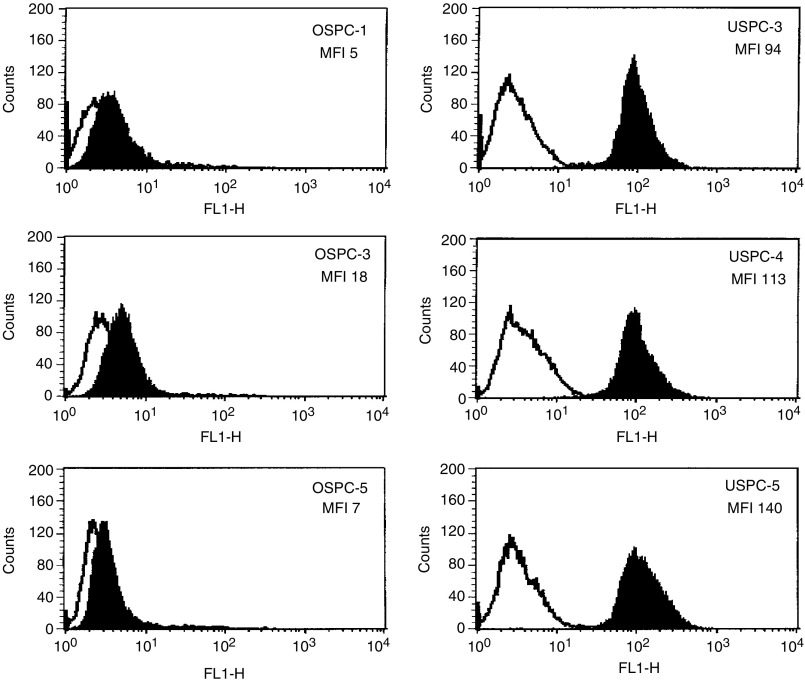
We evaluated HER-2/neu expression by flow cytometry on six primary serous papillary cell lines (three OSPC and three USPC). As positive and negative controls, breast cancer cell lines known to overexpress HER-2/neu (BT-474 and SK-BR-3, American Type Culture Collection), and Epstein – Barr virus-transformed lymphoblastoid cell lines (LCL) established from the same USPC and OSPC patients were also studied. High HER-2/neu receptor expression was found on all three primary USPC cell lines tested (100% positive cells for all three USPC), with mean fluorescence intensity (MFI) ranging from 94 to 140 (Figure 4). In contrast, primary OSPC cell lines were found to express significantly lower levels of HER-2/neu (average MFI was 10-fold lower) than the USPC cells (P<0.001) (Figure 4). These results show that high expression of the c-erbB2 gene product by USPC correlates tightly with high protein expression by the tumour cells. Autologous LCL were consistently negative for HER-2/neu expression, while breast cancer cell lines expressed high levels of HER-2/neu (data not shown).
Figure 4.
FACS analysis of Herceptin staining of three primary OSPC and three USPC cell lines. Data with Herceptin are shown in solid black while isotype control MAb profiles are shown in white. HER-2/neu expression was significantly higher on USPC cell lines compared to OSPC cell lines (P<0.001 by Student's t-test).
Immunohistochemical analysis of HER2/neu expression
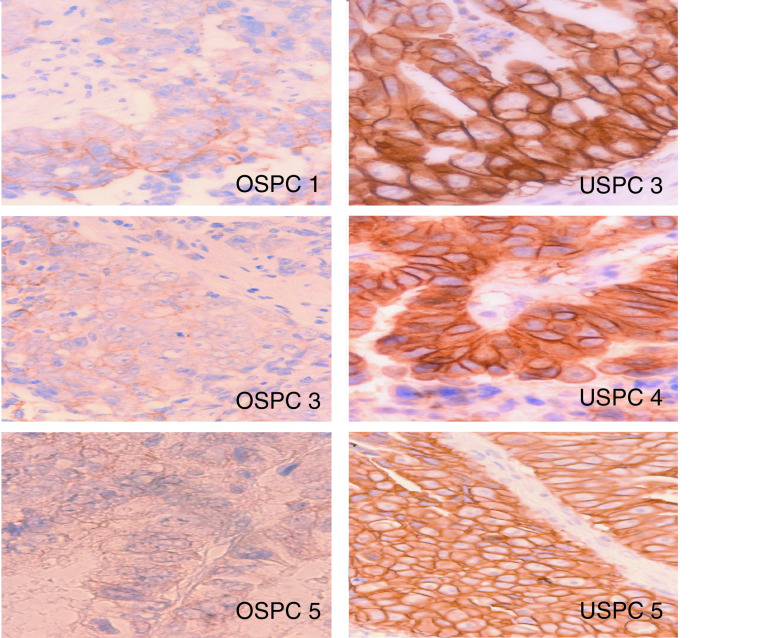
Formalin-fixed tumour tissue blocks from six primary surgical specimens were tested for HER-2/neu expression. Heavy staining for HER-2/neu protein expression (i.e., score 3+) was noted in all three USPC specimens that also overexpressed the c-erbB2 gene product by microarray and flow cytometry, respectively (Figure 5). In contrast, negative or low (i.e., score 0 or 1+) staining was found in all three representative OSPC samples (Figure 5). Similarly, when formalin-fixed tumour tissue blocks from 20 independent surgical specimens (i.e., 10 OSPC vs 10 USPC) were tested for HER-2/neu expression, a moderate to heavy staining was found in 70% of USPC (i.e., 70% score 2+ and 3+, 30% score 1+) vs 10% of OSPC (i.e., 10% score 2+ and 90% score 0 to 1+) (P=0.0002 USPC vs OSPC by student's t-test).
Figure 5.
Immunohistochemical staining for HER-2/neu expression on three paraffin-embedded OSPC3 and three USPC5 specimens from which primary cell lines have been established. OSPC1, OSPC3 and OSPC5 (left panel) showed negative or light (1+) staining for HER-2/neu. USPC3, USPC4 and USPC5 (right panel), showed heavy (3+) staining for HER-2/neu. Original magnification × 400.
DISCUSSION
High-throughput comprehensive technologies for assaying gene expression, such as high-density oligonucleotide and cDNA microarrays, may offer the potential to identify clinically relevant subsets of tumours difficult to distinguish by conventional histopathological assessment (Giordano et al, 2001; Rosenwald et al, 2002; Schwartz et al, 2002). This report represents the first communication of an investigation involving the genome-wide examination of differences in gene expression between serous papillary ovarian cancer (OSPC) and uterine serous papillary carcinoma (USPC), two histologically indistinguishable gynaecologic tumours characterised by a dramatically different biologic behavior and response to chemotherapy.
Advanced and/or metastatic serous papillary gynaecologic tumours, regardless of their ovarian or uterine origin, are currently treated with a combined cisplatinum-based chemotherapy. However, given that: (1) USPC likely arise from metaplastic Mullerian epithelium, while OSPC likely derive from the ovarian surface epithelium, and (2) a dramatic difference in response to standard chemotherapy regimens is commonly reported among these histologically indistinguishable serous carcinomas (Levenback et al, 1992; Sherman et al, 1992; Carcangiu and Chambers, 1995; Nicklin and Copeland, 1996; Kalil and McGuire 2002), a significant diversity in gene expression among these tumours is probable. In agreement with this view, all five USPC patients evaluated in this study either developed progressive disease during chemotherapy or recurred within 6 months from the end of treatment. In contrast, four out of five of the OSPC patients responded completely to standard adjuvant chemotherapy treatment. In this study, we have used short-term primary OSPC and USPC cultures (to minimise the risk of a selection bias inherent in any long-term in vitro growth) to study differential gene expression in highly enriched populations of epithelial tumour cells. Strikingly, we found that hierarchical clustering of the samples and gene expression levels within the samples led to the unambiguous separation of OSPC from USPC. We detected 116 genes differentially expressed between OSPC and USPC whose average change in expression level between the two groups was at least two-fold. Of the 116 genes that yielded P<0.05 via WRS, all 116 were among the genes found significant by SAM. Our study offers therefore the first persuasive support that the dramatically different biologic behaviour and response to treatment commonly reported in OSPC compared to USPC may be dictated by a profound genetic diversity among these histological indistinguishable serous neoplasms. It is therefore likely that a molecular classification based on gene expression profiles may thus potentially identify gynaecologic serous tumours associated with aggressive behaviour and poor prognosis and should allow therapeutic approaches to be better tailored to the biologic and genetic characteristic of each serous tumour type. These novel findings have thus the potential to significantly refine diagnosis and possibly alter management of these cancer patients. Of interest, OSPC2, the only OSPC with mixed clear cell features included in our analysis, clustered with USPC. These data are congruent with a recent report that clear cell ovarian tumours present a distinctive molecular signature from pure high-grade OSPC (Schwartz et al, 2002). Thus, our findings add to previous knowledge showing that clear cell tumours, a variant of ovarian cancer with a particularly unfavourable prognosis, express a molecular signature closer to that of the more aggressive USPC.
A sizeable number of genes differentially expressed in OSPC compared with USPC have been identified through our analysis. Some of these may prove to be useful diagnostic and therapeutic markers for these histologically similar diseases. For example, elevated serum levels of lysophosphatidic acid (LPA) are found in more than 90% of ovarian cancer patients and the level of LPA in plasma has been proposed as a potential biomarker for this disease (Budnik and Mukhopadhyay, 2002). In addition, LPA signalling may have a role in the progression of ovarian cancer cells through stimulation of cellular proliferation, enhanced cellular survival and suppression of apoptosis (Contos et al, 2000). It seems therefore likely that the higher LPA receptor expression found in OSPC relative to USPC may represent a distinctive marker that plays a role in transduction of growth-promoting signals from high local concentrations of LPA (Contos et al, 2000; Budnik and Mukhopadhyay, 2002). Consistent with this view, phospholipase C, another gene that is differentially overexpressed in OSPC relative to USPC has been previously reported to contribute to LPA production in ovarian cancer cells (Budnik and Mukhopadhyay, 2002).
Several reports have shown that plasminogen activator inhibitor-2 (PAI-2), a protein capable of inhibiting invasion (Andreasen et al, 2000), may represent a molecular biomarker for several human tumours including ovarian carcinomas. Consistent with our findings, however, overexpression of PAI-2 in epithelial ovarian cancer has been previously identified as a favourable prognostic factor (Chambers et al, 1997). Indeed, high PAI-2 expression in invasive ovarian tumours seem to be limited to a group of OSPC patients which experience a more prolonged disease free and overall survival (Chambers et al, 1997). These data are therefore consistent with the view that high expression of PAI-2 in OSPC compared to USPC may be a marker indicating a biologically less aggressive disease.
Membrane-associated heparan sulphate proteoglycans are thought to play important roles in many aspects of cell behaviour, including cell – cell and cell – extracellular matrix adhesion and growth factor signalling (David, 1993). Two families of polypeptides appear to carry the majority of heparan sulphate on mammalian cells: glypicans, which are attached to the plasma membrane via glycosylphosphatidylinositol (GPI) anchors, and syndecans, which are transmembrane proteins (David, 1993). Convincing evidence has recently been provided that glypican-1 can interact with FGF-2 and stimulate signalling of the FGF receptor (Steinfeld et al, 1996). Importantly, high glypican-1 and FGF receptor 2 gene expression were found differentially expressed in OSPC when compared to USPC, while syndecan-2 gene expression was significantly higher in USPC when compared USPC. These data therefore support a major difference in the expression of heparan sulphate proteoglycans between these two subsets of histologically indistinguishable serous tumours. Furthermore, because bFGF is produced by OSPC and can bind to FGF receptor 2 expressed on these tumours (Steinfeld et al, 1996), it is likely that the combined overexpression of glypicans and FGF receptor 2 genes found in OSPC may represent a common molecular abnormality with important functional consequences for the progression of OSPC.
Insulin receptor has been previously reported overexpressed on OSPC and to be able to mediate a proliferative response in ovarian cancer cells (Kalli et al, 2002). In our study, consistent with previous reports, OSPC were found to differentially overexpress the insulin receptor gene when compared to USPC. These results therefore support a role for insulin receptor in the growth and regulation of OSPC, but not in USPC.
Unlike OSPC, there have been remarkably few studies aimed at identifying molecular markers characteristic of USPC. Because of the common poor response to standard salvage treatment modalities for advanced or recurrent USPC, the identification of a number of USPC specific markers may lay the groundwork for future studies testing some of these biomarkers for clinical utility in the treatment of these highly aggressive and intrinsically chemotherapy resistant tumours. Of great interest at this regard, c-erbB2 gene was found to be the most highly differentially expressed gene in USPC with over 17-fold upregulation compared with OSPC. Furthermore, the growth factor receptor-bound protein 7 (GRB7), a gene tightly linked to c-erbB2 and previously reported coamplified and coexpressed with this gene in several cancer types (Janes et al, 1997) was also highly differentially expressed in USPC compared to OSPC. The striking overexpression of the c-erbB2 gene as well as of its gene expression product on USPC may therefore represent a distinctive molecular marker for these serous tumours and also provide insights into the disproportionately poor prognosis of USPC relative to OSPC. Consistent with this view, previous studies have reported that the amplification of this gene in a subset of ovarian cancer patients is associated with resistance to chemotherapeutic drugs and shorter survival (Berchuck et al, 1990). On the light of our micrarrays data it is tempting to speculate that some if not all of these highly HER2/neu overexpressing and chemotherapy resistant serous tumours may likely have arisen from metaplastic mesothelial cells and therefore present a genetic fingerprint more similar to USPC than OSPC. Regardless of the histologic site of origin, however, high overexpression of the c-erbB2 gene provides support for the notion that trastuzumab (Herceptin), a humanised anti-HER-2/Neu antibody that is showing great promise for treatment of metastatic breast cancer patients overexpressing HER-2/Neu protein (Slamon et al, 2001), may be a novel, potentially highly effective therapy against this aggressive variant of serous papillary carcinomas. Consistent with this view, our group has recently shown high sensitivity of USPC to the killing activity mediated by natural killer (NK) cells when triggered by anti-HER-2/Neu-specific antibody in vitro (Santin et al, 2002b).
Taken all together, our data demonstrate that OSPC and USPC, two diseases where further molecular characterisation is needed to improve differential diagnosis and therapeutic strategies, can be readily discriminated solely by gene expression profiles. These findings suggest that global gene expression signatures can be an important adjunct to the morphology based classification schemes for serous papillary tumours currently used. Finally, the identification of c-erbB2 as the most highly differentially expressed gene in USPC suggest that targeting HER-2/neu by rhuMAb anti-HER-2 (Herceptin) may be potentially highly beneficial against these biologically aggressive and chemotherapy-resistant variants of endometrial cancer.
Acknowledgments
We wish to thank Eric Siegel for the statistical analysis of the data.
Footnotes
This work was supported in part by grants from the Angelo Nocivelli and the Camillo Golgi Foundation, Brescia, Italy.
References
- Andreasen PA, Egelund R, Petersen HH (2000) The plasminogen activation system in tumour growth, invasion, and metastasis. Cell Mol Life Sci 57: 25–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchuck A, Kamel A, Whitaker R, Kerns B, Olt G, Kinney R, Soper JT, Dodge R, Clarke-Pearson DL, Marks P (1990) Overexpression of HER-2/neu is associated with poor survival in advanced epithelial ovarian cancer. Cancer Res 50: 4087–4091 [PubMed] [Google Scholar]
- Budnik LT, Mukhopadhyay AK (2002) Lysophosphatidic acid and its role in reproduction. Biol Reprod 66: 859–865 [DOI] [PubMed] [Google Scholar]
- Carcangiu ML, Chambers JT (1992) Uterine papillary serous carcinoma: a study on 108 cases with emphasis on prognostic significance of associated endometrioid carcinoma, absence of invasion, and concomitant ovarian cancer. Gynecol Oncol 47: 298–305 [DOI] [PubMed] [Google Scholar]
- Carcangiu ML, Chambers JT (1995) Early pathologic stage clear cell carcinoma and uterine papillary serous carcinoma of the endometrium, comparison of clinicopathological features and survival. Int J Gynecol Pathol 14: 30–38 [DOI] [PubMed] [Google Scholar]
- Chambers SK, Ivins CM, Carcangiu ML (1997) Expression of plasminogen activator inhibitor-2 in epithelial ovarian cancer: a favorable prognostic factor related to the actions of CSF-1. Int J Cancer 74: 571–575 [DOI] [PubMed] [Google Scholar]
- Contos JJ, Ishii I, Chun J (2000) Lysophosphatidic acid receptors. Mol Pharmacol 58: 1188–1196 [DOI] [PubMed] [Google Scholar]
- David G (1993) Integral membrane heparan sulfate proteoglycans. FASEB J 7: 1023–1030 [DOI] [PubMed] [Google Scholar]
- Deligdisch L, Holinka CF (1987) Endometrial carcinoma: two diseases? Cancer Detect Prev 10: 237–246 [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Bostein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Aca Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano TJ, Shedden KA, Schwartz DR, Kuick R, Taylor JM, Lee N, Misek DE, Greenson JK, Kardia SL, Beer DG, Rennert G, Cho KR, Gruber SB, Fearon ER, Hanash S (2001) Organ-specific molecular classification of primary lung, colon, and ovarian adenocarcinomas using gene expression profiles. Am J Pathol 159: 1231–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff BA, Kato D, Schmidt RA, Ek M, Ferry JA, Muntz HG, Cain JM, Tamimi HK, Figge DC, Greer BE (1994) Uterine papillary serous carcinoma: pattern of metastatic spread. Gynecol Oncol 54: 264–268 [DOI] [PubMed] [Google Scholar]
- Hough CD, Cho KR, Zonderman AB, Schwartz DR, Morin PJ (2001) Coordinately up-regulated genes in ovarian cancer. Cancer Res 61: 3869–3876 [PubMed] [Google Scholar]
- Ismail RS, Baldwin RL, Fang J, Browning D, Karlan BY, Gasson JC, Chang DD (2000) Differential gene expression between normal and tumour-derived ovarian epithelial cells. Cancer Res 60: 6744–6749 [PubMed] [Google Scholar]
- Janes PW, Lackmann M, Church WB, Sanderson GM, Sutherland RL, Daly RJ (1997) Structural determinants of the interaction between the erbB2 receptor and the Src homology 2 domain of Grb7. J Biol Chem 272: 8490–8497 [DOI] [PubMed] [Google Scholar]
- Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ (2003) Cancer Statistics 2003. CA Cancer J Clin 53: 5–26 [DOI] [PubMed] [Google Scholar]
- Kalil NG, McGuire WP (2002) Chemotherapy for advanced epithelial ovarian carcinoma. Best Pract Res Clin Obstet Gynaecol 16: 553–571 [DOI] [PubMed] [Google Scholar]
- Kalli KR, Falowo OI, Bale LK, Zschunke MA, Roche PC, Conover CA (2002) Functional insulin receptors on human epithelial ovarian carcinoma cells: implications for IGF-II mitogenic signaling. Endocrinology 143: 3259–3267 [DOI] [PubMed] [Google Scholar]
- Levenback C, Burke TW, Silva E, Morris M, Gershenson DM, Kavanagh JJ, Wharton JT (1992) Uterine papillary serous carcinoma (USPC) treated with cisplatin, doxorubicin, and cyclophosphamide (PAC). Gynecol Oncol 46: 317–321 [DOI] [PubMed] [Google Scholar]
- Nicklin JL, Copeland LJ (1996) Endometrial papillary serous carcinoma: pattern of spread and treatment. Clin Obstet Gynecol 39: 686–695 [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM (2002) The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 346: 1937–1947 [DOI] [PubMed] [Google Scholar]
- Santin AD, Hermonat PL, Ravaggi A, Bellone S, Pecorelli S, Cannon MJ, Parham GP (2000) In vitro induction of tumour-specific human lymphocyte antigen class I restricted CD8+ cytotoxic T lymphocytes by ovarian tumour antigen-pulsed autologous dendritic cells from patients with advanced ovarian cancer. Am J Obstet Gynecol 183: 601–609 [DOI] [PubMed] [Google Scholar]
- Santin AD, Bellone S, Ravaggi A, Roman JJ, Pecorelli S, Parham GP, Cannon MJ (2002a) Induction of tumour specific CD8+ cytotoxic T lymphocytes by tumour lysate-pulsed autologous dendritic cells in patients with serous papillary cancer. Brit J Cancer 86: 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin AD, Bellone S, Gokden M, Palmieri M, Dunn D, Agha J, Roman JJ, Hutchins L, Pecorelli S, O'Brien T, Cannon MJ, Parham GP (2002b) Overexpression of HER-2/neu in uterine serous papillary cancer. Clin Cancer Res 8: 1271–1279 [PubMed] [Google Scholar]
- Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, Misek DE, Wu R, Zhai Y, Darrah DM, Reed H, Ellenson LH, Giordano TJ, Fearon ER, Hanash SM, Cho KR (2002) Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res 62: 4722–4729 [PubMed] [Google Scholar]
- Sherman ME, Bitterman P, Rosenshein NB, Delgado G, Kurman RJ (1992) Uterine serous carcinoma. A morphologically diverse neoplasm with unifying clinicopathological features. Am J Surg Pathol 16: 600–610 [DOI] [PubMed] [Google Scholar]
- Slamon DL, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L (2001) Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 344: 783–792 [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumour subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfeld R, Van Den Berghe H, David G (1996) Stimulation of fibroblast growth factor receptor-1 occupancy and signaling by cell surface-associated syndecans and glypican. J Cell Biol 133: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM (2001) Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Aca Sci USA 98: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, Sanderson R, Yang Y, Wilson C, Zangari M, Anaissie E, Morris C, Muwalla F, van Rhee F, Fassas A, Crowley J, Tricot G, Barlogie B, Shaughnessy Jr J (2002) Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood 99: 1745–1757 [DOI] [PubMed] [Google Scholar]