Abstract
Dramatic clinical responses were observed in patient 888 following the adoptive transfer of autologous tumor-infiltrating lymphocytes (TIL). Previously, extensive analysis of the specificity of class I-restricted T cells from patient 888 TIL has revealed that these T cells recognize a mutated, as well as several nonmutated tumor Ags. Additional studies that were conducted on TIL from patient 888 indicated that they contained CD4-positive T cells that recognized the autologous tumor that had been induced to express HLA class II molecules. Tumor-reactive CD4-positive T cell clones were isolated from TIL and tested for their ability to react with Ags that are recognized by HLA class I-restricted, melanoma-reactive T cells. Using this approach, T cell clones were identified that recognized an epitope expressed in both the tyrosinase-related protein 1 and tyrosinase-related protein 2 Ags in the context of the HLA-DRβ1*1502 class II gene product. Additional clones were found to recognize an epitope of gp100 in the context of the same HLA-DR restriction element. These observations provide an impetus to develop strategies directed toward generating HLA class II-restricted tumor-reactive T cells.
In a high percentage of patients with melanoma, as well as in a lower frequency of patients bearing additional tumor histologies, such as squamous, renal, and ovarian carcinomas, in vitro stimulation of PBMC results in the generation of tumor-reactive, HLA class I-restricted T cells. In about one-third of melanoma patients, cultures of tumor-infiltrating lymphocytes (TIL)2 cultured with IL-2 alone have been found to contain tumor-reactive CTL (1). Tumor-reactive CD4-positive Th cells are naturally present in a significant percentage of melanoma-specific TIL, and in one study tumor-reactive CD4-positive T cells appeared to be present in four of the five TIL cultures that contained tumor-reactive class I-restricted CD8-positive T cells (2).
Analysis of melanoma-reactive CTL present in TIL, as well as T cells generated in mixed lymphocyte tumor cultures has indicated that responses directed against melanocyte differentiation Ags (MDA) gp100 and MART-1 are immunodominant in HLA-A2-positive melanoma patients (3). Melanoma-reactive T cells have also been shown to recognize epitopes of the MDAs tyrosinase, tyrosinase-related protein (TRP) 1, TRP-2, P. polypeptide, and OA-1 in the context of a variety of HLA class I gene products (4).
Recent studies have begun to explore the Ag specificity of HLA class II-restricted, melanoma-reactive T cells. A significant percentage of melanomas constitutively express cell surface HLA class II molecules, and treatment with IFN-γ can induce class II expression on the majority of melanomas that do not normally express significant levels of these gene products (5). In a number of additional tumor cell lines that do not normally express HLA class II molecules, such as ovarian and breast carcinomas, expression of these gene products can be up-regulated by IFN-γ treatment (5). Screening of a cDNA library with a T cell clone that was generated by stimulation with an autologous IFN-γ-treated melanoma cell line resulted in the cloning of a nonmutated Ag that appeared to be overexpressed in tumor cells (6). Mutated tumor Ags have also been cloned using tumor-reactive CD4-positive TIL lines (7–9).
This study was initiated to examine populations of TIL that are associated with in vivo antitumor responses for the presence of HLA class II-restricted, tumor-reactive T cells. Two CD4-positive T cell cultures that were isolated from a recurrent tumor recognized autologous and HLA-DR-matched tumor cell lines. When candidate MDAs were tested for their ability to stimulate these cultures, one recognized both TRP-1 and TRP-2 in the context of HLA-DRβ1*1502, and a minimal peptide epitope that was expressed on the closely related TRP-1 and TRP-2 Ags was identified. Additional screening assays demonstrated that another culture recognized gp100 in the context of HLA-DRβ1*1502, but a peptide epitope from this protein could not be identified, suggesting that this epitope might be modified in some as yet undetermined manner. These observations, in conjunction with previous studies, demonstrate that TIL 1290 contains CD8-positive as well as CD4-positive T cells that react with multiple tumor Ags, and provide a further impetus to characterize HLA class II-restricted tumor-reactive T cells that are present in populations of tumor-reactive T cells that are associated with in vivo tumor regression.
Materials and Methods
Cell lines
The analysis of HLA class II-restricted tumor-reactive T cell responses was conducted on melanoma patient 888, who demonstrated dramatic regression of multiple lesions following treatment with the autologous TIL 888. Three years later, a second TIL was established from a pelvic lesion, designated TIL 1290, and treatment with a mixture of TIL 888 and 1290 resulted in the regression of recurrent pelvic tumor masses.
The TIL 1290 cell line was generated by culturing fresh tumor in RPMI 1640 medium containing 10% human serum (BiochemMed, Winchester, VA) plus 120 IU/ml of IL-2 for 10 days. Cloning of T cells was conducted by initially depleting CD8+ T cells from the day 10 culture using anti-CD8-coated magnetic beads (Dynal, Lake Success, NY). The unbound cells were then plated at 2 and 10 cells/well in U-bottom microtiter trays in 0.1 ml in the presence of 3 × 105 1290 melanoma (mel) cells that had been irradiated with 120 Gy, 104 autologous EBV B that had been irradiated with 120 Gy, and 5 × 104 allogeneic PBMC that had been irradiated with 30 Gy, and 1 day later wells were fed with an equal volume of medium containing 240 IU/ml of IL-2. Two to 3 wk later, a cytokine release assay was conducted to identify wells containing T cells that recognized autologous tumor cells, but not autologous EBV B cells. Wells that contained tumor-reactive T cells were then further expanded using OKT3 stimulation in the presence of PBMC feeder cells, as previously described (10). For these expansions, between 5 × 104 and 2 × 105 T cells were incubated in 25 ml of RPMI 1640 medium containing 10% human serum, 5 × 106 autologous EBV B cells, and 2.5 × 107 allogeneic PBMC in the presence of 30 ng/ml OKT3. The following day, 120 IU/ml of IL-2 was added, and 5 days later the cells were washed and resuspended in fresh medium containing 120 IU/ml of IL-2. The plating efficiency of limiting dilution cultures plated at 2 cells/well was ~6%, and at 10 cells/well was ~30%; thus, under these conditions, the majority of the wells would be expected to contain clonal populations of cells. Melanoma and EBV B cell lines were maintained in RPMI 1640 medium containing 5% FBS.
Cytokine assays
Cytokine assays were conducted in AIM-V medium (Invitrogen, Carlsbad, CA.) containing 2% human serum either in the presence or absence of 120 IU/ml of IL-2 by incubating 1–2 × 104 T cells with 5 × 104 stimulator cells. The IL-2 cytokine release assays were conducted in the absence of added IL-2 and in the presence of an anti-human CD25 Ab (anti-Tac) that was kindly provided by T. Waldmann (National Institutes of Health, Bethesda, MD). After 18–24 h, supernatants were assayed for IFN-γ in an ELISA conducted with mAb pairs (Pierce/Endogen, Rockford, IL).
Transfection and DNA constructs
The episomal vector pEAK8 (EdgeBiosystems, Gaithersburg, MD) was digested with HindIII and NotI, and filled in with Klenow. A synthetic oligomer containing the restriction enzyme sites KpnI, NcoI, BstXI, EcoRI, AscI, EcoRV, HindIII, BstXI, XbaI, and NotI was then ligated into the pEAK8 vector, and the resulting plasmid was designated pEAK8.5. A construct encoding the HLA class II transactivator (CIITA) gene that was cloned in the pRC eukaryotic expression vector (11), kindly provided by G. Blanck (University of South Florida School of Medicine, Tampa, FL), was used for transfection studies. A retroviral construct encoding the CIITA gene was also generated by initially isolating the CIITA gene from the pSVK-FLAG-CIITA vector, kindly provided by J. Boss (Emory University School of Medicine, Atlanta, GA), by digestion with the EcoRI and XhoI restriction endonucleases. The pCLRCX retroviral vector was generated by digesting the pCLNCX vector (12) with the EcoRI and BamHI restriction endonucleases, which released the neomycin phosphotransferase gene, and filling in with Klenow to produce a blunt fragment. A cDNA construct encoding the human nerve growth factor receptor that lacked the cytoplasmic activating domain, kindly provided by R. Morgan (National Institutes of Health), was digested with NotI, filled in with Klenow, and then ligated to the pCLNCX vector. The resulting pCLRCX vector was then digested with HindIII, and a multiple cloning site containing the BglII, EcoRI, BamHI, NsiI, HindIII, NotI, and XhoI restriction endonuclease sites was inserted by ligating complementary oligonucleotides to the HindIII-digested pCLRCX vector to yield the pCLRCX4 vector.
To construct a bacterial expression vector encoding the human TRP-2 gene, a PCR was conducted with a plasmid encoding this gene using a pair of primers, TRP-2–5p (5′-CTGCCACATATGCAGGGTCAGTTCCCC CGA-3′), which contained an NdeI site, and TRP-2–3p (5′-AAGGGGCTC GAGTTACCTATCACAGACAGTTTCCCA-3′), which contained an XhoI site. After digestion with NdeI and XhoI and gel purification of the PCR product, the DNA fragment encoding TRP-2 aa residue 22–519 was ligated to pET-28+ prokaryotic expression vector (Novagen, Madison, WI) that had been digested with NdeI and XhoI. The resulting product contained an in-frame fusion of a polyhistidine peptide with the TRP-2 coding sequence. A similar strategy was also used to amplify the TRP-1 gene using the following primer pairs: TRP-1–5p (5′-GGCAGCCATATGCAATTCCCAAGACAGTGTG-3′), which contained an NdeI site, and TRP-1–3p (5′-AAGGGGGCTCGAGTTAGACCACAGACTGATTAG-3′), which contained an XhoI site, resulting in a product that encoded aa residues 25–537 of the hTRP-1 gene product. The plasmids were introduced into the Escherichia coli strain BL21(DE3), and bacteria were grown at 37°C to log phase, then induced for protein production by adding isopropyl β-D-thiogalactoside to a final concentration of 0.5 mM for 3 h at 37°C. Inclusion bodies of the bacterial extract were obtained, and TRP-1 and TRP-2 proteins were initially bound to a Ni2+ affinity column (Qiagen Sciences, Germantown, MD), and then further purified using a preparative SDS gel electropheresis column (Bio-Rad, Hercules, CA).
Lysis assay
A lysis assay was conducted by incubating the target cells with 51Cr for 1 h and plating 2.5 × 104 labeled cells in round-bottom microtiter plates with effector cells. The percentage of specific lysis was calculated using the formula:
Analysis of the expression of HLA-DR and BV region gene products
The expression of cell surface HLA class II molecules was determined by FACS analysis using the FITC-conjugated anti-HLA-DR Ab L243 (BD Biosciences, Franklin Lakes, NJ). Abs directed against the TCR β-chain V region (BV) families 8, 9, 11, 12, 14, 16, 18, 20, 22, and 23 that were labeled with PE, as well as Abs that were directed against BV 2, 3, and 17 that were labeled with FITC were obtained from Beckman Coulter (Brea, CA). Abs directed against BV 5, 6, 7, and 3 that were labeled with FITC were obtained from Pierce/Endogen. Abs that had been conjugated with either FITC or PE were incubated with 1–5 × 105 T cells for 30 min in PBS with 5% FBS and washed, and cell surface expression of these products was analyzed using a FACScan (BD Biosciences).
Peptides
Peptides from the TRP-1, TRP-2, and gp100 proteins were synthesized using a Gilson (Middleton, WI) AMS322 multiple peptide synthesizer, and all were estimated to be greater than 90% pure, as analyzed by mass spectrometry.
HLA typing
Determinations of the HLA haplotypes of cell lines were conducted at the National Institutes of Health HLA Laboratory by analyzing DNA from cell lines through the use of primers that were designed to amplify specific HLA alleles (13). The HLA haplotypes of the cell lines used in this study are indicated in Table I.
Table I.
Cell line HLA haplotypes
| HLA Alleles
|
||||||
|---|---|---|---|---|---|---|
| Cell Line | HLA-A | HLA-B | HLA-C | HLA-DRβ1 | HLA-DQ | HLA-DRβ3–5 |
| 888/1290 | 1,24 | 5201,55 | 11,202 | 15,021,601 | 05*,0601 | β5*0102 or β5*0203 |
| 1011 | 11,32 | 7,60 | 2,3 | 4,15 | 6,7 | β5*0101 |
| 697 | 2,11 | 7,62 | 3,0 | 4,011,501 | 6,8 | β5*0101 |
| 1102 | 2,24 | 55,62 | 3 | 4,011,502 | 3,010,602 | β4*0401,β5*0101 |
| 1558 | 2,31 | 56,0 | 1,0 | 1,011,101 | 3,010,501 | β3*0202 |
| 1300 | 2,24 | 7,39 | 7,0 | 01,15 | 5,6 | |
| 293 | 3 | 7 | 7 | 1501 | 6 | β5*01 |
Results
Identification of tumor-reactive CD4+ T cell clones
Initial studies conducted on TIL isolated from patient 888 resulted in the identification of four Ags that were recognized in the context of the HLA-A24 class I allele (14–17). These studies were then extended by examining the antitumor reactivity of CD4-positive T cells from these TIL. The CD4-positive T cells that were isolated from TIL 888 failed to secrete either IFN-γ or GM-CSF in response to IFN-γ-treated 1290 mel cells (data not shown); however, the only available TIL that were derived from the 888 tumor had been cultured in vitro for 30 days, and fresh uncultured 888 tumor was not available. In an attempt to identify tumor-reactive CD4-positive T cells that may be present in TIL 1290 tumor, but at the same time avoid the potential biases that may be introduced following long-term in vitro culture, uncultured cryopreserved 1290 tumor cells were incubated in vitro for 12 days in 120 IU/ml of IL-2. The CD8-positive T cells that were present in this population were then depleted using immunomagnetic beads, and this depleted population was then stimulated under limiting dilution conditions with 1290 mel cells that had been treated with IFN-γ to up-regulate HLA class II expression. The cultured 888 and 1290 mel cell lines did not constitutively express cell surface HLA class II gene products at significant levels; however, incubation with 500 U/ml of IFN-γ for 3 days resulted in the induction of HLA-DR gene expression on the 1290 mel cells, but only induced a relatively small increase in the expression of HLA-DR in 888 mel cells (Fig. 1).
FIGURE 1.
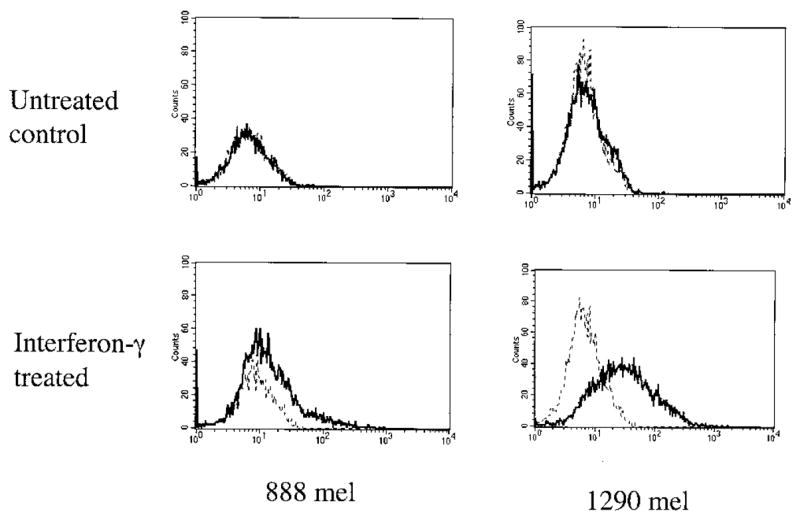
Induction of HLA class II expression on tumor cell lines by IFN-γ treatment. Tumor cells were either left untreated or treated with 1000 U of IFN-γ for 72 h, and the expression of cell surface HLA class II molecules was determined by carrying out FACS analysis using the FITC-conjugated anti-HLA-DR Ab L243. The profiles of untreated cells are represented by the broken line, and the profiles of IFN-γ-treated cells are represented by the solid line.
Twenty-eight putative CD4-positive cultures were expanded using OKT3 stimulation, and tested for their ability to recognize the autologous tumor cells. The T cells from 4 of the 28 cultures, designated C2, C5, C6, and C7, recognized IFN-γ-treated 1290 mel cells, but failed to recognize autologous 888 EBV B cells (Fig. 2). In addition, these cultures recognized 888 EBV B cells that had been pulsed with a tumor cell lysate. The C2 and C6 T cell lines recognized the autologous tumor following stimulation with IFN-γ, but did not respond to untreated tumor cells, whereas the C5 and C7 T cell lines recognized both the treated and untreated tumor cell line, and all of the T cell lines recognized autologous EBV B cells that had been pulsed with lysates of autologous tumor cells. Although the C5 culture recognized unstimulated 1290 mel, the CD8+ T cells that were present in this line may have recognized these tumor cells. The activity of the C5 T cell line did not appear to be stable upon continued culture, however, and this line was not further characterized.
FIGURE 2.

Multiple recognition patterns of autologous cells by CD4-positive T cell clones. The CD4-positive T cell lines (2 × 104 cells/well) were incubated with 105 1290 mel cells that were untreated or that had been treated for 72 h with 500 U/ml of IFN-γ, which are indicated by (+), and extensively washed before addition to the T cell clones. In addition, 2 × 105 EBV B cells were pulsed overnight with 2 × 105 1290 mel cells that were lysed by three successive freeze thaws before addition of T cells. Targets were incubated with 2 × 104 T cells overnight, and the release of IFN-γ was measured in an ELISA.
Analysis of the expression of CD4 and CD8 on the T cell lines indicated that C6 contained 90% CD4- and 4% CD8-positive T cells, C7 contained 98% CD4- and 1% CD8-positive cells, and C2 contained 99% CD4- and less than 1% CD8-positive cells. The T cell lines were then stained with a panel of mAbs directed against TCR-BV families to further evaluate the clonality of these cell lines. Approximately 94% of the cells in the C7 T cell line strongly expressed the BV1 TCR, but some or all of the cells that did not stain with this Ab could have been derived from the allogeneic PBMC feeder cells that were used to expand this culture. Over 99% of the T cells from the C2 T cell line appeared to express the BV17 TCR product, indicating that this represented a T cell clone, and additional experiments are now in progress to identify the Ag recognized by these T cells. The C6 T cell line was not strongly stained with any of the anti-TCR Abs that were tested; however, the available Abs do not detect all of the BV families, and Abs directed against several BV families do not detect all of the sequences within those families.
Assays were then conducted with C6 and C7 T cells to determine the cytokine profile of these T cells. Both the C6 and C7 T cells released GM-CSF in response to autologous tumor cell stimulation (data not shown). In addition, C6 and C7 T cells released 90 and 160 pg/ml of IL-2, respectively, in response to stimulation with the autologous HLA class II-positive tumor cells, whereas undetectable levels of IL-4 and IL-10 were detected in response to these target cells.
Stimulation of T cell clones by allogeneic tumor cell lines
In an attempt to determine whether or not the C6 and C7 T cells recognized shared melanoma Ags, a cytokine assay was conducted with a panel of allogeneic melanomas. Although the 1290, 697, and 1011 mel cell lines failed to constitutively express significant levels of cell surface HLA class II molecules, strong up-regulation of class II expression was observed on these cell lines following treatment with IFN-γ. The C7 T cell line recognized the autologous 1290 mel and the allogeneic 697 and 1011 mel cell lines that had not been treated with IFN-γ, although these tumor cell lines did not appear to express significant levels of cell surface HLA class II molecules (Fig. 3A). Treatment of the 1290 mel cells with IFN-γ resulted in a ~2-fold enhancement of the stimulation of C7 T cells, but did not appear to enhance the recognition of 697 and 1011 mel cells and actually appeared to somewhat diminish recognition of the 1011 mel cells. Stimulation of C7 T cells by the 1290 mel cells was weaker than that stimulated by the allogeneic 697 and 1011 mel cells in this experiment, but in additional experiments the levels of stimulation observed with the autologous and allogeneic mel lines were more comparable (data not shown). A cell line that constitutively expressed HLA class II molecules, 1558 mel, failed to stimulate cytokine release from C7 T cells. In addition, autologous fibroblast cells that demonstrated strong up-regulation of HLA class II expression following IFN-γ treatment and autologous EBV-transformed B cells that constitutively expressed high levels of cell surface HLA class II molecules were not recognized by C7 T cells.
FIGURE 3.

Reactivity of CD4-positive T cells with autologous and allogeneic cell lines indicates recognition of a shared melanoma Ag. A, The response of C7 T cells to autologous 1290 mel, 888 fibroblast, and 888 EBV B cell lines, as well as allogeneic 697, 1011, and 1102 mel cell lines was determined in a cytokine release assay. The melanoma and fibroblast cell lines were either untreated, or incubated with 500 U/ml of IFN-γ for 72 h, which are indicated by (+), and extensively washed before addition to the T cells. Stimulators were plated at 105 cells/well and incubated with 2 × 104 T cells overnight, and IFN-γ release was determined by ELISA. The percentage of cells positive for expression of cell surface HLA-DR was assessed by carrying out FACS analysis. The percentage of each cell population expressing HLA-DR was as follows: 1558 mel, 100; 1011 mel, 1; 1011 mel-IFN-γ, 99; 697 mel, 2; 697 mel-IFN-γ, 47; 1290 mel, 6; 1290 mel-IFN-γ, 81; 888 fibroblast, 0; and 888 fibroblast-IFN-γ, 96. B, The IFN-γ response of the C7-5 T cell clone to untreated and IFN-γ-treated 1290 mel cells was analyzed in an ELISA. C, The IFN-γ response of C6 T cell to the same panel of cell lines used in A was assessed in a cytokine release assay.
The lack of correlation between HLA class II expression and T cell recognition could be a reflection of the sensitivity of T cells to target cells expressing relatively low levels of these molecules. To directly demonstrate that a small percentage of contaminating CD8-positive, tumor-reactive T cells present in the C7 T cell line were not responding to the untreated tumor cell lines, this cell line was subcloned at one cell/well. A T cell clone designated C7-5 that contained less than 0.1% CD8-positive T cells by FACS analysis was then tested for its ability to recognize untreated and IFN-γ-treated 1290 mel cells. The results demonstrate that this clone recognized both the untreated and IFN-γ-treated 1290 mel cells at levels similar to the parental culture (Fig. 3B).
The results of a cytokine assay conducted using the same stimulator cells indicated that C6 strongly recognized the IFN-γ-treated 1290 and 1011 mel cell lines, but failed to recognize the untreated tumor cells (Fig. 3C). The C6 T cell line did not recognize the IFN-γ-treated 697 mel cell line nor the 1558 mel cell line that constitutively expressed HLA class II molecules, and failed to recognize autologous EBV B cells or the autologous IFN-γ-treated fibroblast cell line.
Identification of restriction element used for Ag recognition by T cell clones
Attempts were then made to identify the HLA restriction elements used for tumor recognition by the C6 and C7 T cells. Examination of the HLA haplotypes of the tumor cell lines used in these assays (Table I) demonstrated that the allogeneic 697 mel and 1011 mel cell lines expressed the HLA-DRβ1*1501 class II allele, which is identical with the DRβ1*1502 class II allele expressed by the autologous tumor cell line, with the exception of a single substitution of a valine for a glycine at position 86 of the mature protein. The 1558 mel cell line that did not share expression of any HLA class II gene products with the autologous tumor cell line was not recognized by C6 or C7 T cells. A melanoma cell line that constitutively expressed the HLA-DRβ1*1502 class II allele, 1102, was also recognized by C7, but not C6 T cells (data not shown).
These results indicated that the DRβ1*1501 or 1502 gene products may both represent restriction elements that can be used for Ag recognition by C6 and C7 T cells. The closely linked HLA-DQ6 allele that was shared between the autologous and the 697, 1011, and 1102 mel cell lines, however, could also represent the restriction element used by these T cells. To identify the restriction element used for tumor cell recognition by C6 and C7 T cells, target cells were preincubated with Abs directed against HLA gene products in an attempt to inhibit recognition of these cells (Table II). When IFN-γ-treated 1290 mel cells were preincubated with Ab that recognizes an epitope shared by all HLA-DR molecules, the responses of the C7 and C7-5 T cells to the target cells were reduced by 70– 80%, and the response of C6 T cells was reduced by 98%. Incubation of the target cells with an Ab directed against an epitope shared by all HLA class II molecules also inhibited the response of C6, C7, and C7-5 T cells to the autologous IFN-γ-treated tumor cells, although to a lesser degree than the inhibition seen with the anti-DR Ab. In contrast, incubation of target cells with an Ab directed against a pan-HLA class I epitope had no effect on the response of the C6, C7, or C7-5 T cells. The anti-HLA class I Ab was capable of blocking the response of a CD8-positive T cell clone, which was not blocked by the anti-HLA-DR or pan-class II Abs. These results indicate that C6 and C7 T cells recognize Ags in the context of two closely related restriction elements, HLA-DRβ*1501 and 1502, although the tightly linked HLA-DRβ5*01 or 02 gene products could also theoretically represent the restriction element used by these T cells.
Table II.
Ab blocking of T cell response
| T Cell Line
|
|||||
|---|---|---|---|---|---|
| Stimulator | Blocking Aba | C6 | C7 IFN-γ (pg/ml) | C7-5 | G209b |
| Tumor cellsc | None | 500 | 800 | 1600 | 838 |
| Tumor cells | Anti-DR | 9 (98)d | 240 (70) | 320 (80) | 924 (0) |
| Tumor cells | Anti-class II | 159 (68) | 480 (40) | 890 (44) | 711 (15) |
| Tumor cells | Anti-class I | 534 (0) | 990 (0) | 1800 (0) | <8 (>99) |
| None | None | <8 | <8 | <8 | <8 |
The 1290 mel tumor cell line (2 × 104 cells/well) that was treated with 500 U/ml of IFN-γ was incubated with either an Ab directed against HLA class I (W6/32), HLA class II (IVA12), or HLA-DR molecules (HB55) for 2 h prior to incubation with the indicated T cells (2 × 104 cells/well). Targets were incubated with T cells for 18–24 h, and the release of IFN-γ was measured in an ELISA.
G209 is a T cell clone that recognizes the Gp100:209–217 T cell epitope in the context of the HLA-A2 class I allele.
The C6, C7, and C7-5 cells were stimulated with 1290 mel cells that were treated for 72 h with 1000 U/ml of IFN-γ. The G209 T cells were stimulated with the HLA-A2-expressing 1300 mel cell line.
The numbers in parentheses indicate the percent inhibition of the cytokine release observed in the presence of the indicated Abs.
Identification of Ags recognized by C6 and C7 T cells
In an attempt to establish whether or not these T cells recognized previously identified melanoma Ags, the highly transfectable transformed monkey kidney cell line COS was transiently transfected with constructs that encoded MART-1, tyrosinase, gp100, TRP-1, TRP-2, or ESO-1. Autologous EBV B cells that had been pulsed with lysates of the transfected COS cells or autologous tumor cell lysates were then tested for their ability to stimulate cytokine release from the T cell clones. The C6 and C7 T cells responded to autologous EBV B cells that had been pulsed with an autologous tumor cell lysate, and the C7 responded weakly to cells pulsed with lysates derived from COS cells that had been transfected with the TRP-1 and TRP-2 genes (Fig. 4A). The ability of C7 T cells to respond to autologous APCs that were pulsed with rTRP-1 and rTRP-2 proteins was then evaluated. The results indicated that target cells that had been pulsed with either rTRP-1 or rTRP-2 protein stimulated significant IFN-γ release from the C7 T cells (Fig. 4B).
FIGURE 4.

C7 T cells respond to targets pulsed with lysates of TRP-1- and TRP-2-transfected cells and pulsed with rTRP-1 and rTRP-2 proteins. A, The C6 and C7 T cell lines were stimulated with either autologous 1290 mel tumor cells that were treated with 500 U/ml of IFN-γ for 72 h or autologous EBV B cells (2 × 105 cells/well) that were incubated overnight with lysates of COS cells. The COS cells were transfected for 72 h with plasmids encoding the indicated tumor Ags or a control green fluorescent protein construct, and lysates of the transfected cells prepared by three rapid freeze-thaw cycles were pulsed overnight at an equivalent concentration of 2 × 105 tumor cells/well on the autologous EBV B cells. The autologous 888 EBV B cells were pulsed overnight with the rTRP-1 or rTRP-2 proteins at the indicated concentrations. The T cells were incubated with target cells overnight, and the release of IFN-γ was measured in an ELISA.
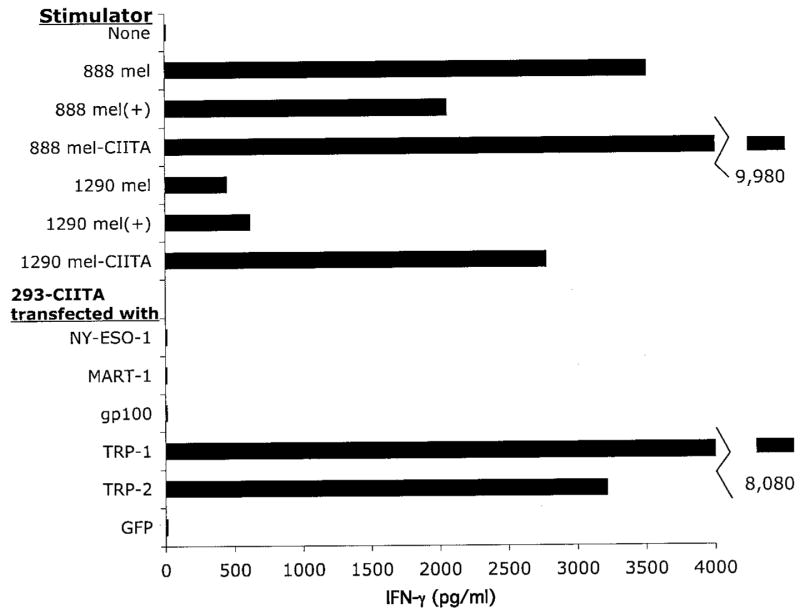
Further analysis of the specificity of T cells was conducted using the highly transfectable 293 cell line, which was genotyped as HLA-DRβ1*1501. The 293 cell line did not appear to constitutively express cell surface HLA class II molecules; however, transfection of the 293 cells with a vector encoding CIITA, a protein that has been shown to play a critical role in regulating expression of many genes that are regulated by IFN-γ (18), resulted in strong up-regulation of HLA class II gene expression. The stably tranfected 293-CIITA cell line was then used to carry out transient transfections of genes encoding potential target Ags and used to stimulate the CD4-positive T cell lines. The results demonstrated that C7 T cells recognized 293-CIITA cells that were transfected with constructs encoding either TRP-1 or TRP-2 (Fig. 5). The autologous 888 and 1290 mel cell lines that were transduced with the CIITA gene expressed high levels of cell surface HLA class II gene product, and were also recognized by C7 T cells. These results provide further evidence that C7 T cells recognize epitopes expressed on TRP-1 and TRP-2 in the context of the HLA-DRβ1*1501 and 1502 class II restriction elements.
FIGURE 5.
C7 T cells specifically recognize TRP-1- and TRP-2-transfected target cells. The 293-CIITA cells (105 cells/well) were transfected with constructs encoding the indicated genes overnight, and the following day C7 T cells (4 × 104 cells/well) were indicated with transfected targets or autologous tumor cells that were either untreated, treated with IFN-γ (500 U/ml) for 72 h, indicated by (+), or transduced with a construct encoding the CIITA gene. The T cells were incubated with target cells overnight, and the release of IFN-γ was measured in ELISA.
Studies were then conducted to identify the Ag recognized by C6 T cells. This cell line, which had previously been found to contain a significant percentage of CD8-positive T cells, was depleted of T cells bearing this marker through the use of anti-CD8-coated beads, resulting in a population of T cells that contained less than 1% CD8-positive T cells. The C6 T cells were then tested for their ability to recognize 293-CIITA cells that were transfected with constructs encoding previously described melanoma Ags (Fig. 6). The results indicated that C6 T cells recognized target cells that were transfected with a gene encoding the gp100 Ag.
FIGURE 6.

C6 T cells specifically recognize gp100-transfected target cells. The 293-CIITA cells (105 cells/well) were transfected with constructs encoding the indicated genes overnight, and the following day C7 T cells (4 × 104 cells/well) were indicated with transfected targets or autologous tumor cells that were either untreated, treated with IFN-γ (500 U/ml) for 72 h, indicated by (+), or transduced with a construct encoding the CIITA gene. The T cells were incubated with target cells overnight, and the release of IFN-γ was measured in ELISA.
Identification of additional CD4+ T cells reactive with TRP-1, TRP-2, and gp100
A number of CD4-positive T cell clones were then generated from a TIL culture, designated TIL 1541, that was grown from a small recurrent s.c. lesion that was resected from patient 888 3 years following the establishment of TIL 1290. These T cell clones were then examined for their ability to recognize autologous HLA class II-positive tumor cells, as well as TRP-1, TRP-2, or gp100 in an HLA class II-restricted manner. In a preliminary screening assay, 11 of 17 clones recognized 293-CIITA cells that were transfected with TRP-1, and one clone appeared to recognize gp100-transfected cells. The eight clones that were successfully expanded were then tested for their ability to recognize TRP-1, TRP-2, and gp100. Four of the eight clones that could be expanded maintained strong reactivity with 293-CIITA cells that were transfected with cDNAs that encoded either TRP-1 or TRP-2 (Fig. 7A). A CD4-positive T cell clone that recognized 293-CIITA cells that were transfected with a cDNA encoding gp100 was also identified from this TIL (Fig. 7B).
FIGURE 7.
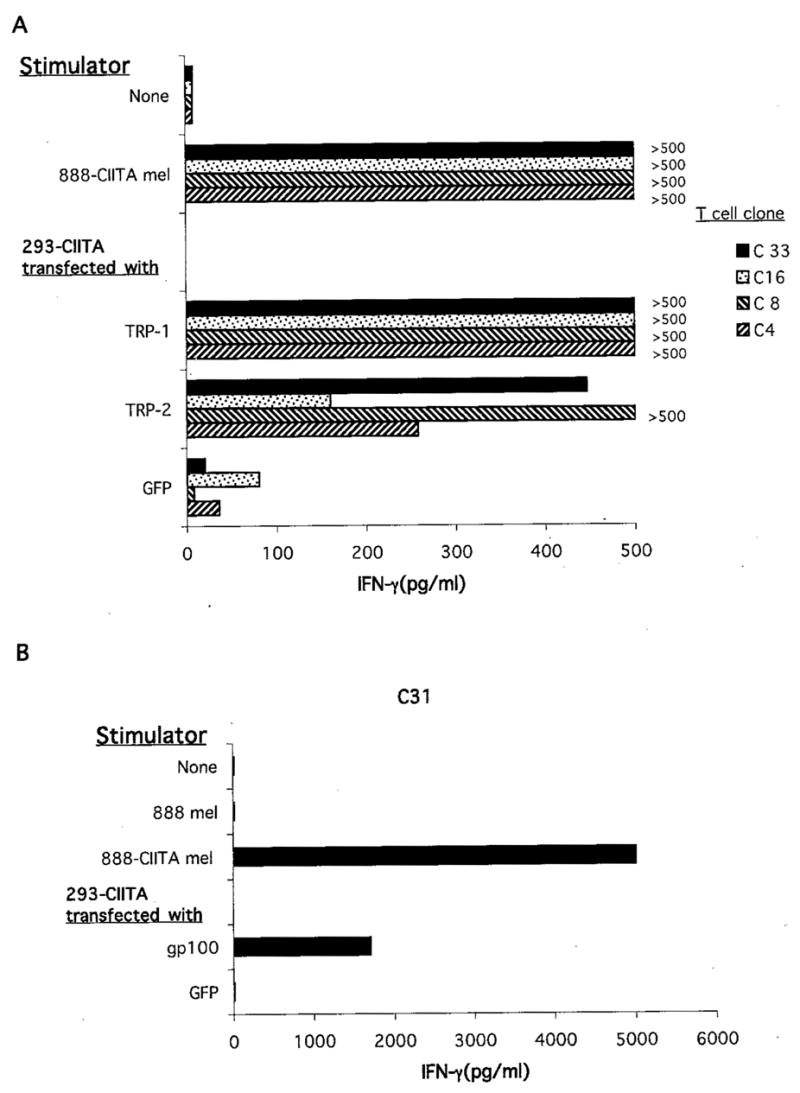
CD4-positive T cell clones derived from TIL 1541 recognize TRP-1, TRP-2, and gp100. The 293-CIITA cells (105 cells/well) were transfected with constructs encoding the indicated gene products overnight, and the following day the T cell clones (2 × 104 cells/well) were incubated with transfected target cells or the autologous 888-CIITA tumor cell line. The T cells were incubated with target cells overnight, and the release of IFN-γ was measured in ELISA.
Identification of HLA class II-restricted T cell epitopes
A set of 20-mer peptides, covering the entire TRP-2 sequence and overlapping by 10 aa, was then synthesized and used to pulse 888 EBV B cells. Target cells that had been pulsed with two of the TRP-2 peptides, corresponding to aa 231–250 (TRP-2231–250) and 241–260 (TRP-2241–260), stimulated the release of 330 and 440 pg/ml of IFN-γ from clone 7 T cells, respectively. Only background levels of IFN-γ (<10 pg/ml) were released from C7 T cells that were stimulated with target cells that had been incubated individually with the other peptides in this panel. A 10-mer peptide corresponding to aa 241–250 of the TRP-2 protein was shared between the two peptides that were positive in the initial cytokine assay, indicating that this might represent a minimal T cell epitope. The TRP-2241–250 peptide AFLPWHRYL, as well as the peptide from the corresponding region of TRP-1, TRP-1245–254, were then pulsed on autologous EBV B cells and tested for their ability to stimulate C7 T cells (Table III). Target cells that were incubated with the 10-mers TRP-2251–260 and TRP-1245–254 stimulated significant levels of cytokine release from C7 and C8 T cells, although enhanced recognition by C7 and C8 T cells was observed with some of the peptides containing extensions at the amino and carboxyl termini of the 10-mer peptide. Variants that lacked a single amino acid at either the amino or carboxyl terminus of the TRP-2251–260 were similar to the 10-mer peptide in their ability to stimulate cytokine release from C7 T cells, whereas peptides that lacked additional amino- or carboxyl-terminal amino acids failed to stimulate significantly cytokine release from C7 T cells (data not shown). Thus, while aa 242–249 of the TRP-2 molecule comprise the minimal T cell epitope recognized by C7 T cells, the additional amino acids at the amino and carboxyl terminus may either stabilize the interaction with the DR15 MHC class II gene product or interfere with degradation of the peptide epitope.
Table III.
Dose response of C7 and C8 T cells to peptides from TRP-1 and TRP-2
| Peptide
(μg/ml) |
IFN-γ (pg/ml)
|
||||||
|---|---|---|---|---|---|---|---|
| QB1a | GC1 | A1 | B1 | E1 | E4 | F4 | |
| C7 T Cell Response | |||||||
| 50 | 1652 | 1272 | 2628 | 1984 | 480 | 684 | 336 |
| 12 | 308 | 464 | 496 | 508 | 316 | 228 | 192 |
| 3.1 | 236 | 324 | 240 | 324 | 264 | 156 | 56 |
| 0.8 | 64 | 160 | 100 | 88 | 108 | 8 | 8 |
| 0.2 | 8 | 116 | 48 | 8 | 60 | 8 | 8 |
| C8 T Cell Response | |||||||
| 50 | 888 | 680 | 1840 | 5540 | 785 | 940 | 610 |
| 12 | 610 | 285 | 826 | 932 | 353 | 330 | 45 |
| 3.1 | 46 | 32 | 62 | 81 | 48 | 31 | <8 |
| 0.8 | <8 | <8 | <8 | 31 | 12 | 14 | <8 |
| 0.2 | <8 | <8 | <8 | <8 | <8 | <8 | <8 |
Autologous EBV B cells (2 × 105) were incubated with peptides for 2 h, followed by incubation with 2 × 104 C6 or C7 T cells, and IFN-γ release was determined 18–24 h later. Target cells were incubated with the following peptides, with the amino acid positions indicated in parentheses, from either TRP-2, QB1-ALPYWNFATGRRNECDVCTDQ (241–260), GC1-LQR LIGNESFALPYWNFATG (231–250), A1-ALPYWNFATGRNECDV (241–256), E1-NESFALPWNFATG (237–250), F4-AL PYWNFATG (241–250), or TPR-1, B1-SLPYWNFATGKNVCDI (245–254) and E4-SLPYWNFATG (245–254).
An attempt was then made to identify the peptide epitope from gp100 that was recognized by C6 T cells. A total of 68 peptides that were either 17 or 21 aa in length and that overlapped by 10 aa were then synthesized and pulsed on autologous EBV B cells at a concentration of 50 μg/ml. None of the target cells that were incubated individually with these peptides, however, induced significant cytokine release from C6 T cells, indicating that these T cells may recognize a peptide that contains an as yet unidentified modification.
Lysis of tumor cells by CD4-positive T cells
Autologous tumor cell targets that had been induced to express HLA class II gene products were then examined for their ability to be lysed by C6 and C7 T cells. The targets included 1290 mel that had been treated with IFN-γ, as well as 1290 mel cells that had been transfected with the CIITA gene. Significant lysis of the CIITA-treated 888 and 1290 mel was observed with both the C6 (Fig. 8A) and C7 (Fig. 8B) T cells, and a low level of lysis of the IFN-γ-treated 1290 mel cells that reached a maximum of 9% was observed with the C6 T cells. Less than 5% lysis of the autologous EBV B cells was observed with either the C6 or C7 T cells, and lysis of the tumor cell lines appeared to be correlated with HLA class II expression, indicating that these results reflect specific tumor Ag recognition by the T cell clones.
FIGURE 8.

Lysis of tumor cell lines by CD4-positive T cell clones. The C6 (A) and C7 (B) T cells were incubated with the indicated target cells in a 4-h chromium release assay. Target cells were either untreated 888 and 1290 mel cells, 888 and 1290 mel cells that were stably transduced with CIITA, or 1290 mel cells that were treated with 1000 U/ml of IFN-γ for 3 days to up-regulate HLA class II expression, designated as 1290 mel (+).
Discussion
A significant percentage of melanoma-reactive TIL contain both tumor-reactive CD8-positive and CD4-positive T cells. These observations led to attempts to identify tumor-reactive CD4-positive cells in populations of TIL that are associated with in vivo anti-tumor responses. In previous studies, T cells generated in a mixed lymphocyte tumor culture appeared to recognize a shared Ag in the context of the HLA-DR15 restriction element, although the Ag was not identified (19). Human CD4+ HLA class II-restricted, tumor-reactive T cells that recognize multiple epitopes from the gp100 (20–22) and NY-ESO-1 Ags (23–25) have also been identified. Two of the genes encoding Ags recognized by tumor-reactive CD4-positive T cells from melanoma patients 1363 and 1359 were identified using 293 cells that were stably transfected with genes that encode HLA class II gene products as well as genes involved with class II Ag processing (8, 9). Another tumor Ag has recently been identified (6) by screening a tumor cell cDNA library in 293 cells that were cotransfected with the gene that encoded the HLA-DRβ1*1101 molecule and a construct encoding CIITA.
For these studies, CD4-positive T cell clones were isolated from patient 888, who demonstrated dramatic responses to adoptive immunotherapy with autologous TIL. Adoptive transfer of TIL 888 along with TIL 1290 resulted in the regression of pelvic lesions that had recurred following the initial regression of multiple lesions that were observed following treatment with TIL 888 alone. Although tumor-reactive CD8-positive T cells present in these TIL may alone have been responsible for in vivo tumor regression, tumor-reactive, CD4-positive T cells present in TIL 1290 may also have contributed to this response.
To characterize the response of tumor-reactive CD4-positive T cells from TIL 1290, limiting dilution cultures were generated from this TIL. Screening of candidate Ags revealed that multiple tumor-reactive CD4-positive T cell cultures from patient 888 recognized a shared epitope expressed on the TRP-1 and TRP-2 melanocyte differentiation Ags in the context of HLA-DRβ1*1501 and 1502. The 10-mer peptide that appeared to represent a minimal epitope from TRP-1 and TRP-2 only differed at the second position, in which the TRP-1 peptide contained a serine residue and the TRP-2 peptide contained an alanine residue. The TRP-1 and TRP-2 proteins are ~50% identical at the amino acid level, suggesting that additional epitopes that are shared between these proteins may exist. Additional CD4-positive T cell clones from patient 888 TIL recognized an epitope expressed on the gp100 melanocyte differentiation Ag in the context of HLA-DRβ1*1501 and 1502.
One unexpected observation made in this study was that C7 T cells that recognized TRP-1 and TRP-2 recognized untreated 888 and 1290 mel cells that did not appear to express significant levels of cell surface HLA-DRβ1*1502 class II molecules. At the same time, CD4-positive gp100-reactive T cells from the same patient did not appear to recognize the untreated tumor cells. There are several possible explanations for these findings. First, the relatively low levels of HLA class II gene expression that are needed to present the TRP-1 and TRP-2 epitopes to CD4-positive T cells may not have been detected by flow cytometry. Another possibility is that a low level of stimulation by the tumor cells may result in the induction of IFN-γ, which then results in the subsequent up-regulation of HLA class II expression on the tumor cell surface, further amplifying T cell recognition. Previous studies have also demonstrated that the intact TRP-1 molecule is expressed at low levels on the surface of melanoma cell lines, and is detected in medium harvested from tumor cell lines, suggesting that this may be shed or secreted by these cells (26). A soluble fusion product of the human low density lipoprotein receptor appears to be directly presented by activated human CD4-positive T cells, which express high levels of cell surface HLA class II molecules (9). Therefore, C7 T cells could take up TRP-1 protein that is released from tumor cells, process, and present this Ag to other T cells in the culture. The failure of the gp100-reactive CD4-positive C6 T cells to recognize the unstimulated 888 or 1290 mel cells indicates that the mechanism that is responsible for mediating recognition of these tumor cells by C7 T cells may not play a role in the recognition of additional Ags, such as gp100 by CD4-positive T cells.
Previous studies of HLA class II-restricted responses to foreign Ags as well as tumor Ags have indicated that class II-restricted Ags can be processed and presented through an endosomal pathway. In one study, CD4-positive T cells isolated from a population of melanoma-reactive TIL were initially found to recognize tumor cell lysates that were pulsed on EBV-transformed B cells. These T cells were screened by examining their ability to secrete cytokines in response to APC that were pulsed with lysates of target cells that were transfected with genes encoding Ags recognized by HLA class I-restricted tumor-reactive T cells. Examination of the ability of the CD4-positive T cells to recognize the pulsed target cells indicated that they recognize the MDA tyrosinase in the context of HLA-DRβ1*0401 (27).
The observation that the TRP-1, TRP-2 Ags can be processed through an exogenous pathway (Figs. 2 and 4) suggests that uptake of tumor cells or Ag that has been released from tumor cells by professional APCs such as dendritic cells may be involved with Ag recognition in vivo. These Ags also, however, appear to be efficiently processed through an endogenous pathway (Figs. 5 and 6), and a number of HLA class II-restricted epitopes derived from these melanosomal Ags that appear to be processed through an endogenous pathway have now been identified (21, 22, 28). Additional studies have provided evidence that a melanosomal targeting sequence present near the carboxyl terminus of TRP-1 (29) as well as other MDAs (30) can target this protein to an endosomal compartment.
Tumor-reactive, CD4-positive T cells have been shown to mediate Ag-specific lysis of target cells, and several examples of tumor-reactive CD4-positive T cells that are either directly lytic for sarcoma cells (31) or melanoma cells (2, 19, 32, 33) have been described. In two of these reports, T cells appeared to recognized unknown melanoma Ags in the context of HLA-DR15 (32) (19). The results presented by Zennadi et al. indicate that there was a discordance between results of the lysis and cytokine release assays, in that target cells that expressed low levels of HLA-DR15 stimulated cytokine release from CD4-positive T cells, but were not lysed by these cells. Similarly, untreated 888 and 1290 mel cells that were not lysed by the C6 and C7 T cells nevertheless stimulated cytokine release from these T cells. These results imply that the levels of MHC-peptide complexes on the surface of target cells that are required for the stimulation of cytokine release may in some cases be lower than those required to mediate the lysis of the same target cells by Ag-reactive CD4-positive T cells.
Although target cell lysis has generally been associated with CD8-positive T cells, several examples of lytic CD4-positive T cells have been reported. It has been reported that CD4-positive T cells that predominantly produce IFN-γ and IL-2 in response to specific stimulation, termed Th1 cells, are generally lytic, whereas only a proportion of Th2 cells, which produce IL-4, but not IFN-γ or IL-2, is capable of lysing target cells (34). For some CD4-positive T cells, the perforin/granzyme pathway may be predominantly responsible for target cell lysis (35), whereas Fas/Fas ligand interactions appear to play a primary role in the target cell lysis mediated by at least some Th2 cells (32). Additional studies of human CD4-positive T cell clones should help to determine whether or not Th cell phenotypes are associated with the mechanisms used by these cells for lysis.
The identification of a large number of Ags recognized by melanoma-reactive T cells has provided the opportunity to explore the issue of which of these play an important role in mediating tumor regression. Studies conducted in mouse model systems have provided direct evidence that tumor regression can result from immune responses directed against TRP-1 or TRP-2. Immunization of mice with human or murine rTRP-1 protein resulted in the induction of autoantibodies and resulted in tumor protection in C57BL/6 mice against the growth of the murine TRP-1 expressing melanoma B16 (36). Immunization with a recombinant vaccinia virus expressing murine TRP-1 also provided protection against the growth of the B16 murine melanoma in C57BL/6 mice that was dependent on CD4-, but not CD8-positive T cells (37). Direct evidence for the role of Abs in tumor protection was demonstrated by the use of a mAb directed against the murine TRP-1 protein, which resulted in tumor protection (38). Additional data indicate that adoptive transfer of CD8-positive T cells directed against an immunodominant HLA class I peptide from TRP-2 can mediate regression of the murine B16 melanoma (39).
The correlation that was noted between development of CD8-positive T cell responses directed against gp100 in TIL and clinical responses to adoptive immunotherapy (40) provided evidence that T cells directed against this Ag may play an important role in tumor regression. Immunization of mice with naked DNA encoding the human gp100 protein (41) as well as a recombinant vaccinia virus encoding human gp100 (42) has been shown to elicit CD8-positive T cells that can protect mice from growth of the murine B16 melanoma. A number of class II-restricted T cell epitopes have now also been identified for gp100 (21, 22, 28, 43). The identification of HLA class II-restricted T cell epitopes of gp100, as well as the epitopes from TRP-1 and TRP-2 described in this study, should allow a determination of whether or not immunization with HLA class II-restricted peptides derived from tumor Ags can further enhance in vivo antitumor responses.
Acknowledgments
We thank Dr. Maria Parkhurst and John Riley for their help with production of synthetic peptides.
Footnotes
Abbreviations used in this paper: TIL, tumor-infiltrating lymphocyte; BV, β-chain V region; CIITA, HLA class II transactivator; MDA, melanocyte differentiation Ag; mel, melanoma; TRP, tyrosinase-related protein.
References
- 1.Schwartzentruber DJ, Hom SS, Dadmarz R, White DE, Yannelli JR, Steinberg SM, Rosenberg SA, Topalian SL. In vitro predictors of therapeutic response in melanoma patients receiving tumor-infiltrating lymphocytes and interleukin-2. J Clin Oncol. 1994;12:1475. doi: 10.1200/JCO.1994.12.7.1475. [DOI] [PubMed] [Google Scholar]
- 2.Markus NR, Rosenberg SA, Topalian SL. Analysis of cytokine secretion by melanoma-specific CD4+ T lymphocytes. J Interferon Cytokine Res. 1995;15:739. doi: 10.1089/jir.1995.15.739. [DOI] [PubMed] [Google Scholar]
- 3.Kawakami Y, Dang N, Wang X, Tupesis J, Robbins PF, Wang RF, Wunderlich JR, Yannelli JR, Rosenberg SA. Recognition of shared melanoma antigens in association with major HLA-A alleles by tumor infiltrating T lymphocytes from 123 patients with melanoma. J Immunother. 2000;23:17. doi: 10.1097/00002371-200001000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Renkvist N, Castelli C, Robbins PF, Parmiani G. A listing of human tumor antigens recognized by T cells. Cancer Immunol Immunother. 2001;50:3. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghton AN, Thomson TM, Gross D, Oettgen HF, Old LJ. Surface antigens of melanoma and melanocytes: specificity of induction of Ia antigens by human γ-interferon. J Exp Med. 1984;160:255. doi: 10.1084/jem.160.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiari R, Hames G, Stroobant V, Texier C, Maillere B, Boon T, Coulie PG. Identification of a tumor-specific shared antigen derived from an Eph receptor and presented to CD4 T cells on HLA class II molecules. Cancer Res. 2000;60:4855. [PubMed] [Google Scholar]
- 7.Pieper R, Christian RE, Gonzales MI, Nishimura MI, Gupta G, Settlage RE, Shabanowitz J, Rosenberg SA, Hunt DF, Topalian SL. Biochemical identification of a mutated human melanoma antigen recognized by CD4+ T cells. J Exp Med. 1999;189:757. doi: 10.1084/jem.189.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang RF, Wang X, Atwood AC, Topalian SL, Rosenberg SA. Cloning genes encoding MHC class II-restricted antigens: mutated CDC27 as a tumor antigen. Science. 1999;284:1351. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 9.Wang RF, Wang X, Rosenberg SA. Identification of a novel major histocompatibility complex class II-restricted tumor antigen resulting from a chromosomal rearrangement recognized by CD4+ T cells. J Exp Med. 1999;189:1659. doi: 10.1084/jem.189.10.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter EA, Greenberg PD, Gilbert MJ, Finch RJ, Watanabe KS, Thomas ED, Riddell SR. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995;333:1038. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 11.Lu Y, Boss JM, Hu SX, Xu HJ, Blanck G. Apoptosis-independent retinoblastoma protein rescue of HLA class II messenger RNA IFN-γ inducibility in non-small cell lung carcinoma cells: lack of surface class II expression associated with a specific defect in HLA-DRA induction. J Immunol. 1996;156:2495. [PubMed] [Google Scholar]
- 12.Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Player MA, Barracchini KC, Simonis TB, Rivoltini L, Arienti F, Castelli C, Mazzocchi A, Belli F, Parmiani G, Marincola FM. Differences in frequency distribution of HLA-A2 subtypes between North American and Italian white melanoma patients: relevance for epitope specific vaccination protocols. J Immunother Emphasis Tumor Immunol. 1996;19:357. doi: 10.1097/00002371-199609000-00005. [DOI] [PubMed] [Google Scholar]
- 14.Robbins PF, El-Gamil M, Kawakami Y, Stevens E, Yannelli J, Rosenberg SA. Recognition of tyrosinase by tumor infiltrating lymphocytes from a patient responding to immunotherapy. Cancer Res. 1994;54:3124. [PubMed] [Google Scholar]
- 15.Robbins PF, El-Gamil M, Li YF, Topalian SL, Rivoltini L, Sakaguchi K, Appella E, Kawakami Y, Rosenberg SA. Cloning of a new gene encoding an antigen recognized by melanoma-specific HLA-A24 restricted tumor-infiltrating lymphocytes. J Immunol. 1995;154:5944. [PubMed] [Google Scholar]
- 16.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, Rosenberg SA. A mutated β-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins PF, El-Gamil M, Li YF, Fitzgerald EB, Kawakami Y, Rosenberg SA. The intronic region of an incompletely spliced gp100 gene transcript encodes an epitope recognized by melanoma-reactive tumor-infiltrating lymphocytes. J Immunol. 1997;159:303. [PubMed] [Google Scholar]
- 18.Chang CH, Flavell RA. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J Exp Med. 1995;181:765. doi: 10.1084/jem.181.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi T, Chapman PB, Yang SY, Hara I, Vijayasaradhi S, Houghton AN. Reactivity of autologous CD4+ T lymphocytes against human melanoma: evidence for a shared melanoma antigen presented by HLA-DR15. J Immunol. 1995;154:772. [PubMed] [Google Scholar]
- 20.Li K, Adibzadeh M, Halder T, Kalbacher H, Heinzel S, Muller C, Zeuthen J, Pawelec G. Tumor-specific MHC-class-II-restricted responses after in vitro sensitization to synthetic peptides corresponding to gp100 and Annexin II eluted from melanoma cells. Cancer Immunol Immunother. 1998;47:32. doi: 10.1007/s002620050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Touloukian CE, Leitner WW, Topalian SL, Li YF, Robbins PF, Rosenberg SA, Restifo NP. Identification of a MHC class II-restricted human gp100 epitope using DR4-IE transgenic mice. J Immunol. 2000;164:3535. doi: 10.4049/jimmunol.164.7.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapointe R, Royal RE, Reeves ME, Altomare I, Robbins PF, Hwu P. Retrovirally transduced human dendritic cells can generate T cells recognizing multiple MCH class I and class II epitopes from the melanoma antigen glycoprotein 100. J Immunol. 2001;167:4758. doi: 10.4049/jimmunol.167.8.4758. [DOI] [PubMed] [Google Scholar]
- 23.Zeng G, Touloukian CE, Wang X, Restifo NP, Rosenberg SA, Wang RF. Identification of CD4+ T cell epitopes from NY-ESO-1 presented by HLA-DR molecules. J Immunol. 2000;165:1153. doi: 10.4049/jimmunol.165.2.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jager E, Jager D, Karbach J, Chen YT, Ritter G, Nagata Y, Gnjatic S, Stockert E, Arand M, Old LJ, Knuth A. Identification of NY-ESO-1 epitopes presented by human histocompatibility antigen (HLA)-DRB4*0101–0103 and recognized by CD4+ T lymphocytes of patients with NY-ESO-1-expressing melanoma. J Exp Med. 2000;191:625. doi: 10.1084/jem.191.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarour HM, Storkus WJ, Brusic V, Williams E, Kirkwood JM. NY-ESO-1 encodes DRB1*0401-restricted epitopes recognized by melanoma-reactive CD4+ T cells. Cancer Res. 2000;60:4946. [PubMed] [Google Scholar]
- 26.Xu Y, Setaluri V, Takechi Y, Houghton AN. Sorting and secretion of a melanosome membrane protein, gp75/TRP1. J Invest Dermatol. 1997;109:788. doi: 10.1111/1523-1747.ep12340971. [DOI] [PubMed] [Google Scholar]
- 27.Topalian SL, Gonzales MI, Parkhurst M, Li YF, Southwood S, Sette A, Rosenberg SA, Robbins PF. Melanoma-specific CD4+ T cells recognize nonmutated HLA-DR-restricted tyrosinase epitopes. J Exp Med. 1996;183:1965. doi: 10.1084/jem.183.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi H, Lu J, Celis E. Identification of helper T-cell epitopes that encompass or lie proximal to cytotoxic T-cell epitopes in the gp100 melanoma tumor antigen. Cancer Res. 2001;61:7577. [PubMed] [Google Scholar]
- 29.Wang S, Bartido S, Yang G, Qin J, Moroi Y, Panageas KS, Lewis JJ, Houghton AN. A role for a melanosome transport signal in accessing the MHC class II presentation pathway and in eliciting CD4+ T cell responses. J Immunol. 1999;163:5820. [PubMed] [Google Scholar]
- 30.Calvo PA, Frank DW, Bieler BM, Berson JF, Marks MS. A cytoplasmic sequence in human tyrosinase defines a second class of di-leucine-based sorting signals for late endosomal and lysosomal delivery. J Biol Chem. 1999;274:12780. doi: 10.1074/jbc.274.18.12780. [DOI] [PubMed] [Google Scholar]
- 31.Heike M, Schlaak J, Schulze-Bergkamen H, Heyl S, Herr W, Schmitt U, Schneider PM, Meyer zum Buschenfelde KH. Specificities and functions of CD4+ HLA class II-restricted T cell clones against a human sarcoma: evidence for several recognized antigens. J Immunol. 1996;156:2205. [PubMed] [Google Scholar]
- 32.Zennadi R, Abdel-Wahab Z, Seigler HF, Darrow TL. Generation of melanoma-specific, cytotoxic CD4+ T helper 2 cells: requirement of both HLA-DR15 and Fas antigens on melanomas for their lysis by Th2 cells. Cell Immunol. 2001;210:96. doi: 10.1006/cimm.2001.1809. [DOI] [PubMed] [Google Scholar]
- 33.Zarour HM, Kirkwood JM, Kierstead LS, Herr W, Brusic V, Slingluff CL, Jr, Sidney J, Sette A, Storkus WJ. Melan-A/MART-1(51–73) represents an immunogenic HLA-DR4-restricted epitope recognized by melanoma-reactive CD4+ T cells. Proc Natl Acad Sci USA. 2000;97:400. doi: 10.1073/pnas.97.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Del Prete GF, De Carli M, Ricci M, Romagnani S. Helper activity for immunoglobulin synthesis of T helper type 1 (Th1) and Th2 human T cell clones: the help of Th1 clones is limited by their cytolytic capacity. J Exp Med. 1991;174:809. doi: 10.1084/jem.174.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khanolkar A, Yagita H, Cannon MJ. Preferential utilization of the perforin/granzyme pathway for lysis of Epstein-Barr virus-transformed lymphoblastoid cells by virus-specific CD4+ T cells. Virology. 2001;287:79. doi: 10.1006/viro.2001.1020. [DOI] [PubMed] [Google Scholar]
- 36.Naftzger C, Takechi Y, Kohda H, Hara I, Vijayasaradhi S, Houghton AN. Immune response to a differentiation antigen induced by altered antigen: a study of tumor rejection and autoimmunity. Proc Natl Acad Sci USA. 1996;93:14809. doi: 10.1073/pnas.93.25.14809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overwijk WW, Lee DS, Surman DR, Irvine KR, Touloukian CE, Chan CC, Carroll MW, Moss B, Rosenberg SA, Restifo NP. Vaccination with a recombinant vaccinia virus encoding a “self” antigen induces autoimmune vitiligo and tumor cell destruction in mice: requirement for CD4+ T lymphocytes. Proc Natl Acad Sci USA. 1999;96:2982. doi: 10.1073/pnas.96.6.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hara I, Takechi Y, Houghton AN. Implicating a role for immune recognition of self in tumor rejection: passive immunization against the brown locus protein. J Exp Med. 1995;182:1609. doi: 10.1084/jem.182.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeh HJ, III, Perry-Lalley D, Dudley ME, Rosenberg SA, Yang JC. High avidity CTLs for two self-antigens demonstrate superior in vitro and in vivo antitumor efficacy. J Immunol. 1999;162:989. [PubMed] [Google Scholar]
- 40.Kawakami Y, Eliyahu S, Jennings C, Sakaguchi K, Kang X, Southwood S, Robbins PF, Sette A, Appella E, Rosenberg SA. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J Immunol. 1995;154:3961. [PubMed] [Google Scholar]
- 41.Hawkins WG, Gold JS, Dyall R, Wolchok JD, Hoos A, Bowne WB, Srinivasan R, Houghton AN, Lewis JJ. Immunization with DNA coding for gp100 results in CD4 T-cell independent antitumor immunity. Surgery. 2000;128:273. doi: 10.1067/msy.2000.107421. [DOI] [PubMed] [Google Scholar]
- 42.Overwijk WW, Tsung A, Irvine KR, Parkhurst MR, Goletz TJ, Tsung K, Carroll MW, Liu C, Moss B, Rosenberg SA, Restifo NP. gp100/pmel 17 is a murine tumor rejection antigen: induction of “self”-reactive, tumoricidal T cells using high-affinity, altered peptide ligand. J Exp Med. 1998;188:277. doi: 10.1084/jem.188.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kierstead LS, Ranieri E, Olson W, Brusic V, Sidney J, Sette A, Kasamon YL, Slingluff CL, Jr, Kirkwood JM, Storkus WJ. gp100/pmel17 and tyrosinase encode multiple epitopes recognized by Th1-type CD4+ T cells. Br J Cancer. 2001;85:1738. doi: 10.1054/bjoc.2001.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]