Abstract
In bacteria, many genes involved in the biosynthesis of cofactors such as thiamine pyrophosphate (TPP) are regulated by ribo switches, regions in the 5′ end of mRNAs to which the cofactor binds, thereby affecting translation and/or transcription. TPP riboswitches have now been identified in fungi, in which they alter mRNA splicing. Here, we show that addition of thiamine to cultures of the model green alga Chlamydomonas reinhardtii alters splicing of transcripts for the THI4 and THIC genes, encoding the first enzymes of the thiazole and pyrimidine branches of thiamine biosynthesis, respectively, concomitant with an increase in intracellular thiamine and TPP levels. Comparison with Volvox carteri, a related alga, revealed highly conserved regions within introns of these genes. Inspection of the sequences identified TPP riboswitch motifs, and RNA transcribed from the regions binds TPP in vitro. The THI4 riboswitch, but not the promoter region, was found to be necessary and sufficient for thiamine to repress expression of a luciferase-encoding reporter construct in vivo. The pyr1 mutant of C. reinhardtii, which is resistant to the thiamine analogue pyrithiamine, has a mutation in the THI4 riboswitch that prevents the THI4 gene from being repressed by TPP. By the use of these ribo switches, thiamine biosynthesis in C. reinhardtii can be effectively regulated at physiological concentrations of the vitamin.
Keywords: regulation of metabolism, Chlamydomonas reinhardtii, luciferase reporter gene, alternative splicing of mRNA
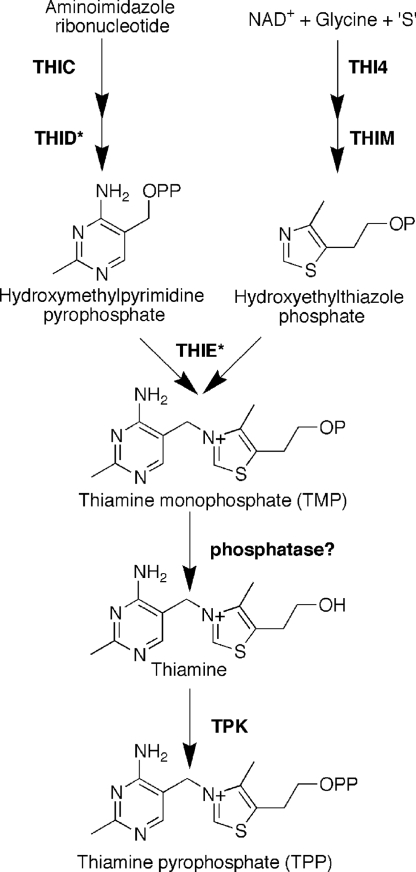
Thiamine (vitamin B1) is synthesized via a branched pathway (Fig. 1) from the condensation of a thiazole and a pyrimidine moiety to make thiamine monophosphate (TMP). This is then phosphorylated to make thiamine pyrophosphate (TPP), the active cofactor, either directly in prokaryotes, or by dephosphorylation followed by pyrophosphorylation in eukaryotes. Like many cofactors, the level of TPP within the cell is low, the majority of it being associated with its cognate enzymes, which are part of the central respiratory pathways. The biosynthetic pathway is carefully regulated to ensure that TPP production meets cellular demand, usually by regulation of gene expression for one or more of the enzymes, and in bacteria and fungi exogenous thiamine severely represses gene expression (1, 2).
Fig. 1.
TPP biosynthetic pathway in C. reinhardtii. As in all organisms, TMP is generated from the condensation of 5-hydroxyethyl-4-methylthiazole phosphate and 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate by the action of TMP synthase (THIE). In C. reinhardtii, THI4 catalyses the first committed step in the formation of the thiazole moiety from NAD+, glycine, and an unknown sulfur donor. The pyrimidine moiety is synthesized from aminoimidazole ribonucleotide, an intermediate in histidine and purine biosynthesis, via the action of THIC and THID. In C. reinhardtii, THID and THIE enzyme activities are found in a single, bifunctional enzyme. To generate the active cofactor, TPP, TMP is first dephosphorylated by an unknown phosphatase, and then subsequently pyrophosphorylated by thiamine pyrophosphokinase (TPK).
In bacteria, an important TPP regulatory mechanism is mediated via riboswitches in one or more of the thiamine biosynthesis genes. Riboswitches are short sequences in mRNAs that bind metabolites directly, without the need for intermediary proteins. Binding of the ligand alters the secondary structure of the RNA, thereby regulating expression of the gene, typically by premature transcription termination and/or initiation of translation (3, 4). Until recently, most of the work on riboswitches was carried out in prokaryotic systems. Now, TPP riboswitches have been found in the 5′ UTR of genes for thiamine biosynthetic enzymes in the fungi Aspergillus oryzae and Neurospora crassa, where they appear to operate by causing alternative splicing of the transcripts (5, 6). In the higher plant Arabidopsis thaliana, a TPP riboswitch was identified in the 3′ UTR, and this has been characterized structurally (7, 8). However, the presence of riboswitches in other eukaryotes has not been established. We have taken advantage of recent genome sequencing projects in two related green algal species, Chlamydomonas reinhardtii and Volvox carteri (9, 10), to discover TPP riboswitches in these organisms by sequence comparison. We characterize the function of these riboswitches in vivo, thereby enabling their role in the regulation of thiamine biosynthesis to be established. By using the same comparative approach, we have also identified possible TPP riboswitches in other photosynthetic eukaryotes.
Results
Identification of Riboswitch Sequences in C. reinhardtii and V. carteri.
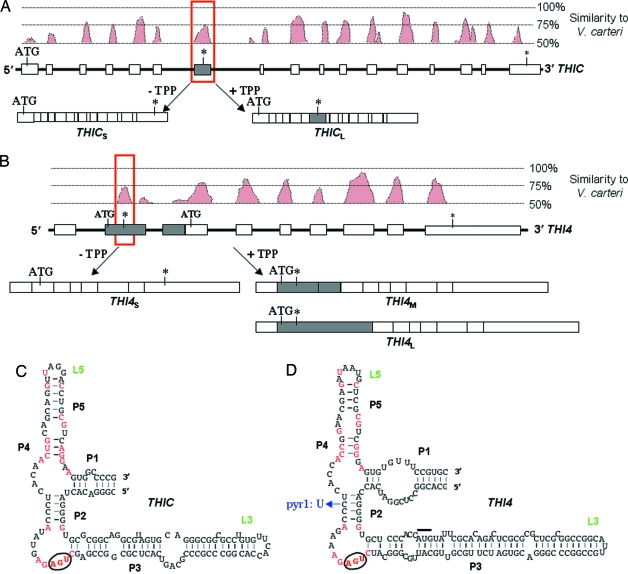
Within the genes of C. reinhardtii and V. carteri, exons usually show a high degree of sequence similarity, but the introns are much less well conserved. We reasoned that, if TPP riboswitches were present in thiamine biosynthetic genes in C. reinhardtii, they would be conserved in V. carteri. Analysis of the five genes involved in thiamine biosynthesis in C. reinhardtii (11) revealed that, in two of them, THIC and THI4, there are sequences within an intron that show a high degree of similarity to the corresponding position in V. carteri [Fig. 2 A and B, and supporting information (SI) Figs. 6 and 7]. In THIC, encoding the first enzyme of the pyrimidine branch, there is a region in the sixth intron that has 75% similarity to V. carteri (Fig. 2A). In THI4, which encodes the enzyme responsible for the synthesis of the thiazole moiety, a region of similarity is seen in the first intron in the 5′ UTR (Fig. 2B). Closer inspection of these conserved sequences in the two algae revealed that they contained 20 of the 22 nucleotides that are conserved in bacterial TPP riboswitches (12), and that they could form the characteristic stem-loop structure (Fig. 2 C and D) determined by inline probing and structural studies (4, 8).
Fig. 2.
Identification of TPP riboswitches in the THIC and THI4 genes of C. reinhardtii. (A) The intron/exon structure of the THIC gene from C. reinhardtii below a plot showing sequence similarity between C. reinhardtii and V. carteri. Introns are depicted by thick black lines, and the exons are depicted by open rectangles. The shaded rectangle indicates an alternatively spliced exon that encodes an in-frame stop codon (indicated by an asterisk). THICS and THICL are the two alternatively spliced transcripts that are produced in a TPP-dependent manner. The red box delineates the sequence that forms the TPP riboswitch shown in C. (B) The organization of the THI4 gene from C. reinhardtii is depicted in the same manner as in A. The start codon for the functional protein is in the second exon. The two shaded rectangles indicate alternatively spliced exons, the first of which encodes a small uORF of 27 aa. THI4S, THI4M, and THI4L correspond to the alternatively spliced transcripts. The red box delineates the sequence that forms the TPP riboswitch shown in D. (C) Detailed analysis of the sequence from the THIC gene that is shown in the red box in A. It can adopt the same secondary structure of stems (P) and loops (L) as TPP riboswitches and contains 20 of the 22 nucleotides that are conserved in >90% of TPP riboswitches (highlighted in red). The circled nucleotides provide the stop codon in the alternative exon. (D) Detailed analysis of the alternative exon from the THI4 gene similarly shows that it conforms to the TPP riboswitch consensus. The overlined nucleotides indicate the start codon for the uORF, and the circled nucleotides indicate the stop codon. In the pyr1 mutant of C. reinhardtii, a single base mutation alters base pairing in stem P2.
Binding of TPP to the Putative Riboswitches.
The putative riboswitch regions identified here by sequence analysis were tested for their ability to bind TPP in vitro, to verify that they were likely to be functional in vivo. They were cloned from the C. reinhardtii THIC and THI4 genes, transcribed into RNA in vitro by using T7 RNA polymerase, and then the binding of [32P]TPP to the RNA was analyzed by equilibrium dialysis. Both putative riboswitch sequences bound TPP, and the Kd for that in THIC was estimated to be 400 nM (SI Fig. 8A). However, the affinity of the THI4 riboswitch was too tight to be able to determine a Kd with this technique, so instead, isothermal titration calorimetry (ITC) was used. The Kd was found to be 30–40 nM (SI Fig. 8B). The affinity of the C. reinhardtii riboswitches for TPP is similar to that reported for Escherichia coli TPP riboswitches (4).
Effect of Exogenous Thiamine on Expression of THIC and THI4.
The presence of the THIC and THI4 riboswitches in introns within the two genes suggested that they may play a role in alternative splicing, as has been shown in N. crassa (5). We first tested whether thiamine affected levels of the transcripts. When C. reinhardtii cells are grown without thiamine, transcripts can be detected by RT-PCR that correspond to THICS and THI4S (Fig. 3 A and C, t = 0, and SI Fig. 9), which encode the functional proteins. A longer transcript for THIC, which might be a splice variant, is also apparent at low intensity. However, if cells are grown in medium supplemented with thiamine, then the THI4 transcript is not detectable, and that for THIC is greatly reduced (SI Fig. 9). At the same time, the longer THIC transcript becomes more abundant. Both THIC transcripts were sequenced and the longer one was found to correspond to THICL in which the conserved sequence was included as an additional exon (Fig. 2A). The additional exon results in the insertion of an in-frame stop codon, which would prevent expression of a full-length THIC protein of 637 residues normally expressed from the THICS transcript. The truncated protein translated from THICL would be missing 385 aa, which include the majority of the conserved residues. Interestingly, the riboswitch actually spans the alternatively spliced exon (Fig. 2A), and the premature stop codon that is recruited into THICL is derived from the conserved CUGAGA pyrimidine-binding domain of the riboswitch (ref. 7; Fig. 2C and SI Fig. 6).
Fig. 3.
The effect of thiamine on the expression of genes containing TPP riboswitches. (A) RT-PCR showing the time course of expression of the THIC transcripts after the addition of 10 μM thiamine to C. reinhardtii cultures by using transcript-specific primers. After the addition of thiamine at time 0, the THICS transcript level declines, whereas that for THICL increases. Primers to the constitutively expressed actin gene (ACT) were used as a control. (B) Levels of thiamine (triangles) and its esters TMP (circles) and TPP (squares) in cultures of C. reinhardtii grown over the same time course. (C) RT-PCR showing the time course of expression of THI4 transcripts after the addition of 10 μM thiamine to C. reinhardtii cultures. Only THI4S is detectable at time 0, whereas THI4M and THI4L appear after ≈2 h. (D) RT-PCR of THI4 transcripts in the pyr1 mutant of C. reinhardtii. THI4S is the only transcript present throughout the time course. (E) Luciferase activity in transgenic C. reinhardtii cells expressing the THI4 riboswitch-luc construct (shown below the graph) from the constitutive PSAD promoter. Squares indicate the activity in cells grown without thiamine, and diamonds indicate the activity in cells to which 10 μM thiamine was added at t = 0.
To investigate this further, we designed separate primers for RT-PCR that would detect either THICS or THICL and analyzed the expression of the gene over a time course after addition of thiamine to the growth medium (Fig. 3A). The THICS transcript declines steadily over 6 h, whereas levels of THICL remain more or less constant. We determined the levels of thiamine, thiamine monophosphate, and TPP in the cells over the same time period (Fig. 3B). All three rise, with the greatest increase seen for TPP, although this then plateaus after 3 h. The similarity in the time dependent accumulation of thiamine and its esters to that of the THICS transcript indicates strongly that the reduction in THICS levels is related to the intracellular TPP content and therefore due to the action of the riboswitch.
A similar observation is made for the THI4 transcript. On addition of thiamine to the medium, THI4S starts to decline after ≈2 h (Fig. 3B), concomitant with the increase in intracellular TPP, and two additional transcripts are detected. Sequencing established that they corresponded to THI4M and THI4L, which contain an additional exon corresponding to the conserved sequence identified by comparison with V. carteri (Fig. 2B). The additional exon contains an upstream start codon, which would drive the expression of a short, 27-aa peptide. These so-called upstream ORFs (uORFs) have been described in several different eukaryotes including yeast (13) and C. reinhardtii (14), where they have been demonstrated to regulate translation. In the C. reinhardtii THI4 gene, it now appears that a TPP riboswitch might control the incorporation of a uORF in the mature mRNA. Interestingly, as with the THIC riboswitch, the stop codon at the end of the THI4 uORF is derived from the conserved CUGAGA pyrimidine-binding domain of the riboswitch (Fig. 2), indicating that, in both cases, the same conserved part of the riboswitch has been recruited to terminate translation.
Further support for the action of the THI4 TPP riboswitch in vivo comes from analysis of the pyr1 mutant of C. reinhardtii (15). This mutant is resistant to pyrithiamine, an antagonist of thiamine, which has been shown to bind to TPP riboswitches in bacteria (16) and fungi (6), thereby preventing expression of thiamine biosynthesis genes. The pyr1 mutation has been mapped to chromosome IV, as has the THI4 gene (15). Accordingly, we amplified the region of the THI4 gene containing the riboswitch from this mutant line and found a single base change of C→U, which would disrupt base pairing in the P2 stem (Fig. 2D). A mutation at the same position has been found in a pyrithiamine-resistant mutant of Bacillus subtilis (16). Moreover, RT-PCR analysis revealed that only a single THI4S transcript is present in the pyr1 mutant whether or not it is grown in the presence of thiamine (Fig. 3D), and the level does not change even in cultures grown continuously in medium supplemented with thiamine. These data confirm that this region in the 5′ UTR is a functional riboswitch, and is responsible for the regulation of transcript splicing by thiamine.
The Riboswitches, but Not the Promoters, Respond to Intracellular TPP.
Levels of the THICL transcript do not vary significantly after the addition of thiamine to the growth medium (Fig. 3A). This indicates that the THIC promoter activity is not regulated by thiamine, but rather the expression of this gene is controlled by the riboswitch. In contrast, the levels of all three THI4 transcripts are much reduced in the presence of thiamine, compared with those seen in its absence. It is possible therefore that, in addition to the riboswitch, the THI4 promoter itself is regulated by thiamine, as it is in Saccharomyces cerevisiae (17). Accordingly, we made reporter constructs in which the THI4 riboswitch was cloned upstream of the Renilla reniformis luciferase-encoding gene (luc; codon optimized for C. reinhardtii) (18). When this construct was expressed from the PSAD promoter, which is constitutive in C. reinhardtii, addition of thiamine to the cultures resulted in a reduction in luminescence in five independently transformed lines. A time course for one of these lines is shown in Fig. 3E. The reduction of the introduced transcript, the reduction in endogenous THI4S transcript (Fig. 3C), and the increase in levels of TPP (Fig. 3B), clearly exhibit similar kinetics. In contrast, when the R. reniformis luc gene was linked directly to the PSAD promoter, without the THI4 riboswitch, the addition of thiamine to the culture medium had no effect on the levels of luminescence from the cells (SI Fig. 10). Thus, the TPP riboswitch is necessary and sufficient for thiamine modulation of gene expression.
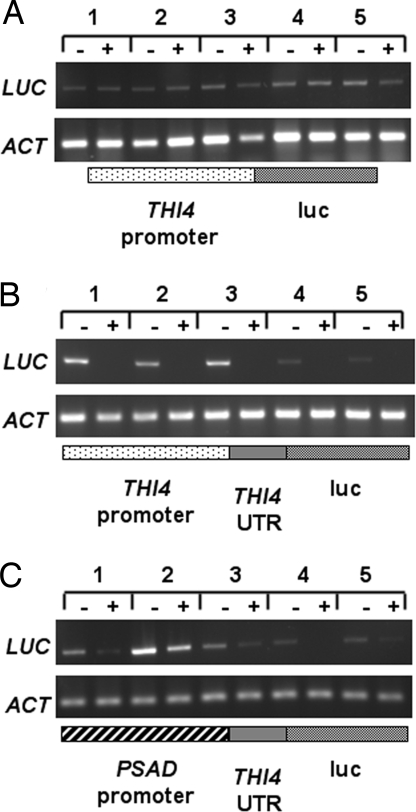
We also tested the luminescence from transgenic lines expressing the luc gene from the THI4 promoter, to determine whether or not it shows any degree of thiamine-responsive gene expression, but the THI4 promoter activity was insufficient to allow luciferase activity to be detected. Instead, we used RT-PCR with primers specific for the luc gene. Fig. 4A shows the results from five independently transformed lines expressing luc from the THI4 promoter; there is essentially no difference whether or not the cells were grown with exogenous thiamine. In contrast, when the THI4 riboswitch was included, all of the lines showed thiamine responsiveness (Fig. 4B). The absolute level of expression varied between transformants, as expected because of the random insertion of transgenes into the nuclear genome of C. reinhardtii (19). This is also seen for the lines expressing the THI4 riboswitch-luc construct from the PSAD promoter (Fig. 4C), where line 2 has particularly high levels of expression. Nevertheless, for all lines, the transcript amount was lower in cells grown in thiamine compared with those grown in its absence.
Fig. 4.
The effect of the THI4 promoter on gene expression in vivo. RT-PCR analysis of luc transcript abundance in five independent lines of C. reinhardtii transformed with the luc gene under the control of the THI4 promoter (shown below the gel image) (A), a THI4 riboswitch-luc construct under the control of the THI4 promoter (B), or a THI4 riboswitch-luc construct under the control of the constitutive PSAD promoter (C). Cells were grown in the presence (+) or absence (−) of 10 μM thiamine. Primers were to the luc gene (LUC) or a constitutively expressed actin gene (ACT). Only those constructs with the THI4 riboswitch (B and C) showed thiamine responsiveness.
Discussion
Model of TPP-Mediated Alternative Splicing.
We have demonstrated that two genes in C. reinhardtii undergo alternative splicing mediated by TPP-responsive riboswitches. It has been demonstrated for bacterial riboswitches that binding of the ligand causes alteration in secondary structure of the mRNA, such that expression of the transcript is altered, for example by masking a Shine–Dalgarno sequence necessary for translation initiation (4). The committed step in splicing in eukaryotes is the formation of a lariat loop between the splice site at the 5′ end of the intron and an acceptor adenine located 15–40 nucleotides from the 3′ end (20). Examination of the sequence of the THIC intron containing the riboswitch reveals that there are two potential splicing acceptor sites, one close to the start of the seventh exon, and another 5′ to the alternative exon; this second site resides in a region (P1″) that is complementary to nucleotides in the P1 stem of the riboswitch (Fig. 5). When TPP is bound to the riboswitch, this acceptor site is available, so that two splicing events occur to produce the THICL transcript containing the additional exon (Fig. 5A). When TPP is not bound, structural modulation may occur, as it does in bacteria, so that the region base pairs with P1′, sequestering the splice acceptor site, and only the downstream acceptor site near the seventh exon is available for splicing (Fig. 5B). This allows complete removal of the intron and production of the THICS transcript encoding a functional protein.
Fig. 5.
Proposed mechanism of TPP-mediated alternative splicing of the C. reinhardtii THIC transcript. (A) Diagram of the sixth intron of the THIC gene, which contains the riboswitch, showing the intron boundaries (GU/AG), the splice acceptor sites (A), and the alternatively spliced exon in red. The green lines show which regions would be joined to form lariat loops. The thick lines depict regions that can form stems. At high TPP concentrations, TPP will be bound to the riboswitch, which means that base pairing occurs between P1 and P1′. Within the intron, there are then two splice acceptor sites available, so that two splicing events occur on either side of the riboswitch. This produces the THICL transcript that encodes a prematurely truncated protein. (B) When there is no TPP bound to the riboswitch (as might be predicted at low intracellular TPP concentrations), it is possible for alternative base pairing to form between P1′ and the region containing the 5′ splice acceptor site (P1″), which is colored blue. There is now only one splice acceptor site available at the 3′ end of the intron, and so the entire intron is removed to form the functional THICS transcript. The nucleotide sequence in Fig. 2C corresponds to the sequence from P1′ to P1.
Identification of TPP Riboswitches in Other Photosynthetic Organisms.
The availability of genome sequences from two related green algae provided us with the means to identify conserved noncoding regions that are riboswitches. We have extended this approach to a group of evolutionarily distinct, photosynthetic eukaryotes, the diatoms, marine algae with complex plastids that are estimated to be responsible for a significant proportion of global carbon fixation (21). The genome sequences for Thalassiosira pseudonana (21) and Phaeodactylum tricornutum (http://genome.jgi-psf.org/Phatr2/Phatr2.home.html) have recently been completed. The THIC genes from these organisms share a highly conserved 3′ UTR, which contains a TPP riboswitch-like motif (SI Fig. 11), suggesting that these algae may also use a riboswitch to regulate the expression of THIC. It is interesting that, in these diatoms, the putative riboswitch is located in the 3′ UTR, as has been reported for the higher plant A. thaliana (7, 8), rather than within an internal intron. However, to date, it is not known how a riboswitch in this position can influence expression of the gene. Unlike green algae and higher plants, diatoms do not encode a homologue of THI4, and thus presumably make the thiazole moiety via a different route. Nonetheless, as the number of genome sequences from related organisms continues to grow, sequence comparison of noncoding regions in likely candidate genes, in a manner similar to that used here, opens the way to identify further classes of riboswitch that regulate metabolism in eukaryotic organisms.
Implications for Regulation of Thiamine Biosynthesis.
Sequence comparison of the other three thiamine biosynthetic genes, THIM, THID/E, and TPK (11), from C. reinhardtii with their counterparts in V. carteri, reveal that they do not share sequence similarity in the noncoding regions. Although the C. reinhardtii THIM 3′ UTR contains some riboswitch-like motifs, none of them is conserved in V. carteri. We cloned the THIM 3′ UTR from C. reinhardtii, and carried out equilibrium dialysis with TPP on the corresponding RNA transcribed in vitro. No binding was observed by equilibrium dialysis (SI Fig. 8A), and moreover, the THIM transcript remains unchanged when cells are grown in thiamine (SI Fig. 9). These data indicate that this region is not a functional riboswitch. Although THIM, THID/E, and TPK may be regulated by other mechanisms, it is interesting that the first gene in each branch is controlled by a riboswitch.
From our analysis, and that carried out in previous studies (5, 6), there now appears to be a general pattern emerging for the control of thiamine biosynthesis in eukaryotes by riboswitches; they regulate the first enzymes in the thiazole and/or pyrimidine branches, which are the committed steps. The TPP content within C. reinhardtii cells grown without thiamine is ≈5 μM, which is greater than the Kd of the riboswitches measured in vitro. However, the latter is determined for a short region of RNA containing the riboswitch only, which may behave differently in the context of the complete mRNA in vivo. Moreover, the affinity of most TPP-dependent enzymes is ≈1 μM (22), so to ensure that they have sufficient bound cofactor, the level at which thiamine biosynthesis is repressed must be at or above this concentration. Our results thus indicate that, in the context of metabolic regulation, riboswitch regulation of gene expression allows fine-tuning of biosynthetic pathways, rather than acting as simple on/off switches.
Materials and Methods
Identification of Riboswitch Sequences.
The sequences of the C. reinhardtii thiamine biosynthesis genes were compared with the equivalent regions of the V. carteri genomic DNA by a combination of local and global sequence alignment techniques to find conserved regions between them (10). Regions in introns that showed >50% sequence similarity, were analyzed in detail for conserved TPP riboswitch elements previously described in ref. 7.
Strains, Media, and Plasmids.
E. coli DH5α was grown in LB medium, and used in all cloning steps. C. reinhardtii cells were grown in Tris-acetate phosphate medium with or without the addition of 10 μM thiamine, under continuous light, at 22°C. For time course experiments, a 50-ml aliquot was removed from the culture every hour, and the cells were immediately centrifuged at 4,000 × g for 5 min. Primer sequences used are listed in SI Table 1, as are conditions for amplification by PCR of the putative riboswitch regions and the THI4 promoter, and details of all cloning procedures and generation of C. reinhardtii transformants.
TPP–RNA Binding Studies.
RNA was produced by in vitro transcription using T7 RNA polymerase (E. coli thiM and C. reinhardtii THIC and THIM) and SP6 RNA polymerase (C. reinhardtii THI4). For ITC of THI4, PCR primers were used to generate a template with a T7 transcription start site immediately upstream of the 5′ end of the riboswitch aptamer domain. Qualitative equilibrium dialysis was performed by using fast microequilibrium dialysers (Harvard Apparatus). In vitro transcription reactions were buffer exchanged by centrifugal concentration (Vivaspin 20; Sartorius) into 50 mM Tris·HCl (pH 8.0), 20 mM MgCl2. The RNA at a final concentration of ≈200 nM was equilibrated overnight against 500 nM [32P]TPP. Equilibrated samples were diluted into scintillation fluid (Optiphase “Hisafe 3”) and quantified by using a Packard Tri-Carb 2100TR liquid scintillation counter. 32P-labeled TPP was generated catalytically from [γ-32P]ATP and thiamine by using thiamine pyrophosphokinase as described in ref. 23. ITC was performed by using a VP-ITC (Microcal). In a typical experiment, 70 μM TPP was titrated into a nominal 10 μM concentration of RNA. Data were fitted in Origin 7.0 by using a single-site binding model.
Determination of Thiamine and Its Esters.
For analysis, mid-log-phase C. reinhardtii cells were extracted in trichloroacetic acid (for details, see SI Methods). The extract was separated on a Finnigan Surveyor HPLC coupled to a Finnigan LCQDECA XP mass spectrometer with an electrospray ionization source (Thermo Fisher Scientific). Chromatographic separation of a 10-μl sample was performed by using a linear gradient elution from a Hypercarb column (150 × 2.1 mm; 4-μm particle diameter) as follows: 0 min, 95% buffer A, 5% acetonitrile (MeCN); 34 min, 75% MeCN; 35 min, 75% MeCN; 36 min, 5% MeCN (A: 250 mM ammonium acetate, pH 5; flow rate was 0.2 ml min−1), followed by mass spectrometry in positive-ion mode by using single ion monitoring (SIM)-MS of [M+H] ions at m/z 265, 324.9, and 424.9 for thiamine, thiamine monophosphate, and TPP, respectively. (MS-MS parameters were as follows: capillary temperature, 300°C; capillary voltage, 3.0 kV; and collision energy level, 70%.) Data were analyzed by using Xcalibur (Thermo Fisher Scientific). Quantification was carried out by using the linear range of a standard curve constructed with known amounts of thiamine, thiamine monophosphate, and TPP (Sigma).
RT-PCR.
Total RNA was extracted from C. reinhardtii as described in ref. 24, and stored under ethanol at −80°C. A 5-μg sample of RNA was treated with RNase-free DNase (Promega) for 30 min at 37°C to remove contaminating DNA. First-strand cDNA was synthesized by using SuperScript II reverse transcriptase (Invitrogen) with dT17 primers as described by the manufacturer's instructions. cDNA was amplified by using TaqDNA polymerase (Bioline). For each primer pair to different regions of the C. reinhardtii THI genes, PCR amplification was carried out for 21–43 cycles, to determine the optimal cycle number for comparisons between samples (SI Figs. 12 and 13). For all of them, 31 cycles was in the linear range, and this cycle number was used throughout. For luc linked to the PSAD promoter, 26 cycles was optimal (SI Fig. 14).
Luciferase Assays.
C. reinhardtii cell pellets were resuspended in 3 ml of 10 mM sodium phosphate, pH 7.5, and distributed into three microfuge tubes containing 300 mg of 0.3-mm glass beads (Sigma). The cells and glass beads were vortexed at maximum speed for 2 min in a Genie Vortex. The resulting suspension was centrifuged at 13,000 × g for 1 min, and 200 μl of the supernatant was used for the luminescence assay in a 96-well plate format by using a BMG Labtech Flurostar plate reader. A total of 10 μl of coelenterazine (10 mM; Lux Biotech) was added to each sample, and the luminescence signal was recorded for 1 min. The luminescence levels were normalized to the total protein concentration, which were measured with Bradford's reagent with BSA as standard.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Prof. Chris Abell, Dr. Finian Leeper, and all members of the 3B Group, Departments of Chemistry and Plant Sciences, University of Cambridge, for helpful and stimulating discussions; the U.S. Department of Energy Joint Genome Institute for providing access to the C. reinhardtii and V. carteri genome sequences for use in this article; and Emmanuel College (Cambridge, U.K.) and the Biotechnology and Biological Sciences Research Council of the United Kingdom for financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705786105/DC1.
References
- 1.Miranda-Rios J, Navarro M, Soberon M. Proc Natl Acad Sci USA. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Praekelt UM, Byrne KL, Meacock PA. Yeast. 1994;10:481–490. doi: 10.1002/yea.320100407. [DOI] [PubMed] [Google Scholar]
- 3.Mandal M, Boese B, Barrick JE, Winkler W, Breaker RR. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 4.Winkler W, Nahvi A, Breaker RR. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 5.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 6.Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 7.Sudarsan N, Barrick JE, Breaker RR. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thore S, Leibundgut M, Ban N. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 9.Grossman AR, Croft M, Gladyshev VN, Merchant SS, Posewitz MC, Prochnik S, Spalding MH. Curr Opin Plant Biol. 2007;10:1–9. doi: 10.1016/j.pbi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croft MT, Warren MJ, Smith AG. Eukaryot Cell. 2006;5:1175–1183. doi: 10.1128/EC.00097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winkler W, Breaker RR. Ann Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 13.Miller PF, Hinnebusch AG. Biochim Biophys Acta. 1990;1050:151–154. doi: 10.1016/0167-4781(90)90157-w. [DOI] [PubMed] [Google Scholar]
- 14.Moseley JL, Page MD, Alder NP, Eriksson M, Quinn J, Soto F, Theg SM, Hippler M, Merchant S. Plant Cell. 2002;14:673–688. doi: 10.1105/tpc.010420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth RD, Martinek GW, Ebersold WT. J Bacteriol. 1975;124:1615–1617. doi: 10.1128/jb.124.3.1615-1617.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Nosaka K. Appl Microbiol Biotechnol. 2006;72:30–40. doi: 10.1007/s00253-006-0464-9. [DOI] [PubMed] [Google Scholar]
- 18.Fuhrmann M, Hausherr A, Ferbitz L, Schodl T, Heitzer M, Hegemann P. Plant Mol Biol. 2004;55:869–881. doi: 10.1007/s11103-004-2150-6. [DOI] [PubMed] [Google Scholar]
- 19.Harris EH. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- 20.Tazi J, Durand S, Jeanteur P. Trends Biochem Sci. 2005;30:469–478. doi: 10.1016/j.tibs.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 22.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. Nucleic Acids Res. 2004;32:431–433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nosaka K, Onozuka M, Nishino H, Nishimura H, Kawasaki Y, Ueyama H. J Biol Chem. 1999;274:34129–34133. doi: 10.1074/jbc.274.48.34129. [DOI] [PubMed] [Google Scholar]
- 24.Witman GB, Carlson K, Berliner J, Rosenbaum JL. J Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.