Abstract
The study of pathway disruption is key to understanding cancer biology. Advances in high throughput technologies have led to the rapid accumulation of genomic data. The explosion in available data has generated opportunities for investigation of concerted changes that disrupt biological functions, this in turns created a need for computational tools for pathway analysis. In this review, we discuss approaches to the analysis of genomic data and describe the publicly available resources for studying biological pathways.
Background
The development of cancer involves the accumulation of genetic and epigenetic alterations. Genetic events such as chromosomal rearrangements, changes in gene dosage, and sequence mutations can influence gene expression patterns, which contribute to the hallmark phenotypes of cancer1,2. The interaction between pathways and the involvement of pathways in multiple phenotypes complicate the interpretation of gene expression patterns. For example, the epidermal growth factor receptor (EGFR, HER1, ERBB-1) signaling pathway plays a role in specific phenotypes including resistance to apoptosis, increased proliferation, mitogenesis, transcription of numerous target genes, and actin reorganization, in several cancers (Fig. 1)1,3,4. In order to decipher the interaction within and between pathways, computational tools are necessary to annotate components, to identify co-regulated expression, and to identify sets of genes or pathways which are statistically over/under-represented within a dataset.
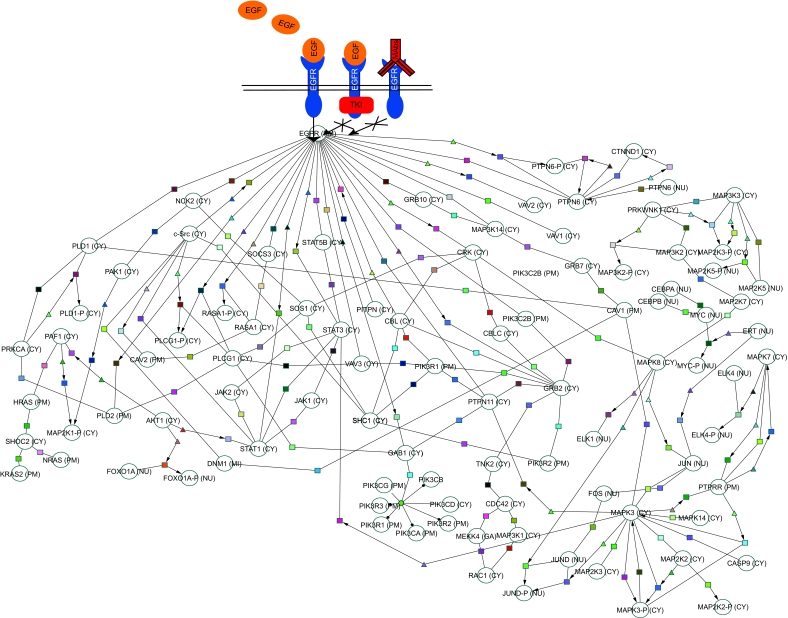
Figure 1.
Example of EGFR-mediated signaling changes, a commonly disrupted pathway in lung cancer. The EGFR pathway could be disrupted by an increased expression of growth factor ligands. By targeting EGFR with tyrosine kinase inhibitors (TKIs) and MAb (monoclonal antibodies), EGFR activity can be eliminated. However, a downstream factor (e.g. MAPK signaling pathway) may also be activated to disrupt the pathway, thus making TKIs ineffective. Pathway data was obtained and selected from the Cancer Cell Map database and drawn using Cytoscape.
Methods for Gene Classification
A major analytical step to mine large microarray data is sample classification or identification of gene sets with characteristic biological function. Entrez Gene at the National Center for Biotechnology Information (NCBI) provides unique identifiers for genes, and is a searchable database providing gene-specific information and links to external databases, including the Gene Ontology (GO) consortium, KEGG and Reactome5. A limitation of Entrez Gene is that genes are searched individually, which could be time consuming. Here, we describe the Gene Ontology (GO), a structural language to annotate gene functions for batch processing, and also methods of clustering analysis. The algorithmic basis of clustering identifies a pattern associated in a data set, which could be subsequently followed by GO analysis to identify its underlying biology.
Gene Ontology annotation
The Gene Ontology (GO) Consortium was established in 2000 to provide a controlled vocabulary for annotating homologous gene and protein sequences in different organisms6,7. GO classifies genes and gene products based on three hierarchical structures that describe a given entry’s biological processes, cellular components, and molecular functions, and organizes them into a parent-child relationship6. Through easy on-line access (http://www.geneontology.org), the genome databases are being unified to expedite the process of retrieving information on genes and proteins based on shared biology among multiple organisms. Several software tools, including GoMiner8,9, MAPPFinder10, and Onto-Express11,12, have been developed to explore the GO relationships among high-throughput data. However, the biological functions of genes/proteins are often complex and annotating them into categories may oversimplify their biology. The flat-format output does not convey the richness of GO’s hierarchical structure. Nevertheless, this established system of nomenclature of genes and proteins is important for the interoperability of databases, batch processing, and the future design of pathway databases.
Clustering
The biological system is integrative with tightly regulated processes, and genes with similar functions often exhibit coordinated expression patterns13–16. Transcriptional profiling studies typically aim to identify patterns of change among clinically related samples or to classify subgroups of samples15–17. Clustering of microarray data is widely used to identify groups of genes that display coordinated expression patterns performed in a supervised or unsupervised manner (Fig. 2)13,14,17–21. Unsupervised clustering is to classify data without a priori labeling of samples, whereas supervised clustering classifies data based on knowledge of samples type (e.g. cancer subtype)21–24. Clustering techniques are generally classified into two types: hierarchical and partitional25,26. Hierarchical clustering is constructed by either agglomerative (bottom-up) or divisive (top-down) approaches25. Agglomerative algorithms begin with separate clusters and merge them into successively larger clusters, while divisive algorithms begin with the whole dataset and divide the data into smaller clusters successively25. The output of agglomerative clustering is a tree of clusters called a dendrogram, in which each branch represents group of genes that have a higher order relationship (Fig. 2B)25,27. Partitional clustering directly reduces the dataset into a set of non-overlapping clusters26. Representative algorithms of partitional clustering include k-means clustering and self-organizing maps (SOM)25. k-means clustering requires the user to define k number of clusters26,28, and SOM partitions data into a two dimensional grid of clusters13,29,30. However, hierarchical clustering is more frequently used17–20,30. Detailed reviews of clustering algorithms are available and this topic will not be discussed further in this review26,31–33.
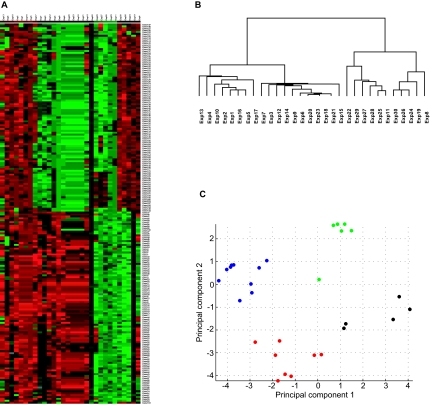
Figure 2.
Graphical output display of heatmap, hierarchical clustering, and principal component analysis. A: An example of a heatmap representation of 30 simulated profiles helps the user to easily visualize four groups of samples along the x-axis with distinct characteristics expression patterns for 300 genes. Heatmap facilitates the grouping of altered genes and sample clusters, but does not convey any spatial relationship between clustered samples. B: An example of a dendrogram generated from hierarchical clustering of the simulated data represented in figure 2A. A dendrogram is a tree diagram consisting of many U-shaped lines connecting objects to represent hierarchical clusters. In this dendrogram, four clusters of samples are formed based on distinct expression signatures. C: A two-dimensional graphical visualization of principal components analysis (PCA) based on the simulated data shown in figure 2A. Samples are color-coded based on the four clusters observed by hierarchical clustering in 2B.
Dimensionality reduction
Dimensionality reduction of data is used to minimize the number of input variables for finding coherent patterns of gene expression in an efficient manner25,34,35. Algorithms like principle component analysis (PCA) and multi-dimensional scaling (MDS) both employ this technique for classification procedures25,34,36,37. PCA visualizes multidimensional datasets by projecting data into a sub-space with 2 or 3 dimensions (Fig. 2C)34,35,37,38. The three-dimensional graphical display of MDS can be useful to portray relationships among the data points but might be complex to interpret and require subjective judgments.
Classification analysis may provide some pattern to the experimental datasets. Subsequently, the identified pattern may be further evaluated for biological interpretation using tools such as GO and/or Entrez Gene. However, the inherent limitation of pre-processed databases is subjective to the interpretation of the curator. Therefore, further validation should be considered. In a study that was conducted under the hypothesis that members in the same cluster would share related biological annotations, the majority of the clusters generated by three different clustering algorithms do not correspond well with known biology39. Furthermore, there is a need to improve the different clustering algorithms to enhance consistency of the results39,40. It is crucial to associate biological functions or regulatory pathways with each identified cluster of genes in order to deduce biological significance to each sample group41.
Construction of Pathway Database
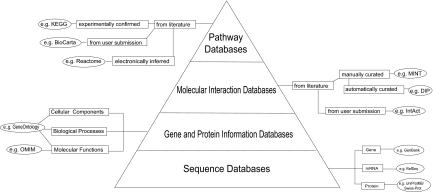
A remarkable number of published articles have collectively yielded thousands of molecular interactions for human and for model species. The challenge is to extract these individual interactions from the literature and to comprehend the dynamics of the interlocking networks as a whole. In recent years, massive efforts have been devoted to managing, integrating, and interpreting the available scientific information in a meaningful manner (i.e. building interactomes or networks of genes and pathways)42,43. Three categories of information are essential for the construction of interactome databases: gene and protein sequences, gene and protein biological information, and molecular interaction resources (Fig. 3). The major repositories of genes and protein sequences are listed in Table 1. Examples of nucleotide sequence databases include NCBI GenBank, EMBL, and DDBJ, all of which are part of the International Nucleotide Sequence Database Collaboration to facilitate data exchange and enhance accuracy44–47. The major databases for gene and protein biological information are listed in Table 2. Gene Ontology (GO), OMIM, Entrez Gene, and Universal Protein Resource Knowledgebase (UniProtKB) are the foundation for building these hierarchical databases5,7,48,49. The main publicly available molecular interaction databases are listed in Table 3. Currently, DIP, IntAct, MINT, HPRD, and MIPS all support the Human Proteome Organization (HUPO) Proteomics Standards Initiative Molecular Interaction (PSI-MI) standard format50–55. This is a unified data standard to represent molecular interaction data in a controlled vocabulary, which facilitates data comparison, exchange, and linking queries together51.
Figure 3.
Biological knowledgebases contain a myriad of specific information on each gene/protein. Sequence databases are the basis for gene and protein information. Gene and protein information is further extracted and their inter-relationships are experimentally identified, building molecular interaction databases. All of this information is the foundation of pathway databases.
Table 1.
Gene and protein databases.
| Database | Full name | Comments | Website | Ref |
|---|---|---|---|---|
| NCBI Gen-Bank | NIH genetic sequence database | An international DNA sequence database | www.ncbi.nlm.nih.gov/Genbank | [45] |
| EMBL Nucleotide Sequence Database/EMBL-Bank | European Molecular Biology Laboratory Nucleotide Sequence Database | Collection of DNA and RNA sequences in Europe and is synchronized with GenBank at NCBI and DDBJ in Japan. | www.ebi.ac.uk/embl | [46] |
| DDBJ | DNA Data Bank of Japan | Nucleotide sequence database in Japan and in collaboration with EMBL and NCBI GenBank | www.ddbj.nig.ac.jp | [47] |
| Entrez Gene | - | NCBI database that focuses on gene-to-sequence relationship and provides gene-specific information. | www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene | [5] |
| RefSeq | NCBI Reference Sequences | NCBI collection of non-redundant set of DNA, RNA, and protein sequences. | www.ncbi.nlm.nih.gov/RefSeq | [72] |
| UniGene | NCBI UniGene | Partitions GenBank sequences into sets of transcript sequences that are likely to represent distinct genes. | www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene | [73] |
| Ensembl | - | A source for comparative chordate genome sequences and gene annotation at EBI/Sanger. | www.ensembl.org | [74] |
| UCSC Genome Browser Database | University of California Santa Cruz Genome Browser Database | Human genome assembly and customizable track browsers at UCSC. | genome.ucsc.edu/bestlinks.html | [75] |
| UniProtKB/TrEMBL | Universal Protein Resource Knowledgebase/Translated European Molecular Biology Laboratories | Computer-curated protein sequence database containing translations of all coding sequences in EMBL/GenBank/DDBJ and also other protein sequences from the literature. | www.ebi.ac.uk/trembl | [49] |
| UniProtKB/Swiss-Prot Protein Knowledge-base | Universal Protein Resource Knowledgebase | Manually-curated protein sequence database providing publicly available information about protein sequences. | www.ebi.ac.uk/swis-sprot | [49] |
Table 2.
Gene and protein information databases.
| Database | Full name | Comments | Website | Ref |
|---|---|---|---|---|
| GO | Gene Ontology | Provides a controlled vocabulary to describe gene and gene product attributes in many organisms. | www.geneontology.org | [7] |
| Entrez Gene | - | NCBI database that focuses on gene-to-sequence relationship and provides gene-specific information. | www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene | [5] |
| OMIM | Online Mendelian Inheritance in Man | Collection of human genes information and genetic disorders. | www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM | [48] |
| HomoloGene | - | Homolog detection among annotated genes of several eukaryotic genomes. | www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=homologene | - |
| iHOP | Information Hyperlinked Over Proteins | Convert PubMed literature into a navigable resource. | www.ihop-net.org/UniPub/iHOP | [76] |
| SCOP | Structural Classification of Proteins | Classifies proteins of known structure based on their evolutionary and structural relationships. | scop.mrc-lmb.cam.ac.uk/scop | [77] |
| RCSB PDB | Research Collaboratory for Structural Bioinformatics Protein Data Bank | Resource for studying biomacromolecular structures and their relationships to sequence, function, and disease. | www.rcsb.org/pdb/Welcome.do;jsessionid=SvJzsMMl-0lENd1T-yXr7Q** | [78] |
| PIR | Protein Information Resource | A resource to identify and interpret protein sequence information. | pir.georgetown.edu | [79] |
| IntEnz | Integrated relational Enzyme database | Contains enzyme data curated and approved by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology | www.ebi.ac.uk/intenz | [80] |
| ENZYME nomenclature database | - | Database of information related to enzyme nomenclature. | ca.expasy.org/enzyme | [81] |
| BRENDA | BRaunschweig ENzyme DAtabase | Collection of enzyme functional data. | www.brenda.uni-koeln.de | [82] |
| Module Map | - | Collection and tools for the analysis of microarray data in 22 tumor types. | ai.stanford.edu/~erans/cancer | [83] |
| Cancer Gene Census | - | Catalogue of cancer-related genes. | www.sanger.ac.uk/genetics/CGP/Census | - |
| Cancer Gene Data Curation Project | - | Catalogue gene-disease and gene-drug relationships in cancer. | ncicb.nci.nih.gov/NCICB/projects/cgdcp | - |
| Tumor Gene Database | - | Database of tumor genes with a standard set of information. | www.tumor-gene.org/TGDB/tgdb.html | - |
| GEO | Gene Expression Omni-bus | A public archive for data submission and provides mining tools to query and download data. | www.ncbi.nlm.nih.gov/geo | [84] |
| CGED | Cancer Gene Expression Database | Database with graphical display of gene expression and clinical data on different tumor types. | cged.hgc.jp | [85] |
| Cancer Genes Resequencing Resource | - | Searchable database of cancer genes. | cbio.mskcc.org/cancergenes | - |
| SMD | Stanford Microarray Database | Database for storage and tools for processing and analyzing microarray data. | smd.stanford.edu/ | [86] |
| Progenetix | - | A public database that collects information about chromosomal alterations in cancer. | http://www.progenetix.de/~pgscripts/progenetix/index.html | [87] |
| ArrayExpress | - | Public repository for microarray data. | http://www.ebi.ac.uk/microarray-as/aer/?#ae-main[0] | [88] |
| CGAP | Cancer Genome Anatomy Project | Database which relates chromosomal alterations to tumor characteristics. | http://cgap.nci.nih.gov/Chromosomes/Mitelman | [89] |
Table 3.
Molecular interaction database.
| Database | Full name | Comments | Visualization capability | Website | Ref |
|---|---|---|---|---|---|
| IntAct | EBI protein intearction database | Protein interaction database by literature curation or user submissions. | HierarchView | www.ebi.ac.uk/intact/site/index.jsf | [52] |
| DIP | Database of Interacting Proteins | Curated both manually and automatically to combine experimentally determined protein-protein interactions. | Y | dip.doe-mbi.ucla.edu | [50] |
| MINT | Molecular INTeractions Database | Curated manually, experimentally verified protein interactions from literature. | MINT Viewer | mint.bio.uniroma2.it/mint/Welcome.do | [53] |
| HPRD | Human Protein Reference Database | Manually curated based on experimental evidence and contains information on domain architecture, post-translational modifications, interaction networks and disease association. | GenMAPP | www.hprd.org | [54] |
| HomoMINT | - | Molecular interactions discovered in model organisms are mapped to orthologs in Homo sapiens. | MINT Viewer | mint.bio.uniroma2.it/HomoMINT | [90] |
| Domino | Domain peptide interactions database | Protein interactions of domain peptides. | MINT Viewer | mint.bio.uniroma2.it/domino | [91] |
| PDZBase | - | Experimentally determined protein-protein interactions involving the PDZ-domains. | N | icb.med.cornell.edu/services/pdz/start | [92] |
| BOND | Biomolecular Object Network Databank | An interaction database that includes high-throughput data submissions and manually curated information from literature. | Cytoscape | bond.unleashedinformatics.com | [93] |
| BioGRID | General Repository for Interaction Datasets | A repository for protein and genetic interactions contributed by the community. | Osprey | www.thebiogrid.org | [94] |
| OPHID | Online Predicted Human Interaction Database | Database with known protein-protein interactions from human and predicted protein-protein interactions from model organisms. | Y | ophid.utoronto.ca/ophid | [95] |
| PIP | Potential Interactions of Proteins | Predicted protein-protein interactions derived from homology with experimentally known interactions from other species. | Y | bmm.cancerre-searchuk.org/~pip | [96] |
| MPPI | MIPS mammalian protein-protein interaction database | Published experimental protein interaction data in mammals | Y | mips.gsf.de/proj/ppi | [97] |
| HPID | Human Protein Interaction Database | Human protein interaction information and infer interactions between submitted proteins. | WebInter-Viewer | www.hpid.org or wilab.inha.ac.kr/hpid | [98] |
| InterDom | Database of Interacting Domains | Putative protein domain interactions information. | N | interdom.lit.org.sg | [99] |
| STRING | Search Tool for the Retrieval of Interacting Proteins | Database of known and predicted protein-protein interactions. | Y | string.embl.de | [100] |
The wealth of biological resources can complicate the construction of pathway databases (Fig. 4). When assembling information into a pathway database, developers must be cautious to distinguish those interactions that are deduced from hypothetical situations from those that have been experimentally confirmed. Within the latter group, care must also be taken to determine whether interactions have been confirmed in a single direct experiment or a high-throughput experiment. Furthermore, the use of natural language processing (NLP) systems to automate the extraction of information from published articles and to identify relationships between gene and protein names or interactions must be reviewed for biological relevance56,57. This method is useful as a first-pass tool for mining and extracting the knowledge in the literature. However, the constantly advancing nature of research, the further refinement of biological knowledge associated with each gene or protein further refining, the incompletion of the annotation database, and the complexity of entity names in the biological domain often makes it challenging for NLP to be high-quality with huge successes.
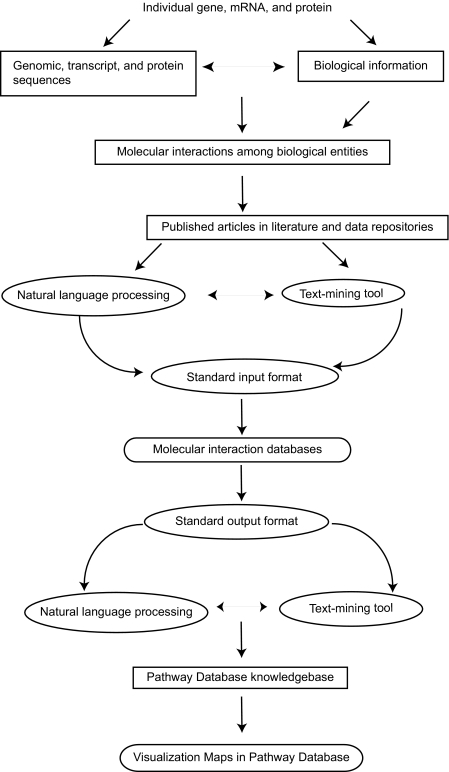
Figure 4.
An approach to building pathway databases. Biological knowledgebases are represented as rectangles with squared edges. Computational tools for text-mining and language control are represented as ellipses. Molecular interaction and pathway databases are represented by rectangles with rounded edges.
Descriptions of Specific Pathway Database
Pathway databases facilitate the data mining process for cancer researchers. The major pathway databases are listed in Table 4. A collection of biological pathway and network databases is summarized in Table 5, including Pathguide: The Pathway Resource List (http://www.pathguide.org)58. This website is updated regularly and currently about 224 biological pathway resources are accessible through the Pathguide website. Here, we focus on a subset of databases that are publicly available.
Table 4.
Pathway databases.
| Pathway databases | Full name | Comments | Cost | Visualization capability | Website | Ref |
|---|---|---|---|---|---|---|
| KEGG pathway | Kyoto Encyclopedia of Genes and Genomes Pathway | Manually drawn pathway maps with different organisms. | Free | Y | www.genome.ad.jp/kegg/kegg2.html | [59] |
| The Cancer Cell Map | - | Ten human cancer-related signaling pathways. | Free | Cytoscape | cancer.cellmap.org/cellmap | N/A |
| Reactome | - | Biological pathways that include experimentally confirmed, manually inferred, and electronically inferred reactions. | Free | Skypainter | www.reactome.org | [62] |
| HPRD | Human Protein Reference Database | Ten human cancer signaling pathways and 10 immune system signaling pathway. | Free | GenMAPP | www.hprd.org | [54] |
| BioCarta | Charting Pathways of Life | Graphical display of known and suggested pathways. | Free | Y | www.biocarta.com/genes/index.asp | N/A |
| STKE | Signal Transduction Knowledge Environment | Database of cellular signaling pathways. | Free | SVG | stke.sciencemag.org | [101] |
| PharmGKB | The Pharmacogenetics and Pharmacogenomics Knowledge Base | Database to explore relationships among drugs, diseases and genes, including their variations and gene products. | Free | Y | www.pharmgkb.org | [102] |
| Panther Classification System | Protein Analysis Through Evolutionary Relationships | Predict protein function and contains over 139 pathways mapped to protein sequences. | Free | CellDesigner | www.pantherdb.org/pathway | [103] |
| MetaCyc | Metabolic Encyclopedia of enzymes and pathways | Non-redundant, experimentally determined pathways from more than 900 different organisms. | Free | Y | metacyc.org | [104] |
| aMAZE | - | Molecular interactions and cellular processes. | Free | N | www.scmbb.ulb.ac.be/amaze | [105] |
| CGAP | Cancer Genome Anatomy Project | Pathways are from BioCarta and KEGG. | Free | Y | cgap.nci.nih.gov/Pathways | [106] |
| INOH Pathway Database | Integrating Network Objects with Hierarchies | Pathway database of different organisms which organize pathway objects in an ontology-based system. | Free | INOH Client tool | www.inoh.org | N/A |
Table 5.
Collection of databases.
| Databases | Full name | Comments | Cost | Website | Ref |
|---|---|---|---|---|---|
| Pathguide | The Pathway Resource List | List about 222 biological pathway databases. | Free | www.pathguide.org | [58] |
| UBiC Bioinformatics Links Directory | UBC Bioinformatics Centre | Curated links to molecular resources, tools, and databases. | Free | bioinformatics.ubc.ca/resources/links_directory | [107] |
| NAR Molecular Biology Database Collection | Nucleic Acids Research online Molecular Biology Database Collection | Provide external links to sequence, structures, and pathway data-bases. | Free | www3.oup.co.uk/nar/database/subcat/6/25 | [108] |
KEGG
The KEGG (Kyoto Encyclopedia of Genes and Genomes) database has been established since 1995 and has been one of the most popular knowledge databases to date59. The KEGG PATHWAY database consists of manually assembled pathway maps based on inspection of published literature. Pathway maps are grouped into metabolism, genetic information processing, environmental information processing, cellular processes, human diseases, and drug development. Most of the pathways associated with cancer are listed in the environmental information processing section, which is further subdivided into membrane transport, signal transduction, and signaling molecules and interaction. Beside human databases, information from other model organisms such as chimpanzee, mouse, rat, dogs, cows, and pigs is also available. KEGG pathway maps can be manipulated through the KEGG Markup Language (KGML) files, which provide graphical information to customize pathways.
The Cancer Cell Map
The Cancer Cell Map (http://cancer.cellmap.org) is the only database that focuses on signaling pathways implicated in cancer. This resource contains ten cancer-related pathways and each pathway has approximately 100 to 400 interactions. Interactions are manually curated and reviewed for biological validity. Extensive information is provided in each pathway, including the cellular locations of the proteins, the types of physical interactions including molecular interaction, biochemical reaction, catalysis and transport, and post-translational protein modifications. The original citations, experimental evidences, and links to other databases are also listed. Gene expression data can also be visualized in the context of Cancer Cell Map pathways using the Cytoscape network visualization and analysis software60.
Human Protein Reference Database
The HPRD (Human Protein Reference Database) contains ten cancer signaling pathways and ten immune signaling pathways which are graphically visualized in GenMAPP pathway maps54,61. The HPRD also offers the flexibility for investigators to refine their search of interested protein by multiple criteria, including molecular class from GO, domain name, motif, site of expression, length of protein sequence, molecular weight, and disease association (e.g. ovarian cancer and breast cancer). The protein domain architecture is graphically visualized with description of the domains and motifs within the queried protein. Post-translational modifications, protein interactions, and disease type are linked to PubMed, OMIM, Swiss Prot, Gene-Prot, Entrez Gene, or pathways within the HPRD. Individual genes within the pathway map are also linked to biologically relevant databases. Results from pathway analysis and HPRD entries can be readily exported. The use of XML (extensible markup language) for HPRD entries makes this database interoperable with other public databases. As with Cytoscape, the development of GenMAPP allows users to map microarray data onto pathway maps61.
Reactome
Reactome is a publicly available, peer-reviewed resource of human biological pathways.62 Although the primary focus is on H. sapiens, it is now extending human pathways onto other organisms via putative orthologs to make them applicable to 21 model organisms, including mouse, rat, chicken, puffer fish, worm, fly, yeast, and E. coli. All the information in Reactome is cross-referenced with PubMed, GO, and the sequence databases at NCBI, Ensembl, and UniProt. Small molecules are linked to ChEBI (http://www.ebi.ac.uk/chebi), catalyst activities to the GO molecular function ontology, and sub-cellular locations to the GO cellular compartment ontology. The OMIM morbid map can be overlaid into the reaction map to see which genes have been implicated in the disease in the literature. Reactions from direct evidence in the literature and indirect evidence that are inferred via orthology in other species are indicated by color-coding. The Reactome SkyPainter tool facilitates the labeling of genes or proteins in the reaction maps. Thus, quantitative data from microarray experiments can be superimposed on Reactome maps to provide visualization and exploration in a pathway context.
Visualization tools
Cross-talk between pathways can complicate the graphical representation of observed biological interactions. Therefore, visualization tools such as Cytoscape60 and GenMAPP61 have been developed to illustrate molecular interactions intuitively.
Cytoscape
Cytoscape is a software tool for the integration of pathways with expression profiles. It allows the querying of networks by using several filtering tools, and linking a given network to public databases for functional annotations60. An important feature of Cytoscape is its extensible software framework which allows users to implement new algorithms and network computations. In addition to its use by the Cancer Cell Map (described above), Cytoscape can also be used in conjunction with other protein interaction databases or genetic interaction databases63,64. Molecular species are represented as nodes and intermolecular interactions are linked as edges. Different visual properties such as node color, shape, and size can be chosen, and subsets of nodes and edges can be displayed based on the criteria that are selected by the user. Visualization properties and analysis parameters are customizable.
GenMAPP
GenMAPP (Gene Map Annotator and Pathway Profiler; previously called Gene MicroArray Pathway Profiler) is a computer program designed to display gene expression data in the context of biological pathways61. Based on the quantitative data that is loaded, the program will map genes onto relevant pathways and the user can set up criteria to color code the genes accordingly. GenMAPP visualize data in a file format called “MAPPs”, which allow users to organize the genes by their functional component. The user has the choice to download specific pathways or from the archive of MAPPs at www.netpath.org. The MAPPs database is manually curated, with interactions derived from textbooks, review articles, and public pathway databases. Gen-MAPP also has the feature to construct and modify the pathways by the user, a quality that is not possible if analyzing pre-existing pathway databases like EcoCyc, MetaCyc, and KEGG. Gene identification (ID) from GenBank, SWISS-PROT, Gene Ontology, or other known databases is used to link the gene object on the MAPP to public databases like SWISS-PROT or Entrez Gene by selecting the gene of interest. In addition, GenMAPP displays gene expression levels and provides statistical analysis based on the representation of altered genes in a given pathway MAPP.
Software Tools to Analyze HTP Data
GoMiner8,9, MAPPFinder10, and EASE65 are software tools developed to correlate gene expression changes with GO terms to categorize the biological processes, cellular components, or molecular functions that are statistically affected. However, visualization of the pathway networks is challenging and complicated. Many software tools have been developed for microarray researchers to analyze large scale high-throughput data within the context of biological pathways, including the above mentioned Cytoscape and GenMAPP. Some of the most commonly used software tools are listed in Table 6. Here, we describe some of the freely available software tools that provide graphical representations of gene networks.
Table 6.
Software tools.
| Gene ontology (GO) analysis tools | Full name | Comments | Cost | Visualization | Knowledge-base | Website | Ref |
|---|---|---|---|---|---|---|---|
| MAPPFinder | MicroArray Pathway Profiles Finder | View data in the context of Gene Ontology (GO) and GenMAPP biological pathways. | Free | Y | GO | www.genmapp.org/MAPPFinder.html | [10] |
| GoMiner | - | Tool to classify gene onto the Gene Ontology (GO) hierarchy framework. | Free | Y | GO | discover.nci.nih.gov/gominer | [8] |
| EASE | Expression Analysis Systematic Explorer | Statistical tool to analyze gene list by GO. | Free | Y | GO | david.abcc.ncifcrf.gov | [65] |
| Onto-Express | - | Analyze the list of genes into GO hierarchy. | Free | Y | GO | vortex.cs.wayne.edu/Projects.html#Onto-Express | [11] |
| GoSurfer | - | Analyze gene list using GO and visualize them as a hierarchical tree. | Free | Y | GO | bioinformatics.bioen.uiuc. edu/gosurfer | [109] |
| FatiGO | Fast Assignment and Transference of Information | Web-based tool to analyze and compare GO terms in 2 sets of gene list. | Free | Y | GO | fatigo.bioinfo.cipf.es | [110] |
| Pathway analysis tools | Full name | Comments | Cost | Visualization | Knowledgebase | Website | Ref |
|---|---|---|---|---|---|---|---|
| PathwayExplorer | - | Web-based tool to visualize data on publicly available biological pathways. | Free | KEGG, Biocarta, GenMAPP | KEGG, Biocarta, Gen-MAPP | pathwayexplorer.genome. tugraz.at | [68] |
| WholePathway-Scope | - | Pathway-based analysis tool to visualize BioCarta, KEGG, and GO term relationships. | Free | KEGG, Biocarta, GO | KEGG, Bio-carta, GO | www.abcc.ncifcrf.gov/wps/wps_index.php | [67] |
| PathwayExpress | - | Analyze the list of genes in the context of KEGG pathways. | Free | KEGG | KEGG | vortex.cs.wayne.edu/Projects.html#Onto-Express | [12] |
| Pathway Processor | - | Visualization and statistical analysis | Free | KEGG | KEGG | sysbio.harvard.edu/csb | [66] |
| Reactome Sky-painter | - | Tool to calculate statistical significance of affected pathways and visualize pathways. | Free | Skypainter maps | Reactome | www.reactome.org/cgi-bin/skypainter2?DB=gk_current | [62] |
| Oncomine | - | Cancer profiling database and provides web-based tools to analyze data. | Free | Scalable Vector Graphics (SVG) | KEGG, BioCarta, HRD, SOURCE | www.oncomine.org | [111] |
| DAVID | Database for Annotation, Visualization and Integrated Discovery | Offers various functional annotation tools to analyze gene list. | Free | DAVID Pathway Viewer | BioCarta, KEGG | david.abcc.ncifcrf.gov/home.jsp | [112] |
| GenePattern | A platform for integrative genomics | Tools for statistical analysis of data. | Free | heatmaps and other tools. | Multiple tools. | www.broad.mit.edu/cancer/software/genepattern | [113] |
| VisANT | Visualization and analysis tool for biological networks and pathways | A web-based application for the predicted functional links between genes and proteins analysis of biological networks and pathways. | Free | nodes and edges | Predictome, MIPS, BIND | visant.bu.edu | [114] |
| FatiGO+ | Fast Assignment and Transference of Information | Compare distributions of GO or KEGG pathways between two groups of genes. | Free | KEGG | KEGG | babelomics.bioinfo.cipf. es/fatigoplus/cgi-bin/fatigoplus.cgi | [115] |
| PathwayStudio/PathwayAssist | - | Pathway analysis software. | License | Y | Manual curation | www.ariadnegenomics.com/products/pathway | [116] |
| Ingenuity Pathways Analysis | - | Pathway analysis software. | License | Y | Manual curation | www.ingenuity.com/products/pathways_analysis.html | N/A |
| MetaCore | - | Pathway analysis software. | License | Y | Manual curation | www.genego.com | N/A |
| PathArt | - | Pathway analysis software. | License | Y | Manual curation | jubilantbiosys.com/ppa.htm | N/A |
| Visualization tools | Full name | Comments | Cost | Visualization | Knowledge-base | Website | Ref |
|---|---|---|---|---|---|---|---|
| Cytoscape | - | Visualize molecular interactions and integrate gene expression profiles into pathways. | Free | Y | Retrieved from various databases. | www.cytoscape.org | [60] |
| GenMAPP | Gene Microarray Pathway Profiler | Visualize gene expression and genomic data on biological pathway maps. | Free | Y | Retrieved from various databases. | www.genmapp.org | [61] |
| Osprey | - | Graphical representation from Gene Ontology annotated interaction data by The GRID. | Y | The GRID | biodata.mshri.on.ca/osprey/servlet/Index | [117] | |
| CellDesigner | - | Software to draw pathways. | Y | Panther Classification System | www.celldesigner.org | [118] | |
| PathwayArchitect | - | Software for building pathways from databases or from own interaction data. | License | Y | - | www.stratagene.com/tradeshows/feature.aspx?fpId=90 | N/A |
Pathway Processor
Pathway Processor is designed to visualize whole genome microarray data in the framework of metabolic networks and provides statistical significance of the reliability of each differentially expressed gene66. This program displays data based on the information from the KEGG pathway database. Pathway Processor is implemented as two programs: Pathway Analyzer and Expression Mapper. Pathway Analyzer is the portion responsible for the statistical analysis of pathway significance, while Expression Mapper facilitates the visualization of this data on KEGG pathway maps.
Whole Pathway Scope
Whole Pathway Scope (WPS) is a software tool to analyze high-throughput microarray experiments by referencing pathway or gene information from KEGG, BioCarta, and Gene Ontology67. The internal database also includes information from the Genetic Association Database and MedGene Database to allow users to rapidly identify disease-associated genes and highlight them inside their network diagram or select them for further network manipulation. One of the key features is the ability to view multiple experiments simultaneously and color-code the expression value with its p-value. In addition, this software allows users to customize their own metabolic pathway and gene groupings with the option of using statistical analysis.
Pathway Explorer
Pathway Explorer is a web-based service available at https://pathwayexplorer.genome.tugraz.at to map expression profiles of genes onto pathway maps extracted from KEGG, BioCarta, and Gen-MAPP68. This web-based service reduces the local requirement for computational resources. It offers customizable analysis of the data by analyzing in a single or multiple pathways, and a right-tailed Fisher’s exact test and false discovery rate analysis were applied to determine the significance of the different pathways. Multiple experiments can also be displayed simultaneously on a single pathway with corresponding expression values. Data is linked to publicly available biological databases (e.g. the NCBI Entrez cross-database search, OMIM, KEGG pathways). The online accessibility of PathwayExplorer enables visualization of DNA or gene expression profiles within the context of biological pathways in a rapid manner.
Future Considerations
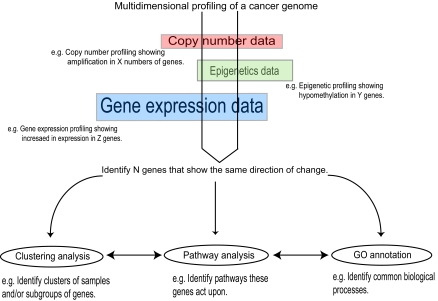
The development of various computational tools to interrogate biological databases is accelerating the process to understand high-throughput genomic studies. However, these new tools pose new challenges, and one must be cautious about the limitations and errors associated with various databases. For example, it has been reported that when a partial Enzyme Commission (EC) number, which is a combination of four digits to annotate enzymatic activities without the fourth digit, is assigned to a gene, several pathway databases have used partial EC number annotations and inaccurately assigned them to a set of reactions that are associated with the same partial EC number under each orthology group69. Pathway database users should be aware of the possible inherent problems associated with any databases due to the variable quality of the published data. Comprehensive examination of the literature, as well as additional experimental validation, should be used to confirm any findings. Cross-platform integrative analysis of genomics, epigenomics, and transcriptional profiling will offer a deeper understanding of the biological complexity underlying disease processes (Fig. 5)70. The current challenge is to incorporate these data together for direct comparison, visualization, and analysis in order to identify salient gene candidates71. Once this is accomplished, the next step will be to place these candidates in the context of their proper signaling pathways for a given cancer type. Ultimately, the software programs used to do this should be intuitive to use, provide accurate information, allow customizable analyses, and offer sophisticated statistical tools. All of these features will be essential for characterization of disrupted gene networks in cancer. This will set the stage for rational therapeutic selection based on the underlying genetic realties of a specific tumor38,41.
Figure 5.
Genome-wide integrative analysis to identify pathways disrupted in cancer. Genome-wide analyses including copy number profiling, epigenetic profiling, and transcription profiling performed on the same cancer sample could narrow down the number of candidate genes, which would in turn help to pinpoint disrupted pathway involved in cancer.
Acknowledgments
The authors thank Bradley P. Coe and Ian M. Wilson for critical discussion. This work was supported by funds from National Institute of Dental and Craniofacial Research (NIDCR) grants R01 DE15965 and R01 DE13124, Genome Canada/Genome British Columbia, and the Canadian Institutes of Health Research. IFLT is supported by the Roman M. Babicki fellowship. RC and TPHB are supported by scholarships from the Canadian Institutes of Health Research and Michael Smith Foundation for Health Research.
Footnotes
Copyright in this article, its metadata, and any supplementary data is held by its author or authors. It is published under the Creative Commons Attribution By licence. For further information go to: http://creativecommons.org/licenses/by/3.0/.
References
- [1].Alfarano C, et al. 2005The Biomolecular Interaction Network Database and related tools 2005 update Nucleic Acids Res 33Database issueD418–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- [3].Al-Shahrour F, et al. FatiGO +: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20(4):578–80. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- [5].Andreeva A, et al. 2004SCOP database in 2004: refinements integrate structure and sequence family data Nucleic Acids Res 32Database issueD226–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bader GD, Cary MP, Sander C.2006Pathguide: a pathway resource list Nucleic Acids Res 34Database issueD504–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28(1):304–5. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Baldi P, Hatfield GW. Cambridge: Cambridge University Press; 2002. DNA Microarrays and Gene Expression: From Experiments to Data Analysis and Modeling. [Google Scholar]
- [10].Barrett T, et al. 2007NCBI GEO: mining tens of millions of expression profiles—database and tools update Nucleic Acids Res 35Database issueD760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Barthelmes J, et al. 2007BRENDA, AMENDA and FRENDA: the enzyme information system in 2007 Nucleic Acids Res 35Database issueD511–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Baudis M, Cleary ML. Progenetix.net: an online repository for molecular cytogenetic aberration data. Bioinformatics. 2001;17(12):1228–9. doi: 10.1093/bioinformatics/17.12.1228. [DOI] [PubMed] [Google Scholar]
- [13].Benson DA, et al. 2007GenBank Nucleic Acids Res 35Database issueD21–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Beuming T, et al. PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics. 2005;21(6):827–8. doi: 10.1093/bioinformatics/bti098. [DOI] [PubMed] [Google Scholar]
- [15].Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. U.S.A. 2001;98(24):13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- [17].Boutros PC, Okey AB. Unsupervised pattern recognition: an introduction to the whys and wherefores of clustering microarray data. Brief Bioinform. 2005;6(4):331–43. doi: 10.1093/bib/6.4.331. [DOI] [PubMed] [Google Scholar]
- [18].Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4(3):R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21(9):2076–82. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- [20].Brown MP, et al. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. U.S.A. 2000;97(1):262–7. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brunak S, et al. Nucleotide sequence database policies. Science. 2002;298(5597):1333. doi: 10.1126/science.298.5597.1333b. [DOI] [PubMed] [Google Scholar]
- [22].Cary MP, Bader GD, Sander C. Pathway information for systems biology. FEBS Lett. 2005;579(8):1815–20. doi: 10.1016/j.febslet.2005.02.005. [DOI] [PubMed] [Google Scholar]
- [23].Caspi R, et al. 2006MetaCyc: a multiorganism database of metabolic pathways and enzymes Nucleic Acids Res 34Database issueD511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ceol A, et al. 2007DOMINO: a database of domain-peptide interactions Nucleic Acids Res 35Database issueD557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chari R, et al. SIGMA: a system for integrative genomic microarray analysis of cancer genomes. BMC Genomics. 2006;7:324. doi: 10.1186/1471-2164-7-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chatr-aryamontri A, et al. 2007MINT: the Molecular INTeraction database Nucleic Acids Res 35Database issueD572–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10(6):529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- [28].Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell. Biol. 2006;7(7):505–16. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- [29].Clare A, King RD. How well do we understand the clusters found in microarray data? In. Silico. Biol. 2002;2(4):511–22. [PubMed] [Google Scholar]
- [30].Cochrane G, et al. 2006EMBL Nucleotide Sequence Database: developments in 2005 Nucleic Acids Res 34Database issueD10–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Coe BP, et al. Differential disruption of cell cycle pathways in small cell and non-small cell lung cancer. Br. J. Cancer. 2006;94(12):1927–35. doi: 10.1038/sj.bjc.6603167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dahlquist KD, et al. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002;31(1):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- [33].Datta S, Datta S. Comparisons and validation of statistical clustering techniques for microarray gene expression data. Bioinformatics. 2003;19(4):459–66. doi: 10.1093/bioinformatics/btg025. [DOI] [PubMed] [Google Scholar]
- [34].Demeter J, et al. 2007The Stanford Microarray Database: implementation of new analysis tools and open source release of software Nucleic Acids Res 35Database issueD766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome. Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- [36].D’Haeseleer P. How does gene expression clustering work? Nat. Biotechnol. 2005;23(12):1499–501. doi: 10.1038/nbt1205-1499. [DOI] [PubMed] [Google Scholar]
- [37].Doniger SW, et al. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome. Biol. 2003;4(1):R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Draghici S, et al. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31(13):3775–81. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fernandez JM, Hoffmann R, Valencia A. iHOP web services. Nucleic Acids Res. 2007. [DOI] [PMC free article] [PubMed]
- [41].Fleischmann A, et al. 2004IntEnz, the integrated relational enzyme database Nucleic Acids Res 32Database issueD434–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fox JA, McMillan S, Ouellette BF.2006A compilation of molecular biology web servers: 2006 update on the Bioinformatics Links Directory Nucleic Acids Res 34Web Server issueW3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Funahashi A, et al. Cell Designer: a process diagram editor for gene-regulatory and biochemical networks. BIOSILICO. 2003;1:159–162. [Google Scholar]
- [44].Galperin MY.2007The Molecular Biology Database Collection: 2007 update Nucleic Acids Res 35Database issueD3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Garcia O, et al. GOlorize: a Cytoscape plug-in for network visualization with Gene Ontology-based layout and coloring. Bioinformatics. 2007;23(3):394–6. doi: 10.1093/bioinformatics/btl605. [DOI] [PubMed] [Google Scholar]
- [46].Garnis C, Buys TP, Lam WL. Genetic alteration and gene expression modulation during cancer progression. Mol. Cancer. 2004;3:9. doi: 10.1186/1476-4598-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Golub TR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- [48].Gough NR, Adler EM, Ray LB. Cell signaling: from beginning to end. Sci. STKE. 2005;2005(305):9. doi: 10.1126/stke.3052005eg9. [DOI] [PubMed] [Google Scholar]
- [49].Green ML, Karp PD. Genome annotation errors in pathway databases due to semantic ambiguity in partial EC numbers. Nucleic Acids Res. 2005;33(13):4035–9. doi: 10.1093/nar/gki711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Grosu P, et al. Pathway Processor: a tool for integrating whole-genome expression results into metabolic networks. Genome. Res. 2002;12(7):1121–6. doi: 10.1101/gr.226602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hamosh A, et al. 2005Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders Nucleic Acids Res 33Database issueD514–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Han K, et al. HPID: the Human Protein Interaction Database. Bioinformatics. 2004;20(15):2466–70. doi: 10.1093/bioinformatics/bth253. [DOI] [PubMed] [Google Scholar]
- [53].Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- [54].Hastie T, et al. Supervised harvesting of expression trees. Genome Biol. 2001;2(1):RESEARCH0003. doi: 10.1186/gb-2001-2-1-research0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hedenfalk I, et al. Gene-expression profiles in hereditary breast cancer. N. Engl. J. Med. 2001;344(8):539–48. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- [56].Hermjakob H, et al. The HUPO PSI’s molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22(2):177–83. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- [57].Hosack DA, et al. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hu Z, et al. 2005VisANT: data-integrating visual framework for biological networks and modules Nucleic Acids Res 33Web Server issueW352–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hubbard TJ, et al. 2007Ensembl 2007 Nucleic Acids Res 35Database issueD610–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jolliffe I. Principal Component Analysis. Springer. 2002:487. [Google Scholar]
- [61].Jonsson PF, et al. Cluster analysis of networks generated through homology: automatic identification of important protein communities involved in cancer metastasis. BMC Bioinformatics. 2006;7:2. doi: 10.1186/1471-2105-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kanehisa M, et al. 2006From genomics to chemical genomics: new developments in KEGG Nucleic Acids Res 34Database issueD354–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kato K, et al. 2005Cancer gene expression database (CGED): a database for gene expression profiling with accompanying clinical information of human cancer tissues Nucleic Acids Res 33Database issueD533–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kerrien S, et al. 2007IntAct—open source resource for molecular interaction data Nucleic Acids Res 35Database issueD561–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Khatri P, et al. Profiling gene expression using onto-express. Genomics. 2002;79(2):266–70. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- [66].Kouranov A, et al. 2006The RCSB PDB information portal for structural genomics Nucleic Acids Res 34Database issueD302–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kuhn RM, et al. 2007The UCSC genome browser database: update 2007 Nucleic Acids Res 35Database issueD668–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Maglott D, et al. 2007Entrez Gene: gene-centered information at NCBI Nucleic Acids Res 35Database issueD26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mewes HW, et al. 2006MIPS: analysis and annotation of proteins from whole genomes in 2005 Nucleic Acids Res 34Database issueD169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mi H, et al. 2007PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways Nucleic Acids Res 35Database issueD247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mishra GR, et al. 2006Human protein reference database—2006 update Nucleic Acids Res 34Database issueD411–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mitelman F, Johansson B, Mertens F, editors. Mitelman Database of Chromosome Aberrations in Cancer 2007. http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- [73].Mlecnik B, et al. 2005PathwayExplorer: web service for visualizing high-throughput expression data on biological pathways Nucleic Acids Res 33Web Server issueW633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ng SK, et al. InterDom: a database of putative interacting protein domains for validating predicted protein interactions and complexes. Nucleic Acids Res. 2003;31(1):251–4. doi: 10.1093/nar/gkg079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nikitin A, et al. Pathway studio—the analysis and navigation of molecular networks. Bioinformatics. 2003;19(16):2155–7. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- [76].Oda K, et al. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst Biol. 2005;1:2005–10. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Pagel P, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21(6):832–4. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- [78].Parkinson H, et al. 2005ArrayExpress—a public repository for microarray gene expression data at the EBI Nucleic Acids Res 33Database issueD553–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- [80].Persico M, et al. HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC Bioinformatics. 2005;6(Suppl 4):S21. doi: 10.1186/1471-2105-6-S4-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Pomeroy SL, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415(6870):436–42. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- [82].Pruitt KD, Tatusova T, Maglott DR.2007NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins Nucleic Acids Res 35Database issueD61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ramaswamy S, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl. Acad. Sci. U.S.A. 2001;98(26):15149–54. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38(5):500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- [85].Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Salwinski L, Eisenberg D. The MiSink Plugin: Cytoscape as a graphical interface to the Database of Interacting Proteins. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btm304. [DOI] [PubMed] [Google Scholar]
- [87].Salwinski L, et al. 2004The Database of Interacting Proteins: 2004 update Nucleic Acids Res 32Database issueD449–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Santos C, Eggle D, States DJ. Wnt pathway curation using automated natural language processing: combining statistical methods with partial and full parse for knowledge extraction. Bioinformatics. 2005;21(8):1653–8. doi: 10.1093/bioinformatics/bti165. [DOI] [PubMed] [Google Scholar]
- [89].Segal E, et al. A module map showing conditional activity of expression modules in cancer. Nat. Genet. 2004;36(10):1090–8. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- [90].Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome. Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Simon RM, et al. Design and Analysis of DNA Microarray Investigations. New York, NY: Springer-Verlag; 2003. [Google Scholar]
- [92].Simon RM, McShane KE, Radmacher LM, Wright MD, Zhao GWY. Design and Analysis of DNA Microarray Investigations. New York, NY: Springer-Verlag; 2003. [Google Scholar]
- [93].Skusa A, Ruegg A, Kohler J. Extraction of biological interaction networks from scientific literature. Brief Bioinform. 2005;6(3):263–76. doi: 10.1093/bib/6.3.263. [DOI] [PubMed] [Google Scholar]
- [94].Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Speed T. Statistical Analysis of Gene Expression Microarray Data. Boca Raton, FL: Chapman and Hall/CRC; 2003. [Google Scholar]
- [96].Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9(12):3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Stark C, et al. 2006BioGRID: a general repository for interaction datasets Nucleic Acids Res 34Database issueD535–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Stekel D. Microarray Bioinformatics. In: Southern E, editor. Cambridge University Press. 2003. p. 280. [Google Scholar]
- [99].Strausberg RL, et al. The cancer genome anatomy project: online resources to reveal the molecular signatures of cancer. Cancer Invest. 2002;20(7–8):1038–50. doi: 10.1081/cnv-120005922. [DOI] [PubMed] [Google Scholar]
- [100].Su AI, et al. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61(20):7388–93. [PubMed] [Google Scholar]
- [101].Sugawara H, et al. 2007DDBJ working on evaluation and classification of bacterial genes in INSDC Nucleic Acids Res 35Database issueD13–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Tamayo P, et al. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. U.S.A. 1999;96(6):2907–12. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Tavazoie S, et al. Systematic determination of genetic network architecture. Nat. Genet. 1999;22(3):281–5. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- [104].The Gene Ontology (GO) project in 2006 Nucleic Acids Res 34Database issueD322–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol. Biol. 2005;311:179–91. doi: 10.1385/1-59259-957-5:179. [DOI] [PubMed] [Google Scholar]
- [106].van Helden J, et al. From molecular activities and processes to biological function. Brief Bioinform. 2001;2(1):81–93. doi: 10.1093/bib/2.1.81. [DOI] [PubMed] [Google Scholar]
- [107].Vastrik I, et al. Reactome: a knowledge base of biologic pathways and processes. Genome. Biol. 2007;8(3):R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat. Med. 2004;10(8):789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- [109].von Mering C, et al. 2007STRING 7—recent developments in the integration and prediction of protein interactions Nucleic Acids Res 35Database issueD358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Warner GJ, Adeleye YA, Ideker T. Interactome networks: the state of the science. Genome. Biol. 2006;7(1):301. doi: 10.1186/gb-2006-7-1-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Wheeler DL, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31(1):28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wu CH, et al. The Protein Information Resource. Nucleic Acids Res. 2003;31(1):345–7. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wu CH, et al. 2006The Universal Protein Resource (UniProt): an expanding universe of protein information Nucleic Acids Res 34Database issueD187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Yan B, et al. Genome-wide identification of novel expression signatures reveal distinct patterns and prevalence of binding motifs for p53, NF-kappaB and other signal transcription factors in head and neck squamous cell carcinoma. Genome. Biol. 2007;8(5):R78. doi: 10.1186/gb-2007-8-5-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Yi M, et al. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zeeberg BR, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4(4):R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zeeberg BR, et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhong S, et al. GoSurfer: a graphical interactive tool for comparative analysis of large gene sets in Gene Ontology space. Appl Bioinformatics. 2004;3(4):261–4. doi: 10.2165/00822942-200403040-00009. [DOI] [PubMed] [Google Scholar]