Abstract
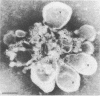
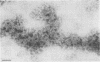
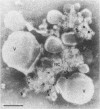
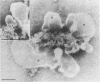
Two populations of membrane-bound replication complexes were isolated from poliovirus-infected HEp-2 cells by sucrose gradient centrifugation. The two fractions show similar ultrastructural features: the replication complex is enclosed in a rosettelike shell of virus-induced vesicles and contains a very tightly packed second membrane system (compact membranes). The vesicular fraction, which bands in 30% sucrose, contains replicative intermediate (RI) and 36S RNA. The fraction banding in 45% sucrose contains only minute amounts of RI and contains mainly 36S RNA, two-thirds of which is encapsidated. In vitro, the two fractions show similar RNA synthesizing capacities and produce 36S plus-strand RNA. Dissolving the membranes within and around synthetically active replication complexes with sodium deoxycholate abolishes the completion of 36S RNA but still allows elongation in the RI. Our findings suggest an architecture of the replication complex that has the nascent plus strands on the RI enclosed in the compact membranes and the replication forks wrapped additionally in protein. Plus-strand RNA can be localized by in situ hybridization with a biotinylated riboprobe between the replication complex and the rosette of the virus-induced vesicles. It was found that the progeny RNA strands are set free soon after completion from the replication complex at the sites where the compact membranes within the replication complex are in close contact with the surrounding virus-induced vesicles.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. Structure of the poliovirus replicative intermediate RNA. J Mol Biol. 1968 Mar 14;32(2):359–368. doi: 10.1016/0022-2836(68)90015-6. [DOI] [PubMed] [Google Scholar]
- Bendayan M., Puvion E. Ultrastructural detection of RNA: complementarity of high-resolution autoradiography and of RNAase-gold method. J Ultrastruct Res. 1983 Jun;83(3):274–283. doi: 10.1016/s0022-5320(83)90134-x. [DOI] [PubMed] [Google Scholar]
- Bienz K., Egger D., Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987 Sep;160(1):220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- Bienz K., Egger D., Rasser Y., Bossart W. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology. 1983 Nov;131(1):39–48. doi: 10.1016/0042-6822(83)90531-7. [DOI] [PubMed] [Google Scholar]
- Bienz K., Egger D., Rasser Y., Bossart W. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology. 1980 Jan 30;100(2):390–399. doi: 10.1016/0042-6822(80)90530-9. [DOI] [PubMed] [Google Scholar]
- Bienz K., Egger D., Troxler M., Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J Virol. 1990 Mar;64(3):1156–1163. doi: 10.1128/jvi.64.3.1156-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Glynias M. J., Merrick W. C. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drzeniek R., Bilello P. Absence of glycoproteins in poliovirus particles. J Gen Virol. 1974 Oct;25(1):125–132. doi: 10.1099/0022-1317-25-1-125. [DOI] [PubMed] [Google Scholar]
- Emerson S. U., McRill C., Rosenblum B., Feinstone S., Purcell R. H. Mutations responsible for adaptation of hepatitis A virus to efficient growth in cell culture. J Virol. 1991 Sep;65(9):4882–4886. doi: 10.1128/jvi.65.9.4882-4886.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison D., Ehrenfeld E. Comparison of replication complexes synthesizing poliovirus RNA. Virology. 1981 May;111(1):33–46. doi: 10.1016/0042-6822(81)90651-6. [DOI] [PubMed] [Google Scholar]
- Girard M. In vitro synthesis of poliovirus ribonucleic acid: role of the replicative intermediate. J Virol. 1969 Apr;3(4):376–384. doi: 10.1128/jvi.3.4.376-384.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Wolf Y. I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990 Mar 12;262(1):145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- Johnson K. L., Sarnow P. Three poliovirus 2B mutants exhibit noncomplementable defects in viral RNA amplification and display dosage-dependent dominance over wild-type poliovirus. J Virol. 1991 Aug;65(8):4341–4349. doi: 10.1128/jvi.65.8.4341-4349.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon S. M., Murphy P. C., Shields P. A., Ping L. H., Feinstone S. M., Cromeans T., Jansen R. W. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991 Apr;65(4):2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. P., Baltimore D. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J Virol. 1990 Mar;64(3):1102–1107. doi: 10.1128/jvi.64.3.1102-1107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. P., Baltimore D. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J Virol. 1988 Nov;62(11):4016–4021. doi: 10.1128/jvi.62.11.4016-4021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax N. B., Yin F. H. Evidence for the role of the P2 protein of human rhinovirus in its host range change. J Virol. 1989 May;63(5):2396–2399. doi: 10.1128/jvi.63.5.2396-2399.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla A., Paul A. V., Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991 Dec 13;254(5038):1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- Pasamontes L., Egger D., Bienz K. Production of monoclonal and monospecific antibodies against non-capsid proteins of poliovirus. J Gen Virol. 1986 Nov;67(Pt 11):2415–2422. doi: 10.1099/0022-1317-67-11-2415. [DOI] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981 Nov 20;214(4523):916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- Takeda N., Kuhn R. J., Yang C. F., Takegami T., Wimmer E. Initiation of poliovirus plus-strand RNA synthesis in a membrane complex of infected HeLa cells. J Virol. 1986 Oct;60(1):43–53. doi: 10.1128/jvi.60.1.43-53.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegami T., Semler B. L., Anderson C. W., Wimmer E. Membrane fractions active in poliovirus RNA replication contain VPg precursor polypeptides. Virology. 1983 Jul 15;128(1):33–47. doi: 10.1016/0042-6822(83)90316-1. [DOI] [PubMed] [Google Scholar]
- Troxler M., Pasamontes L., Egger D., Bienz K. In situ hybridization for light and electron microscopy: a comparison of methods for the localization of viral RNA using biotinylated DNA and RNA probes. J Virol Methods. 1990 Oct;30(1):1–14. doi: 10.1016/0166-0934(90)90039-i. [DOI] [PubMed] [Google Scholar]
- Young D. C., Tuschall D. M., Flanegan J. B. Poliovirus RNA-dependent RNA polymerase and host cell protein synthesize product RNA twice the size of poliovirion RNA in vitro. J Virol. 1985 May;54(2):256–264. doi: 10.1128/jvi.54.2.256-264.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]