Abstract
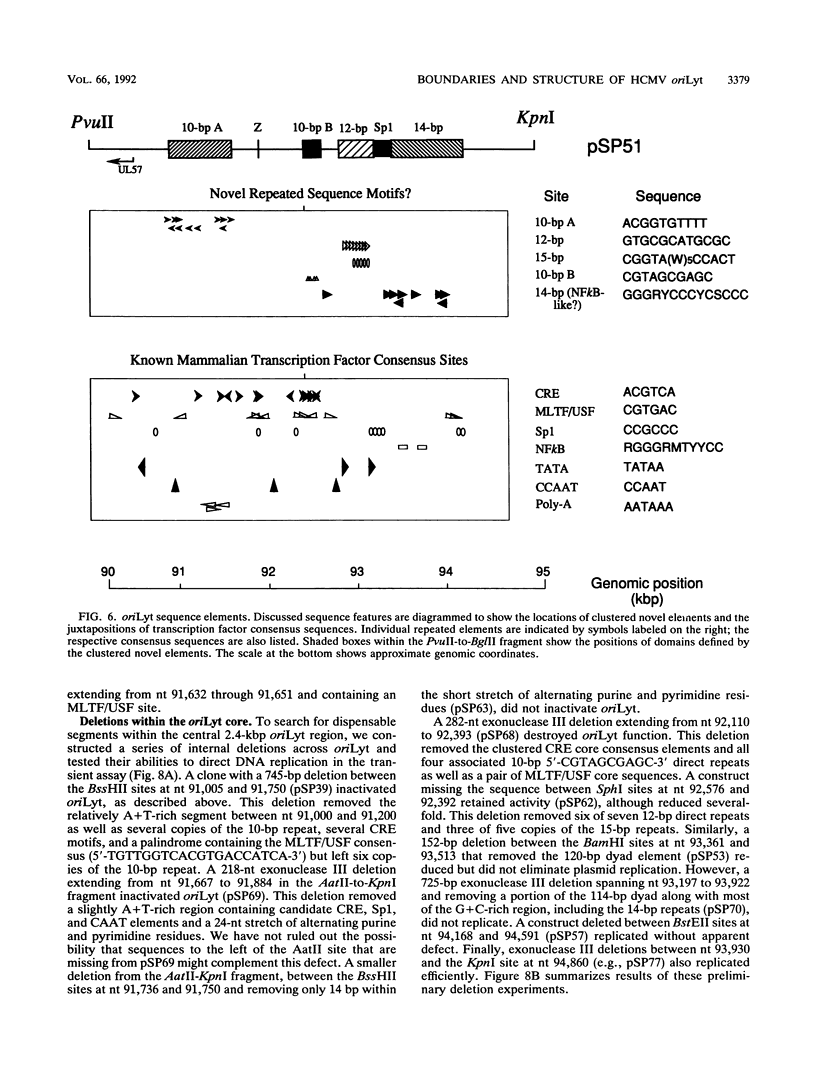
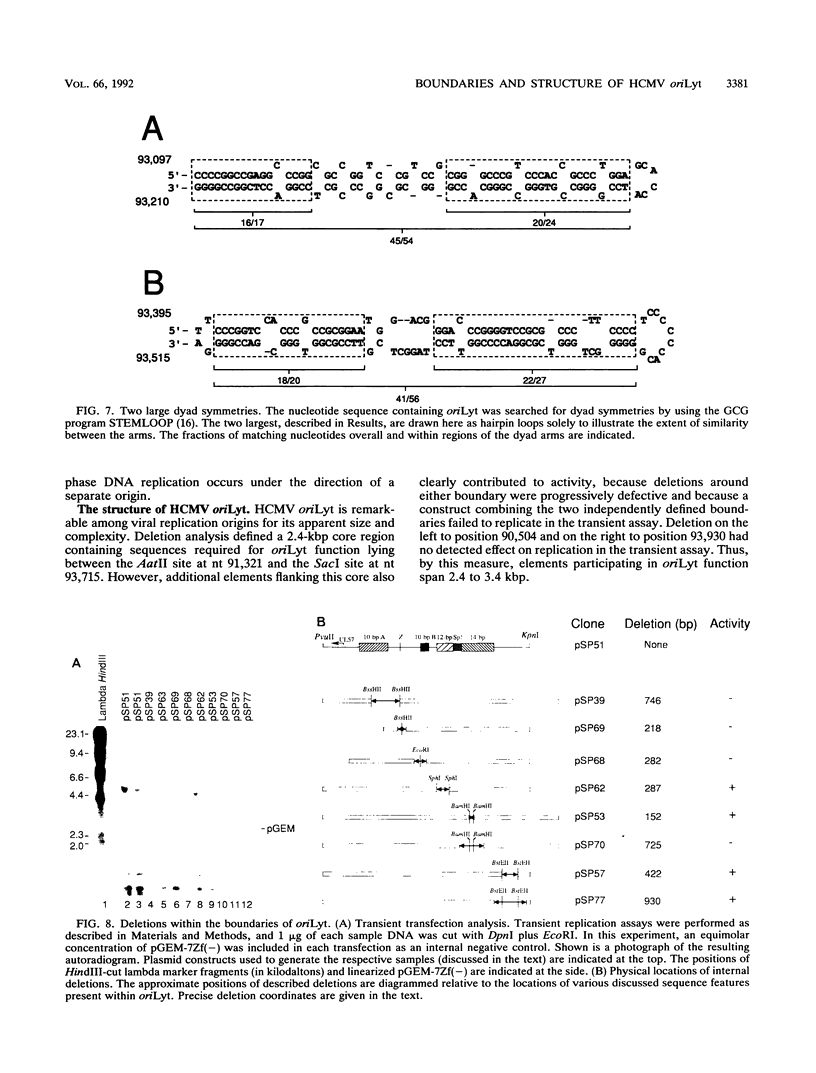
We have localized a cis-acting sequence that promotes initiation of lytic-phase DNA replication (oriLyt) within the HindIII D fragment of the human cytomegalovirus (HCMV) AD169 genome and investigated its sequence requirements by testing the ability of plasmid constructs to mediate DNA replication in a transient transfection-plus-infection assay. Replication of plasmids containing HCMV oriLyt required at least the virus-specified DNA polymerase activity supplied by HCMV infection of transfected cells and was autonomous in that it did not result from recombination with the virus genome. Progeny molecules in the transient assay were high-molecular-weight tandem oligomers, which is consistent with predictions of a rolling-circle model. Experiments testing subclones of HindIII-D defined a core 2.4-kbp region containing elements required for oriLyt function that extended rightward from around 1.0 kbp upstream of UL57 near the middle of the long unique component of the virus genome. Sequences flanking this core also were needed for full activity. The defined region contains at least four clustered sets of repeated sequence elements identical to or candidate counterparts of elements present in the corresponding cytomegalovirus Colburn lytic-phase replication origin. These elements are novel in that they apparently do not correspond to previously characterized motifs. Also present are multiple copies of elements similar to known binding sites for the transcription factors ATF/CREB, MLTF/USF, and Sp1. Preliminary deletion analysis suggests that multiple components within the boundaries of oriLyt cooperate to enable initiation of HCMV lytic-phase DNA synthesis.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anders D. G., Gibson W. Location, transcript analysis, and partial nucleotide sequence of the cytomegalovirus gene encoding an early DNA-binding protein with similarities to ICP8 of herpes simplex virus type 1. J Virol. 1988 Apr;62(4):1364–1372. doi: 10.1128/jvi.62.4.1364-1372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders D. G. Nucleotide sequence of a cytomegalovirus single-stranded DNA-binding protein gene: comparison with alpha- and gammaherpesvirus counterparts reveals conserved segments. J Gen Virol. 1990 Oct;71(Pt 10):2451–2456. doi: 10.1099/0022-1317-71-10-2451. [DOI] [PubMed] [Google Scholar]
- Anders D. G., Punturieri S. M. Multicomponent origin of cytomegalovirus lytic-phase DNA replication. J Virol. 1991 Feb;65(2):931–937. doi: 10.1128/jvi.65.2.931-937.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Baran N., Lapidot A., Manor H. Formation of DNA triplexes accounts for arrests of DNA synthesis at d(TC)n and d(GA)n tracts. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):507–511. doi: 10.1073/pnas.88.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron K. K., Stanat S. C., Sorrell J. B., Fyfe J. A., Keller P. M., Lambe C. U., Nelson D. J. Metabolic activation of the nucleoside analog 9-[( 2-hydroxy-1-(hydroxymethyl)ethoxy]methyl)guanine in human diploid fibroblasts infected with human cytomegalovirus. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2473–2477. doi: 10.1073/pnas.82.8.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Bruckner R. C., Crute J. J., Dodson M. S., Lehman I. R. The herpes simplex virus 1 origin binding protein: a DNA helicase. J Biol Chem. 1991 Feb 5;266(4):2669–2674. [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Challberg M. D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Chee M. S., Bankier A. T., Beck S., Bohni R., Brown C. M., Cerny R., Horsnell T., Hutchison C. A., 3rd, Kouzarides T., Martignetti J. A. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Chee M. The HCMV genome project: what has been learned and what can be expected in the future. Transplant Proc. 1991 Jun;23(3 Suppl 3):174-80, discussion 180. [PubMed] [Google Scholar]
- DePamphilis M. L. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988 Mar 11;52(5):635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias P., O'Donnell M. E., Mocarski E. S., Lehman I. R. A DNA binding protein specific for an origin of replication of herpes simplex virus type 1. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6322–6326. doi: 10.1073/pnas.83.17.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein B., Müller I., Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982 Apr;18(1):39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco M. A., Cullen B. R. Molecular basis of latency in pathogenic human viruses. Science. 1991 Nov 8;254(5033):815–820. doi: 10.1126/science.1658933. [DOI] [PubMed] [Google Scholar]
- Ghazal P., Young J., Giulietti E., DeMattei C., Garcia J., Gaynor R., Stenberg R. M., Nelson J. A. A discrete cis element in the human immunodeficiency virus long terminal repeat mediates synergistic trans activation by cytomegalovirus immediate-early proteins. J Virol. 1991 Dec;65(12):6735–6742. doi: 10.1128/jvi.65.12.6735-6742.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D. A relational database of transcription factors. Nucleic Acids Res. 1990 Apr 11;18(7):1749–1756. doi: 10.1093/nar/18.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W. Protein counterparts of human and simian cytomegaloviruses. Virology. 1983 Jul 30;128(2):391–406. doi: 10.1016/0042-6822(83)90265-9. [DOI] [PubMed] [Google Scholar]
- Gibson W. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology. 1981 Jun;111(2):516–537. doi: 10.1016/0042-6822(81)90354-8. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W., Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988 Nov 4;55(3):427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- Hamzeh F. M., Lietman P. S., Gibson W., Hayward G. S. Identification of the lytic origin of DNA replication in human cytomegalovirus by a novel approach utilizing ganciclovir-induced chain termination. J Virol. 1990 Dec;64(12):6184–6195. doi: 10.1128/jvi.64.12.6184-6195.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronn R., Jahn G., Bürkle A., Freese U. K., Fleckenstein B., zur Hausen H. Genomic localization, sequence analysis, and transcription of the putative human cytomegalovirus DNA polymerase gene. J Virol. 1987 Jan;61(1):119–124. doi: 10.1128/jvi.61.1.119-124.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgstrand E., Eriksson B., Johansson N. G., Lannerö B., Larsson A., Misiorny A., Norén J. O., Sjöberg B., Stenberg K., Stening G. Trisodium phosphonoformate, a new antiviral compound. Science. 1978 Sep 1;201(4358):819–821. doi: 10.1126/science.210500. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Gompels U. A., Barrell B. G., Craxton M., Cameron K. R., Staden R., Chang Y. N., Hayward G. S. Deviations from expected frequencies of CpG dinucleotides in herpesvirus DNAs may be diagnostic of differences in the states of their latent genomes. J Gen Virol. 1989 Apr;70(Pt 4):837–855. doi: 10.1099/0022-1317-70-4-837. [DOI] [PubMed] [Google Scholar]
- Ibanez C. E., Schrier R., Ghazal P., Wiley C., Nelson J. A. Human cytomegalovirus productively infects primary differentiated macrophages. J Virol. 1991 Dec;65(12):6581–6588. doi: 10.1128/jvi.65.12.6581-6588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble G. W., McCormick A. L., Pereira L., Mocarski E. S. A cytomegalovirus protein with properties of herpes simplex virus ICP8: partial purification of the polypeptide and map position of the gene. J Virol. 1987 Oct;61(10):3143–3151. doi: 10.1128/jvi.61.10.3143-3151.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher K. M., Spector D. H. The human cytomegalovirus 2.7-kilobase RNA promoter contains a functional binding site for the adenovirus major late transcription factor. J Virol. 1990 Sep;64(9):4189–4198. doi: 10.1128/jvi.64.9.4189-4198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A., Schwedes J. F., Tegtmeyer P. Herpes simplex virus origin-binding protein (UL9) loops and distorts the viral replication origin. J Virol. 1991 Jun;65(6):3284–3292. doi: 10.1128/jvi.65.6.3284-3292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A., Tegtmeyer P. Characterization of major recognition sequences for a herpes simplex virus type 1 origin-binding protein. J Virol. 1988 Nov;62(11):4096–4103. doi: 10.1128/jvi.62.11.4096-4103.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., Bankier A. T., Satchwell S. C., Weston K., Tomlinson P., Barrell B. G. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J Virol. 1987 Jan;61(1):125–133. doi: 10.1128/jvi.61.1.125-133.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. A., Hai T. Y., SivaRaman L., Thimmappaya B., Hurst H. C., Jones N. C., Green M. R. A cellular protein, activating transcription factor, activates transcription of multiple E1A-inducible adenovirus early promoters. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8355–8359. doi: 10.1073/pnas.84.23.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Dolan A., McNab D., Perry L. J., Taylor P., Challberg M. D. Structures of herpes simplex virus type 1 genes required for replication of virus DNA. J Virol. 1988 Feb;62(2):444–453. doi: 10.1128/jvi.62.2.444-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr I. J., Clark R., Sun S., Androphy E. J., MacPherson P., Botchan M. R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990 Dec 21;250(4988):1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- Montminy M. R., Bilezikjian L. M. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987 Jul 9;328(6126):175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- Olivo P. D., Nelson N. J., Challberg M. D. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5414–5418. doi: 10.1073/pnas.85.15.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge D. S. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci. 1991 Apr;7(2):203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- Reisman D., Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986 Nov;6(11):3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Stow N. D., Davison A. J. Identification of a varicella-zoster virus origin of DNA replication and its activation by herpes simplex virus type 1 gene products. J Gen Virol. 1986 Aug;67(Pt 8):1613–1623. doi: 10.1099/0022-1317-67-8-1613. [DOI] [PubMed] [Google Scholar]
- Stow N. D. Localization of an origin of DNA replication within the TRS/IRS repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1(7):863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988 Feb 26;52(4):559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss J. H., Roizman B. Properties of two 5'-coterminal RNAs transcribed part way and across the S component origin of DNA synthesis of the herpes simplex virus 1 genome. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8454–8458. doi: 10.1073/pnas.85.22.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir H. M., Calder J. M., Stow N. D. Binding of the herpes simplex virus type 1 UL9 gene product to an origin of viral DNA replication. Nucleic Acids Res. 1989 Feb 25;17(4):1409–1425. doi: 10.1093/nar/17.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Spadaro A., Schaffer J. E., Murray A. W., Maxam A. M., Schaffer P. A. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol Cell Biol. 1985 May;5(5):930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. D. Unusual DNA structures. J Biol Chem. 1988 Jan 25;263(3):1095–1098. [PubMed] [Google Scholar]
- Wong S. W., Schaffer P. A. Elements in the transcriptional regulatory region flanking herpes simplex virus type 1 oriS stimulate origin function. J Virol. 1991 May;65(5):2601–2611. doi: 10.1128/jvi.65.5.2601-2611.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. A., Nelson N. J., McGeoch D. J., Challberg M. D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988 Feb;62(2):435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]