Abstract
In general, the transcriptional competence of a chromatin domain is correlated with increased sensitivity to DNase I cleavage. A recent observation that actively transcribing RNA polymerase II piggybacks a histone acetyltranferase activity [Wittschieben, B., Otero, G., de Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., Li, Y., Allis, C. D., Tempst, P. & Svejstrup, J. Q. (1999) Mol. Cell 4, 123–128] implies that the state of histone acetylation, and hence the ability of chromatin to fold, can be altered by a processive mechanism. In this article, it is proposed that tracking-mediated chromatin modification could create and/or maintain an open configuration in a complete chromatin domain including both intra- and extragenic regions. This mechanism suggests a putative functional role for the extragenic transcription observed at the β-globin and other loci in vertebrate cells.
The transcriptional competence of eukaryotic chromatin requires access both for the activating gene-specific factors and for RNA polymerase II and its associated protein complexes. The reorganization of chromatin associated with activation can be mediated by large remodeling complexes and is also strongly correlated with histone hyperacetylation (1–3). The state of histone modification can modulate both the access of the transcription machinery to regulatory regions and the effectiveness of chromatin as a template for RNA synthesis. This modification is believed to stabilize the unfolded state of chromatin by antagonizing internucleosomal interactions (4) and so could facilitate the passage of the transcribing enzyme, especially in long transcription units. This article summarizes recent findings suggesting that the elongating RNA polymerase II may itself directly mediate histone acetylation and consequent chromatin unfolding by piggybacking an acetyltranferase activity. The possible implications of this observation with respect to domain opening are discussed.
Local and Extensive Chromatin Modification.
In Saccharomyces, several studies suggest that the extent of histone modification mediated by transcription factor-targeted histone acetyltranferases and deacetyltranferases, such as Gcn5 and Rpd3, respectively, is restricted to chromatin in the immediate vicinity of regulatory regions (5–7). In these examples, the histones of only one or, at most, two nucleosomes are modified whereas those associated with the downstream transcribed region are unaffected and could still present a barrier to efficient transcription elongation. This localized modification also contrasts with the pattern observed in certain chromatin domains of higher eukaryotes. Functionally a chromatin domain, such as that typified by the chicken β-globin locus, constitutes a region of position-independent gene expression and is delimited by boundary or insulator elements that confer this property (8). Early studies of the avian globin, lysozyme, and ovalbumin loci showed that the transcriptional competence of a domain is correlated with an increased sensitivity to cleavage by DNase I, indicative of a general change of chromatin structure throughout the domain. Studies on the chicken β-globin locus containing four globin genes and a locus control region (LCR) revealed approximate coincidence between DNase I sensitivity and the presence of hyperacetylated core histones (9). Within the domain, variations in the extent of modification at ≈1-kilobase resolution are minor, although the assay does not exclude the presence of short unmodified regions (<0.5 kilobases). Significantly, the 5′ boundary of the hyperacetylated region maps immediately 5′ of the DNase I hypersensitive site located at the 5′ boundary element. In this example, the correlation between accessible chromatin and hyperacetylation is compelling.
Piggybacking: A Mechanism for Coupling Chromatin Modification and Transcription Elongation.
The restriction of local histone modification to regulatory regions leaves unanswered the question of how modification of the chromatin of the transcribed regions or of whole domains is effected. Any mechanism proposed to explain this extensive modification must address the physical reality of the extent of the modified regions. Even in organisms, such as yeast, that lack extensive introns, the longer regions of transcribed chromatin may contain ≈30–50 nucleosomes. In higher organisms, the problem is even more acute. The chicken and human β-globin loci encompass 33 and 70 kilobases, respectively, whereas many other domains, such as the Drosophila Ubx locus, are substantially larger. Even the chicken lysozyme locus, which contains a single gene, is some 20 kilobases (equivalent to ≈100 nucleosomes) in extent. A second consideration is that the structural conversion is from a condensed to an open state of chromatin and, consequently, any modification process must be associated, although not necessarily directly, with the disruption of a higher order structure. Given the immense length of DNA within a domain or transcribed region and the requirement for concomitant disruption of condensed chromatin, it seems implausible that a histone acetyltranferase tethered to a static DNA-binding protein could by itself efficiently access all of the nucleosomes within a domain. A more likely scenario is the coupling of modification to a processive DNA tracking helicase associated with a piggybacking histone acetyltranferase (10). In the case of the longer transcribed regions, this helicase could be RNA polymerase II itself (11). This model also provides a mechanism for the initial disruption of condensed chromatin because the helicase, if itself unable to rotate, would generate in front of it a region of positively supercoiled DNA. Such superhelicity could potentially destabilize the negative supercoils constrained by core nucleosome particles.
In support of these ideas, Wittschieben et al. (12) now provide the first direct evidence that histone modification may be directly coupled to transcription elongation by RNA polymerase II. In vitro pure RNA polymerase II can transcribe naked DNA without substantial impediment, but its ability to transcribe a chromatin template efficiently is severely compromised. However, in vivo, the actively transcribing assembly contains not only polymerase II but also a multisubunit complex, termed elongator, which associates with the hyperphosphorylated C-terminal domain of the largest polymerase subunit (13). One of the components of the elongator complex is a 60-kDa protein, termed Elp3, which Wittschieben et al. (12) show to be homologous to the GNAT superfamily of acetyltransferases. Deletions of the gene impair transcription elongation in vivo to a similar extent to deletons in another elongator subunit, Elp1, as indicated by an enhanced sensitivity to elongation inhibitors. In addition, isolated Elp3 possesses histone acetyltransferase activity, acetylating the N-terminal tails of all four core histones in vitro.
At least two other chromatin-modifying activities have been implicated in chromatin remodeling associated with transcription elongation. Cho et al. (14) show that the transcription factor PCAF, which possesses both histone acetyltranferase activity and a bromodomain, binds preferentially to the hyperphosphorylated elongation-competent form of mammalian polymerase II rather than the holoenzyme containing with a nonphosphorylated C-terminal domain. In addition, enhancer-mediated recruitment of PCAF can stimulate transcription in vivo (15). Taken together, these observations would be consistent with the notion that PCAF might also be piggybacked by polymerase II and so facilitate transcription elongation (15). However, in contrast to Elp3, which is highly conserved from yeast to mammals (12), it has yet to be established that PCAF is associated with actively elongating polymerase II. Thus, an alternative scenario might envisage that PCAF is required to initiate chromatin unfolding downstream of a promoter site whereas Elp3 then propagates this unfolding. A second complex, FACT (16), has been shown to potentiate transcription by polymerase II in vitro by disrupting nucleosome structure. However, this complex has been shown to interact with DNA polymerase α, and it remains unclear whether its biological role is to facilitate transcription and/or DNA replication or simply to act as a more general histone chaperone.
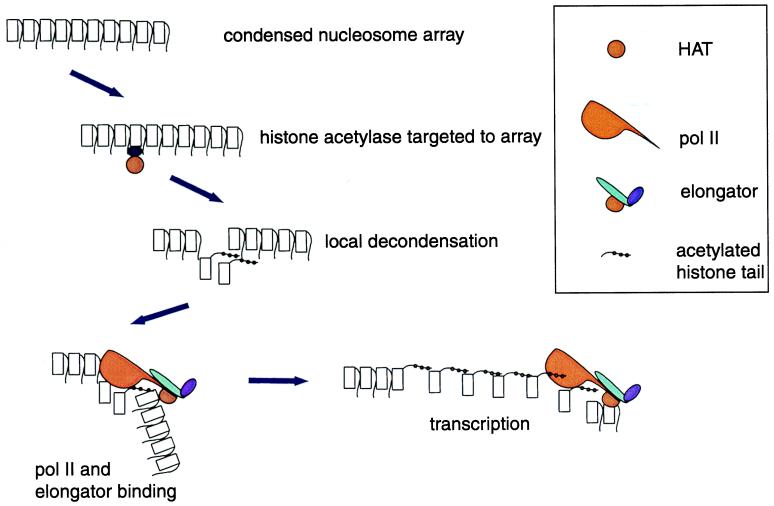
Not only do the experiments of Wittschieben et al. (12) suggest a mechanism for the direct coupling of transcription elongation to chromatin modification (Fig. 1), but they also provide the first evidence for the piggybacking of chromatin modification activities by helicases in general and RNA polymerase II in particular. The concept of piggybacking is not novel because it has already been established that other activities (for example, components of mRNA capping) and polyadenylation machinery (refs. 17–19; reviewed in ref. 20) and possibly proteins involved in premRNA splicing (21, 22), can be piggybacked in a similar fashion.
Figure 1.
Sequential histone acetylation by transcription factor-targeted histone acetyltransferases and by a transcription-coupled histone acetyltransferase. Polymerase II association with the promoter precedes binding of the elongator, which requires phosphorylation of the polymerase II CTD.
Domain Opening Mediated by DNA Tracking?
There are few candidate processes for coupling the opening of a chromatin domain to a processive mechanism. The most obvious possibilities are DNA replication and, again, transcription. Piggybacking chromatin modification to replication has the merit of simplicity and would in principle result in the uniform modification of the whole domain. However, this mechanism, perhaps less plausibly, also requires that the histones recruited to the newly replicated DNA be concomitantly incorporated into the modified structure after the passage of the replication machinery.
Domain opening by piggybacking on the transcription machinery has the potential advantage that transcription could be more discriminatory so that the extent of chromatin modification need not be uniform throughout the domain, nor, moreover, need it necessarily be coupled to replication. However, such a mechanism inevitably requires the transcription of most, if not all, of the DNA contained within the domain and the existence of inter- and extragenic transcripts originating from outside the regions encoding mRNA precursors. Do transcripts of this nature exist? For at least 25 years, there have been persistent reports of the existence of giant transcripts originating from the globin loci of avian and murine erythroblasts, but the nature and precise origin of these transcripts has remained obscure, although two more recent studies have identified transcription within the LCR (23, 24). However, the problem has been revisited by a contemporary study analyzing transcripts of the human β-globin locus using nuclear run-on assays in erythroid cell lines (25). The structure of this locus is similar to that in avian cells and contains an LCR upstream of five erythroid-specific globin genes, organized in the order of their developmental expression (ɛ-Gγ-Aγ-δ-β). The run-on assays reveal the existence of detectable strand-specific extragenic transcripts covering most of the locus in erythroid but not in nonerythroid cells. In contrast to the mRNA transcripts, these novel RNAs are exclusively nuclear. Intriguingly, the extragenic transcription is not uniform, especially in the vicinity of the mRNA encoding regions. Upstream of the ɛ-globin gene, the five DNase I hypersensitive sites (HS1–HS5) constituting the LCR, together with at least the assayed flanking sequences, are all transcribed, as in avian erythroid cells, but the signal abruptly terminates ≈400 bp upstream of the ɛ-globin transcription start point. Similarly, there is a decrease in polymerase density after the poly(A) site of the ɛ- and γ-globin genes followed by higher densities of transcription in the downstream intergenic regions. This distribution of transcripts implies that the LCRs are transcribed but not the regulatory region immediately upstream of the ɛ-globin gene. Similarly, any potential regulatory elements immediately downstream of this gene would only be weakly transcribed. Transcription of the most 5′ enhancer element, HS5, is driven by a promoter in the long terminal repeat of an endogenous retrovirus, ERV-9, located in the apparent 5′ boundary region of the LCR (26). However, other studies indicate that transcripts can be initiated within the more downstream HS2 enhancer element, even in the absence of the ERV-9 long terminal repeat (27). Although these observations support the notion that the extragenic transcripts are initiated at multiple sites, it cannot be assumed that similar retroviral long terminal repeats are present in the β-globin loci of other species.
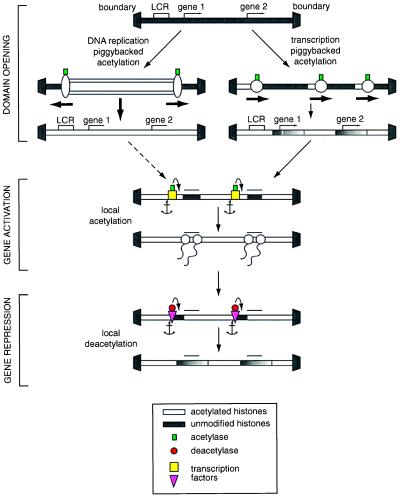
With this pattern of transcripts, any processive piggybacking mechanism for chromatin modification would result in the chromatin structure in the LCR becoming modified and thus accessible to DNA-binding transcription modifiers whereas any regulatory regions in the immediate vicinity of the genes themselves would only be weakly, if at all, modified and would remain inaccessible until activated by gene-specific protein assemblies. At the structural level, this would ensure that domain opening was uncoupled from transcriptional activation and would minimize any undirected transcription of the relevant genes consequent upon a generalized opening of the domain. At the same time, the transcription of the genes would depend on local modification of chromatin structure directed by DNA-tethered modifying complexes. In this way, chromatin structure could be harnessed to repress genes locally before activation in the context of an open domain. Conversely, promoters could be shut down after the required period of expression by tether-directed local modification, such as histone deacetylation. One example of such local modification is the hypoacetylation of histone H4 at the inactive Xist promoter in male mice, contrasting with higher average levels of acetylation in female mice, in which one of the two alleles is active (28). However, in both sexes, similar average levels of histone hyperacetylation are observed over the remainder of the locus. Again, such mechanisms could eliminate inappropriate expression even though the structure of the domain as a whole remains open and would be expected to be utilized when a differentiating lineage reached the end of the line. Very similar considerations would apply to chromatin modification dependent on processive piggybacking on the replication machinery (Fig. 2). Here, however, because of the overwhelming requirement to copy all of the DNA within a domain, even including the boundary DNA itself, it might be necessary for local repression mediated by cytosine methylation and/or local histone deacetylation to be actively reimposed after the passage of the modifying machinery, again to minimize premature expression. Although replication-coupled processive piggybacking is likely to be a once-for-all process, transcription-coupled piggybacking could both change and continuously monitor the state of chromatin modification. In the latter case, the activity could compensate for any nontargeted histone deacetylation.
Figure 2.
Scheme for the domain-wide modification of chromatin structure by piggybacking of histone modifiers on processive helicases. The mechanisms shown for replication- and transcription-coupled modification are not mutually exclusive—for example, a deacetyltranferase could be piggybacked from an SV40 T antigen–retinoblastoma protein complex to either RNA polymerase or to the replication machinery.
Are the properties of the observed extragenic transcripts in the human β-globin locus consistent with any modifying model? There is no evidence as to whether the intergenic transcription is a cause or consequence of domain opening. The apparent strand specificity of transcription and the precision of the apparent location of presumed initiation and termination sites within the intergenic regions argue against this transcription being a simple consequence of a more open chromatin structure. Moreover, the strand specificity implies that chromatin modification should possess directionality. In addition, the tracking model implies that extragenic transcription should be independent of the expression of the genes contained within the locus. Indeed, the transient transfection of an active β-globin class gene into nonerythroid cell lines induces transcription of the LCR and intergenic regions from the chromosomal locus, but, in this situation, the chromosomal β-globin genes themselves remain transcriptionally silent (25). This establishes the independence of intergenic transcription from intragenic transcription. Interestingly, the induction of LCR transcription appears to require the physical association, or at least the close proximity of the transfected plasmid and the chromosomal locus. Such proximity could allow the transfer of polymerase II and any associated modifiers to the chromosomal locus or alternatively could relocate the chromosomal locus to an active area of the nucleus. In either situation, the synthesis of these transcripts would be independent of replication. This system would also provide the optimal experimental test of whether intergenic transcription is involved in chromatin modification.
Abbreviation
- LCR
locus control region
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 2.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 3.Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 4.Tse C, Sera F, T, Wolffe A P, Hansen J C. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadosh D, Struhl K. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo M-H, Zhou J, Jambeck P, Churchill M E, Allis C D. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Nature (London) 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 8.Felsenfeld G. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 9.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Travers A A. Cell. 1992;69:573–575. doi: 10.1016/0092-8674(92)90218-2. [DOI] [PubMed] [Google Scholar]
- 11.Brownell J E, Allis C D. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 12.Wittschieben B, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis C D, Tempst P, Svejstrup J Q. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 13.Otero G, Fellows J, de Bizemont T, Dirac A M G, Gustafsson C M, Erdjument-Bromage H, Tempst P, Svejstrup J Q. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 14.Cho H, Orphanides G, Sun X, Yang X-J, Ogrysko V, Lees E, Nakatani Y, Reinberg D. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krumm A, Madisen l, Madisen l, Yang X-J, Goodman R, Nakatani Y, Groudine M. Proc Natl Acad Sci USA. 1998;95:13501–13506. doi: 10.1073/pnas.95.23.13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orphanides G, LeRoy G, Chang C H, Luse D S, Reinberg D. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 17.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. Nature (London) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 18.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho E J, Takagi T, Moore C R, Buratowski S. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neugebauer K M, Roth M B. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- 21.Cramer P, Pesce C G, Baralle F E, Kornblihtt A R. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirose Y, Tacke R, Manley J L. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collis P M, Antoniou M, Grosveld F. EMBO J. 1990;9:233–240. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuan D, Kong S, Hu K. Proc Natl Acad Sci USA. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashe H L, Monks J, Wijgerde M, Fraser P, Proudfoot N J. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long Q, Bengra C, Li C, Kutlar F, Tuan D. Genomics. 1998;15:542–555. doi: 10.1006/geno.1998.5608. [DOI] [PubMed] [Google Scholar]
- 27.Kong S, Bohl D, Li C, Tuan D. Mol Cell Biol. 1997;17:3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCabe V, Formstone E J, O’Neill L P, Turner B M, Brockdorff N. Proc Natl Acad Sci USA. 1999;96:7155–7160. doi: 10.1073/pnas.96.13.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]