Abstract
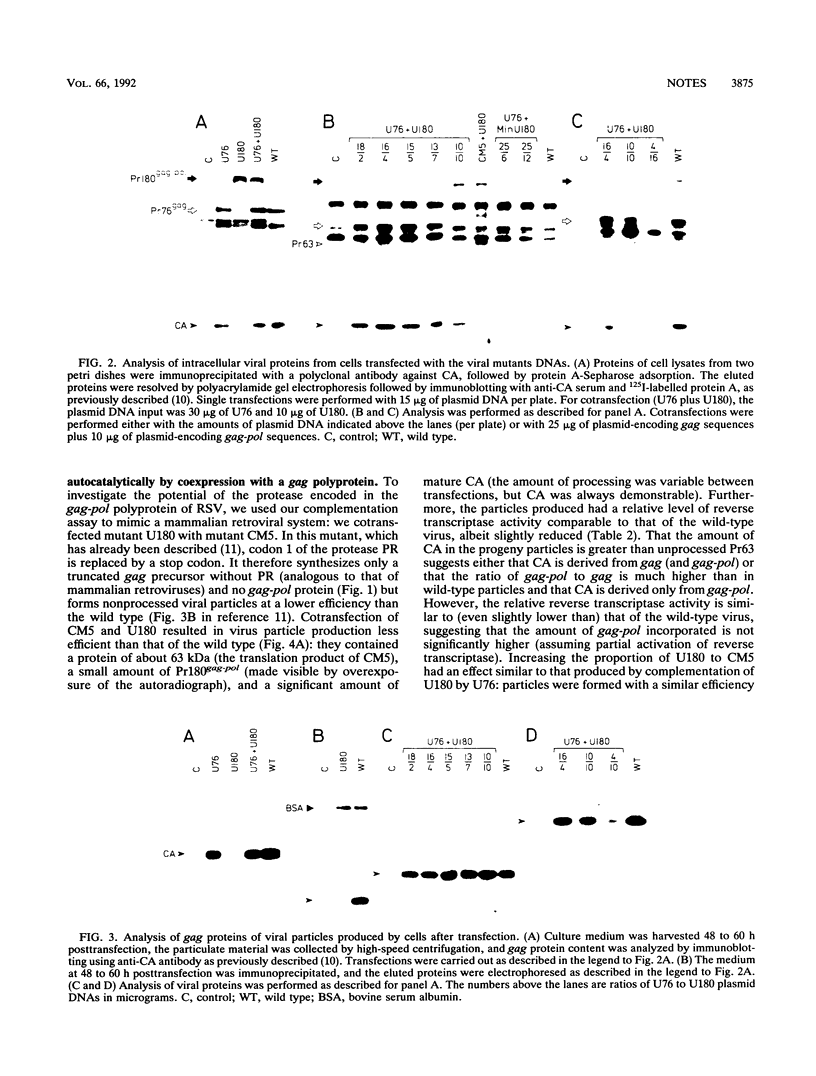
Avian retroviruses (with the notable exception of spleen necrosis virus) express their protease (PR) both in their gag and their gag-pol polyprotein precursors, in contrast to other retroviruses, notably, the mammalian retroviruses, in which PR is encoded in the gag-pol polyprotein or in a separate reading frame as a gag-pro product. The consequence is that the avian PR is expressed in stoichiometric rather than catalytic amounts. To investigate the significance of the particular genome organization of the avian retrovirus prototype Rous sarcoma virus, we developed an assay that measures complementation between the gag and the gag-pol polyproteins by expressing them from two different plasmids in transfected cells. By using this assay, we showed that the protease PR from the gag-pol polyprotein is capable of autocatalytic self-cleavage and -activation when coexpressed with a protease-deficient gag protein and that the PR domain has a role in viral particle assembly. Furthermore, this complementation assay can be used to investigate the role of the gag domain in the gag-pol polyprotein by determining whether it can rescue a defect in the gag polyprotein. We report here the results of such an experiment, which studied a mutation in the N terminus of the gag gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett R. P., Rhee S., Craven R. C., Hunter E., Wills J. W. Amino acids encoded downstream of gag are not required by Rous sarcoma virus protease during gag-mediated assembly. J Virol. 1991 Jan;65(1):272–280. doi: 10.1128/jvi.65.1.272-280.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R. C., Bennett R. P., Wills J. W. Role of the avian retroviral protease in the activation of reverse transcriptase during virion assembly. J Virol. 1991 Nov;65(11):6205–6217. doi: 10.1128/jvi.65.11.6205-6217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein K. M., Goff S. P. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J Virol. 1988 Jun;62(6):2179–2182. doi: 10.1128/jvi.62.6.2179-2182.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Blaug G., Hansen M., Barklis E. Assembly of gag-beta-galactosidase proteins into retrovirus particles. J Virol. 1990 May;64(5):2265–2279. doi: 10.1128/jvi.64.5.2265-2279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotler M., Danho W., Katz R. A., Leis J., Skalka A. M. Avian retroviral protease and cellular aspartic proteases are distinguished by activities on peptide substrates. J Biol Chem. 1989 Feb 25;264(6):3428–3435. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis J., Baltimore D., Bishop J. M., Coffin J., Fleissner E., Goff S. P., Oroszlan S., Robinson H., Skalka A. M., Temin H. M. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988 May;62(5):1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méric C., Spahr P. F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986 Nov;60(2):450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertle S., Spahr P. F. Role of the gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J Virol. 1990 Dec;64(12):5757–5763. doi: 10.1128/jvi.64.12.5757-5763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Morrow C. D. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol. 1991 Sep;65(9):5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepinsky R. B., Vogt V. M. Identification of retrovirus matrix proteins by lipid-protein cross-linking. J Mol Biol. 1979 Jul 15;131(4):819–837. doi: 10.1016/0022-2836(79)90203-1. [DOI] [PubMed] [Google Scholar]
- Stewart L., Vogt V. M. trans-acting viral protease is necessary and sufficient for activation of avian leukosis virus reverse transcriptase. J Virol. 1991 Nov;65(11):6218–6231. doi: 10.1128/jvi.65.11.6218-6231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Weaver T. A., Talbot K. J., Panganiban A. T. Spleen necrosis virus gag polyprotein is necessary for particle assembly and release but not for proteolytic processing. J Virol. 1990 Jun;64(6):2642–2652. doi: 10.1128/jvi.64.6.2642-2652.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J. W., Craven R. C., Weldon R. A., Jr, Nelle T. D., Erdie C. R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991 Jul;65(7):3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]