Abstract
Under physiological conditions, the Escherichia coli cytoplasm is maintained in a reduced state that strongly disfavors the formation of stable disulfide bonds in proteins. However, mutants in which the reduction of both thioredoxins and glutathione is impaired (trxB gor mutants) accumulate oxidized, enzymatically active alkaline phosphatase in the cytoplasm. These mutants grow very poorly in the absence of an exogenous reductant and accumulate extragenic suppressors at a high frequency. One such suppressor strain, FA113, grows almost as rapidly as the wild type in the absence of reductant, exhibits slightly faster kinetics of disulfide bond formation, and has fully induced activity of the transcriptional activator, OxyR. FA113 gave substantially higher yields of properly oxidized proteins compared with wild-type or trxB mutant strains. For polypeptides with very complex patterns of disulfide bonds, such as vtPA and the full-length tPA, the amount of active protein was further enhanced up to 15-fold by co-expression of TrxA (thioredoxin 1) mutants with different redox potentials, or 20-fold by the protein disulfide isomerase, DsbC. Remarkably, higher yields of oxidized, biologically active proteins were obtained by expression in the cytoplasm of E. coli FA113 compared with what could be achieved via secretion into the periplasm of a wild-type strain, even under optimized conditions. These results demonstrate that the cytoplasm can be rendered sufficiently oxidizing to allow efficient formation of native disulfide bonds without compromising cell viability.
The fundamental discovery that a denatured protein, ribonuclease, could assemble correctly in the absence of any catalysts indicated that all of the information for the proper folding of a protein was present in its primary amino acid sequence. Because disulfide bonds are necessary for the proper folding of ribonuclease, these experiments were also taken to mean that disulfide bond formation was independent of enzyme catalysts. Thus, it had been presumed that only the presence of oxygen (or small molecules such as oxidized glutathione) is needed in vivo for disulfide bond formation. This presumption appeared to explain the fact that proteins with structural disulfide bonds are only found in the more oxidizing noncytosolic intracellular compartments or in the extracellular space. According to this view, disulfide bonds do not form in the cytosol simply because the reducing components such as glutathione and thioredoxins keep such bonds reduced.
The first modification of this view of disulfide bond formation and the basis for its compartmentalization came from the finding that disulfide bond formation in Gram-negative bacteria does require the presence of a protein catalyst, DsbA (1–5). This finding not only changed the picture of how disulfide bond formation takes place normally but also raised questions about the basis for the absence of disulfide bonds in cytosolic proteins. Normally, the formation of stable disulfide bonds in the cytoplasm is an exceedingly rare event (6). Transient disulfide bonds that are not required for the stability of the native state have been detected in a few cytoplasmic proteins that include enzymes such as ribonucleotide reductase, the transcription factors OxyR and RsrA, the Hsp33 chaperone, and in a partially folded intermediate of the P22 tailspike endorhamnosidase (7–9). In general, the oxidation of cysteine thiols in cytoplasmic proteins is strongly disfavored for both thermodynamic and kinetic reasons. First of all, the thiol-disulfide redox potential of the cytoplasm is too low to provide a sufficient driving force for the formation of stable disulfides. Second, under physiological conditions, there are no enzymes that can catalyze protein thiol oxidation. The Escherichia coli cytoplasm contains two thioredoxins, TrxA and TrxC, and three glutaredoxins (10, 11). The oxidized form of these proteins can catalyze the formation of disulfide bonds in peptides. However, in the cytosol, both the thioredoxins and the glutaredoxins are maintained in a reduced state by the action of thioredoxin reductase (TrxB) and glutathione, respectively. In E. coli, glutathione is synthesized by the gshA and gshB gene products. The enzyme glutathione oxidoreductase, the product of the gor gene, is required to reduce oxidized glutathione and complete the catalytic cycle of the glutathione-glutaredoxin system.
In a trxB null mutant, stable disulfide bonds can form in normally secreted proteins, such as alkaline phosphatase, when they are expressed in the cytoplasm without a signal sequence (12). Subsequent studies revealed that, in a trxB mutant, the two thioredoxins are oxidized and serve as catalysts for the formation of disulfide bonds (13). Disulfide bond formation was found to be even more efficient in double mutants defective in both the thioredoxin (trxB) and glutathione (gor or gshA) pathways (14). Double mutants, trxB gor or trxB gshA, grow very poorly (doubling time ≈300 min) and require an exogenous reductant such as DTT to achieve a reasonable growth rate. In this paper, we describe the construction of strains that grow normally even though the cytoplasm is highly oxidizing and that favor the formation of disulfide bonds in certain proteins with an efficiency even higher than that of the periplasm. To characterize the formation of disulfide bonds in these strains, we used four model proteins that are normally secreted and exhibit increasingly complex cysteine connectivities: E. coli alkaline phosphatase containing two disulfide bonds linking cysteines that are consecutive in the primary sequence, a property we term linear cysteine connectivity; a version of mouse urokinase with six disulfide bonds, only one of which is linear; a truncated form of the human tissue plasminogen activator (vtPA) consisting of the kringle 2 and protease domains with a total of nine disulfide bonds (one linear); and, finally, the full-length human tPA containing 17 disulfide bonds and one free cysteine. For all of the model eukaryotic proteins, including the highly complex full length tPA, expression in the cytoplasm of our strains resulted in appreciable yields of active protein exceeding that which could be obtained by periplasmic expression. The properties of these strains raise interesting questions about the requirements for efficient folding of disulfide-bonded proteins and about the impact of such an altered redox environment on cytoplasmic proteins in general.
Materials and Methods
Strains and Plasmids.
All strains are derivatives of DHB4 (MC1000 ΔphoA(PvuII) phoR ΔmalF3 F′[lac+(lacIQ) pro]) (15). Strains FA196 and FA222 were constructed as follows: a 1.0-kilobase fragment of DNA upstream of trxB was first amplified by PCR, and a 441-bp fragment was generated by digestion with NsiI. This 441-bp fragment was cloned into the NsiI site of a pBAD33 vector (16) with trxB cloned under the control of the arabinose promoter. The complete construct containing the 441-bp upstream region, the araC repressor gene, and the arabinose-controlled trxB allele was subcloned into the vector pK0V (G. Church laboratory, Harvard Medical School) followed by integration into the chromosome of E. coli DHB4 using the published procedure of Link et al. (17), generating FA196. P1 transduction of the gor522… mini-Tn10 allele (14) to FA196 resulted in strain FA222.
A list of plasmids used is shown in Table 1. The precise amino acid sequence of vtPA is as described (20). The vtPA gene was cloned into plasmid pTrc99A (Amersham Pharmacia) under control of the trc promoter. For periplasmic expression, vtPA as well as the full length tPA were targeted for secretion by fusion to the heat-stable enterotoxin II signal sequence (21). The genes encoding TrxA (thioredoxin 1) and active-site mutants thereof, GrxA (glutaredoxin 1), rat protein disulfide isomerase (PDI), and DsbA and DsbC with or without their signal sequence were all expressed under control of the araB promoter from plasmids derived from pBAD33 (16). TrxA active-site mutants were amplified from the constructs of Huber et al. (22) and Mossner et al. (19, 23). Rat PDI was amplified from a construct of De Sutter et al. (24). Plasmid pBADΔSSdsbC is based on pEJS75 (E. J. Stewart and J.B., unpublished work). Plasmid pΔA-uPA expresses the catalytic domain only of mouse urokinase type plasminogen activator (14).
Table 1.
Plasmids and strains employed in this study
| Plasmid name | Relevant features | Source |
|---|---|---|
| pAID135 | PhoA(Δ2-22) under control of tac promoter | Ref. 18 |
| pΔA-uPa | ΔA-uPA under control of tac promoter | Ref. 14 |
| pFA2 | GrxA in pBAD33 under its own RBS | This work |
| pFA3 | TrxA in pBAD33 under its own RBS | Ref. 19 |
| pFA4 | TrxA(P34S) in pBAD33 | This work |
| pFA5 | TrxA(G33P,P34Y) in pBAD33 | Ref. 19 |
| pfA6 | TrxA(G33P,P34H) in pBAD33 | Ref. 19 |
| pFA7 | TrxA(P34H) in pBAD33 | Ref. 19 |
| pFA8 | TrxA(C35A) in pBAD33 | Ref. 19 |
| pFA21 | TrxB in pBAD33 under its own RBS | This work |
| pFA22 | trxB upstream region in pFA21 at NsiI site | This work |
| pFA28 | trxB upstream region, araC, ParaB-trxB in pKOV | This work |
| pBADdsbA | DsbA in pBAD33 with optimized RBS | This work |
| pBADdsbC | DsbC in pBAD33 with optimized RBS | This work |
| pBADΔSSdsbA | DsbA(Δ2-19) in pBAD33 with optimized RBS | This work |
| pBADΔSSdsbC | DsbC(Δ2-20) in pBAD33 with optimized RBS | This work |
| pBADrPDI | mature rat PDI in pBAD33 with optimized RBS | This work |
| pTrcStIItPA | human tPA with StII leader in pTrc99A | Ji Qiu, University of Texas |
| pTrcStIIvtPA | tPA(Δ6-175) with StII leader in pTrc99A | This work |
| pTrctPA | tPA in pTrc99A | This work |
| pTrcvtPA | tPA(Δ6-175) in pTrc99A | This work |
| pBAD33 | araBAD promoter, CmR, pACYC ori | Ref. 16 |
| pTrc99A | trc promoter, AmpR, ColE1 ori | Amersham Pharmacia |
| pKOV | counter-selectable allele exchange vector | G. Church laboratory, Harvard University |
| Strain | Genotype | Source |
| DHB4 | MC1000 ΔphoA(PvuII) phoR ΔmalF3 F′[lac+(lacIQ) pro] | Ref. 15 |
| WP597 | DHB4 trxB::Km | W. Prinz, Harvard University |
| FA112 | DHB4 gshA20::Tn10Km trxB::Km… Tn10 supp | This work |
| FA113 | DHB4 gor522…mini-Tn10Tc trxB::Km supp | This work |
| FA196 | DHB4 ΔtrxB::ParaB-trxB | This work |
| FA222 | FA196 gor522…mini-Tn10Tc | This work |
Activity Assays.
vtPA and tPA.
Cells expressing either vtPA or full length tPA were grown with shaking at 30°C in LB medium supplemented with antibiotics (50 μg/ml carbenicillin and 25 μg/ml chloramphenicol) as needed. At OD600 = 0.8, arabinose was added to 0.2% wt/vol final concentration; 30 min later, IPTG was added to 1 mM, and the culture was grown an additional 3 hr. Cells were harvested by centrifugation, were resuspended in cold PBS, and were lysed in a French pressure cell (Spectronic Instruments, Rochester, NY). The insoluble fractions were removed by centrifugation (12,000 × g, 10 min, 4°C), and soluble protein was quantified by the Bio-Rad protein assay, using BSA as standard. Plasminogen activation was quantified by an indirect chromogenic assay as follows. In a microtiter plate, 5 μg of soluble protein was added to wells containing 50 mM Tris⋅HCl (pH 7.4), 0.01% Tween 80, 0.04 mg/ml human glu-plasminogen (American Diagnostica, Greenwich, CT), and 0.4 mM Spectrozyme PL (American Diagnostica), with a 260 μl final volume. The plate then was incubated at 37°C, and absorbance at 405 nm was read after 2 or 3 hr. Activity is directly proportional to ΔA405, which is the absorbance after subtracting the background of a strain lacking a vector expressing tPA. Relative activities were normalized to the ΔA405 obtained by expressing vtPA alone in FA113. In some experiments, vtPA and tPA activities were determined by monitoring fibrin clearance as described (21, 25).
Kinetics of Alkaline Phosphatase Oxidation.
Cells were diluted 1:100 from overnight cultures into M63 supplemented with all 18 amino acids except methionine and cysteine and were grown at 37°C. When the cells reached an OD600 of 0.2, IPTG was added to 2 mM to induce expression of alkaline phosphatase. The pulse–chase was started by the addition of [35S]methionine. After 1 min, unlabeled methionine at 0.1% wt/vol (final concentration) was added, and, subsequently, samples were collected at the indicated time points and mixed with 0.1 M iodoacetamide.
In Vivo Redox State.
The in vivo redox states of DsbC, TrxA, and the “Grx-like” TrxA variant were assayed by derivatization of free thiols by 4-acetamido-4′-maleimidyl-stilbene-2,2′-disulfonic acid (Molecular Probes) and Western blotting as described (26). Anti-DsbC antibodies were a gift from John Joly (Genentech); anti-TrxA antibodies were purchased from Sigma.
Results
Isolation and Phenotypic Characterization of trxB gshA supp and trxB gor supp Strains.
E. coli depends for aerobic growth on the presence of either of the two major thiol reduction systems—the thioredoxin and the glutathione-glutaredoxin pathways. When both of these pathways are eliminated by mutation, in a trxB gor or trxB gshA double mutant, the cells grow extremely slowly (14). However, these cells can be rescued by the addition of the reductant DTT to the growth medium. When a trxB gor or trxB gshA strain is grown in medium containing DTT and then transferred to medium lacking DTT, the cytoplasm becomes even more oxidizing than in the trxB strain, resulting in the accumulation of high levels of alkaline phosphatase or mouse urokinase activity (14). Even when grown in the presence of DTT, both the trxB gshA and trxB gor strains give rise to fast growing derivatives at a high frequency. Because the trxB, gshA, and gor alleles in these strains are nonreverting null mutations, the faster-growing derivatives must result from extragenic suppressor mutations.
We examined whether any of the suppressed mutants might still retain the high cytoplasmic oxidizing potential of the parental trxB gshA or trxB gor strains. A fast-growing colony from each genetic background, FA112 and FA113, respectively, was examined in more detail for disulfide bond formation in a variety of model proteins. We first tested MalS, a periplasmic amylase that contains two disulfide bonds. When a signal sequenceless version of MalS was expressed in various genetic backgrounds, enzymatically active protein was detected only in the trxB gor supp strain FA113 (data not shown). Encouraged by this result, we proceeded to study the cytoplasmic expression of mouse urokinase, vtPA and full length tPA, all of which contain multiple disulfide bonds with nonlinear connectivities. Signal sequenceless versions of these proteins were expressed from a strong promoter, and the amount of active protein was determined by zymography (in the case of mouse urokinase) or by fibrin clearance assays (for vtPA and tPA). For all three proteins, substantially higher levels of activity were detected in FA113, compared with the wild type, the trxB mutant, or the trxB gshA supp strain FA112. Representative results showing the fibrin clearance produced by cells expressing vtPA are presented in Fig. 1. Quantitative analysis of the protease activity of vtPA using a coupled assay that measures the activation of plasminogen to plasmin revealed that the level of active vtPA in FA113 is 10-fold higher than when expressed in the wild-type strain DHB4 and 2.5-fold higher than in the trxB gshA supp strain. Because the trxB gor supp strain, FA113, gave the highest yields of active protein, it was selected for more detailed characterization.
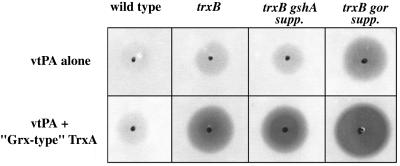
Figure 1.
tPA activity in DHB4 (wild type), WP597 (trxB), FA112 (trxB gshA supp), and FA113 (trxB gor supp) monitored by the fibrinolysis assay. Cells were transformed with plasmids pTrcvtPA, and pFA5 (bottom row only). Soluble protein (10 μg) from induced cultures was spotted onto fibrin/agarose plates and was incubated for 24 hr at 37°C. Clearance zones qualitatively measure biological activity of bacterially produced vtPA.
At 37°C, in rich media, FA113 was found to grow almost as well as the wild-type (DHB4, trxB+gor+) strain with doubling times of 30 and 27 min, respectively (Fig. 2). In contrast, WP778, the trxB gor parent of FA113, grew with a doubling time of 300 min in the absence of DTT (14). It was of interest to compare the formation of protein disulfides in the cytosol of the strain FA113 relative to a strain with the trxB gor phenotype that had not accumulated suppressor mutations. A direct comparison of the yields of disulfide-bonded proteins in FA113 and the parental strain WP778 is not meaningful because of the dramatic difference in the growth rate of the two strains. Therefore, we constructed the strain FA222 in which the trxB gene was placed under the control of the arabinose promoter and that also contained the gor allele of FA113. This strain grows well in the presence of arabinose but exhibits a trxB gor phenotype when transferred to growth media lacking arabinose. Under these conditions, the accumulation of mouse urokinase in the cytosol of FA222 was comparable to that obtained in FA113 (data not shown). Thus, although the suppressor mutation alleviates the growth defect of trxB gor, it does not interfere with disulfide bond formation in the cytoplasm.
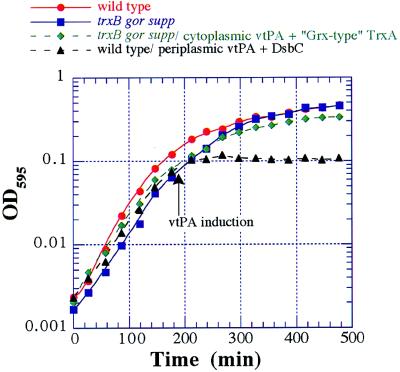
Figure 2.
Growth curves of DHB4 (●); FA113 (■); FA113/pTrcvtPA/pFA5 (♦); and DHB4/pTrcStIIvtPA/pBADdsbC (▴) grown aerobically at 37°C in LB medium in test tubes. Expression of vtPA was induced at late log phase as described in the text. Optical density was measured in a microtiter plate reader.
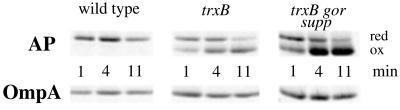
Exposure of E. coli to elevated concentrations of hydrogen peroxide or diamide renders the cytoplasm more oxidizing and, among other things, results in the formation of a disulfide bond in the transcription factor OxyR. The oxidized form of OxyR activates the transcription of trxC encoding thioredoxin 2 and several other genes that play a role in protecting the cell from oxidative damage (27) (D. Ritz, H. Patel, B. Doan, M. Zheng, F. Å., G. Storz, and J.B., unpublished work), personal communication). FA113 exhibited nearly full activation of OxyR as judged by the expression of TrxC and the level of oxyS RNA (data not shown). Pulse–chase experiments were carried out to determine the rate of protein oxidation in signal sequenceless alkaline phosphatase. In FA113, ≈50% of the alkaline phosphatase was oxidized within 1 min and was >95% was complete after 11 min (Fig. 3). The kinetics of disulfide bond formation in the trxB gor supp strain were slightly faster than in a trxB mutant. In contrast, no oxidized alkaline phosphatase accumulated in the wild-type strain, even after 11 min.
Figure 3.
Pulse–chase of alkaline phosphatase. Signal sequenceless alkaline phosphatase (AP) was immunoprecipitated and separated by native PAGE, such that the oxidized form (ox) was distinguished from the reduced form (red). Time is indicated in minutes postchase. Strains used were DHB4 (wild type), WP597 (trxB), and FA113 (trxB gor supp) transformed with plasmid pAID135. OmpA was used as an internal standard.
Effects of Cysteine Oxidoreductase Coexpression.
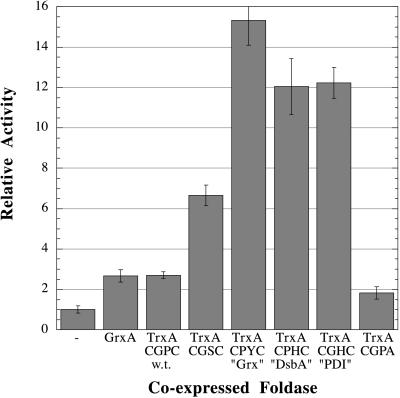
Stewart et al. (13) have shown that disruption of trxB results in an accumulation of oxidized thioredoxins that can then act as oxidases, the reverse of their normal role. Likewise, in FA113, TrxA expressed from the chromosome was present solely in the oxidized form (P.H.B., J. Qiu, and G.G., unpublished data). We examined the effect of high level expression of TrxA and TrxA mutant proteins with varying redox potentials on the folding of the more complex multidisulfide proteins, namely vtPA and tPA. The redox potential of most cysteine oxidoreductases, including TrxA, is strongly influenced by the sequence of the dipeptide within the CXXC active site motif (19, 23, 28). TrxA with a wild-type active site (-CGPC-) and five mutants with varying redox potentials were cloned into plasmids under the control of the araB promoter and were transformed into FA113 together with a compatible expression vector for vtPA or full length tPA synthesis. Cells were induced with arabinose followed by addition of IPTG 30 min later to initiate synthesis of the tPA protein, and the yield of active tPA was analyzed 3 hr later. Western blot analysis revealed that TrxA and the TrxA variants accumulated to the same level at steady state (data not shown). Overexpression of TrxA resulted in a modest increase in the level of active vtPA (Fig. 4). Coexpression of more oxidizing TrxA variants gave significantly higher accumulation of active vtPA. For example, coexpression of a more oxidizing variant with the active site of GrxA (glutaredoxin 1) resulted in active vtPA at levels 15-fold greater than the control. Analysis of the in vivo redox state of overexpressed TrxA revealed that the wild-type enzyme is present primarily in the oxidized form, with a minor fraction in the reduced state. In contrast, the mutant with the Grx-like active site is mainly reduced (data not shown). GrxA co-expression was much less effective than the Grx-like TrxA, presumably a consequence of its lower redox potential and the fact that glutaredoxin is a less efficient catalyst of disulfide bond formation or reduction compared with TrxA (29). Similar relative increases to those reported above were obtained with the full length tPA substrate (data not shown). Interestingly, coexpression of the Grx-like TrxA variant significantly improved disulfide bond formation not only in FA113 but also in the trxB gshA supp strain FA112 and in the trxB mutant WP597 (Fig. 1).
Figure 4.
The effect of coexpression of vtPA and oxidoreductases in the cytoplasm of strain FA113 (trxB gor supp) quantified by an indirect assay for plasminogen activation using a chromogenic plasmin substrate (see text). Activity has been normalized to the value obtained from vtPA expressed alone in FA113 (column 1). Cell lysates used for columns 2–8 were from strains cotransformed with plasmids pFA2-pFA8, respectively. Plasmids pFA3–8 overexpress active site mutants of TrxA. The amino acid sequence of the active site is listed, along with the known oxidoreductase that contains that native active site.
The folding of proteins containing multiple disulfide bonds with nonlinear connectivities is greatly assisted by the addition of catalysts that enhance the rate of disulfide bond isomerization. In the E. coli periplasm, the formation of active urokinase or tPA depends critically on the DsbC disulfide isomerase activity (21, 30). Therefore, we asked whether causing DsbC to be localized to the cytoplasm might enhance the yield of properly assembled disulfide-containing proteins. A version of DsbC without a signal sequence was constructed and placed behind the araB promoter and an optimized ribosome binding site to achieve efficient translation. DsbC overexpressed in the cytoplasm of FA113 was found predominantly in a form in which its structural disulfide had formed, but the active site was reduced (data not shown). A 20-fold increase in vtPA activity was observed, corresponding to the highest accumulation of active protein in this study (Fig. 5). In contrast, coexpression of DsbA under identical conditions actually reduced the accumulation of active vtPA. The effect of the eukaryotic rat PDI expressed in the cytoplasm was also evaluated, but the increase in vtPA activity compared with the control cells without any foldase overexpression was marginal (data not shown).
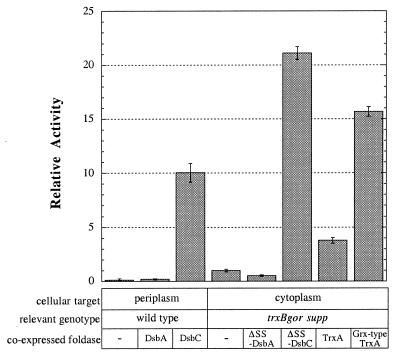
Figure 5.
Plasminogen activation in lysates of cells expressing vtPA secreted to the periplasm of DHB4 (wild type) or in the cytoplasm of FA113 (trxB gor supp). Foldases were coexpressed from plasmids pBADdsbA, pBADdsbC, pBADΔSSdsbA, pBADΔSSdsbC, pFA3, and pFA5.
It is instructive to compare the formation of protein disulfide bonds following secretion in the periplasm of a wild-type strain or in the cytoplasm of FA113. When vtPA was expressed in the cytoplasm of FA113, a higher level of active protein was obtained compared with the periplasm of the wild-type counterpart, DHB4 (Fig. 5, columns 1 and 4). Analysis by Western blotting revealed that the total level of tPA accumulation at steady state was approximately the same in the cytoplasm of FA113 and in the periplasm of DHB4. The amount of active vtPA in the periplasm of DHB4 could be increased by more than two orders of magnitude by coexpression of periplasmic DsbC. Coexpression of a signal sequenceless DsbC with vtPA in the cytoplasm of the trxB gor supp strain resulted in even higher accumulation of correctly folded protein. Under these conditions, the level of active vtPA represented a 2-fold increase relative to periplasmic expression and a 200-fold increase compared with expression in the cytoplasm of DHB4. Moreover, whereas high level expression of DsbC and vtPA in the periplasm resulted in growth arrest, cytoplasmic expression did not have any appreciable effect on cell growth.
Discussion
In this work, we describe strains in which the cytoplasm of E. coli is so altered that disulfide bond formation in proteins localized to the cytoplasm can be more efficient than when these same proteins are localized in the oxidizing environment of the periplasm. The initial step in this study was the isolation of strains carrying mutations that suppress the slow growing phenotype of trxB gor and trxB gshA strains. The suppressor mutations do not moderate the high cytoplasmic thiol-disulfide redox potential conferred by the inactivation of the thioredoxin and glutathione reduction pathways. In fact, the cytoplasm of FA113 was more favorable for the formation of disulfide bonds than that of a trxB mutant for all model substrates tested and resulted in constitutive activation of the transcription factor OxyR.
Stewart et al. (13) have shown that disulfide bond formation in trxB mutants is mediated by thioredoxins, which serve as oxidants instead of their normal role as protein reductants. Because the oxidized thioredoxins are responsible for promoting disulfide bond formation in trxB backgrounds, we asked whether increasing the expression of TrxA (thioredoxin 1) over that achieved by the chromosomal gene would enhance this process. Coexpression of wild-type TrxA (−270 mV) had a modest effect on disulfide bond formation. In contrast, the efficiency of disulfide bond formation in FA113 could be markedly increased by introducing plasmids expressing thioredoxin mutant proteins poised at higher redox potentials. The Grx-like, “DsbA-like,” and the “PDI-like” TrxA mutants resulted in 10- to 15-fold higher levels of active, correctly oxidized proteins. The redox potentials of these mutants have been estimated from the equilibrium constants with glutathione solutions to be −195 mV, −204 mV, and −221 mV, respectively (23).
The most dramatic increase in the yield of active vtPA and full length tPA, both of which contain numerous disulfide bonds in their native state, was obtained by coexpression of a cytoplasmic form of DsbC. The effect of DsbC cannot be solely a consequence of its high redox potential (−130 mV) (31) because coexpression of DsbA, which has a comparable redox potential, was actually detrimental (Fig. 5). The thioredoxins, glutaredoxins, DsbA, and DsbC, are all members of the thioredoxin superfamily. Why is it that the presence of Grx-like TrxA or DsbC renders disulfide bond formation in the cytoplasm so much more efficient compared with wild-type TrxA, GrxA, or DsbA? To understand the differing properties of these proteins in our system, it is necessary to consider the various steps that must occur to allow the proper assembly of complex disulfide bond-containing proteins. First, a protein catalyst of disulfide bond formation must be able to accumulate in oxidized form in the cytoplasm. Second, the oxidized form of the protein catalyst must be efficient at transferring its disulfide to a substrate protein. Third, a protein capable of catalyzing disulfide bond isomerization must be provided and maintained in a reduced form. We found that, in the trxB gor supp mutant, TrxA accumulates predominantly in the oxidized state. Oxidized TrxA will promote disulfide bond formation in substrate proteins (32). Thus, it can fulfill the first and second functions outlined above. Increasing substantially the level of wild-type TrxA only marginally increases correct disulfide formation indicating that oxidation is already as efficient as it will get, and isomerization (or reduction) of incorrect disulfide bonds has become the limiting step. Because thioredoxin is fully oxidized, it cannot catalyze disulfide bond rearrangements. In contrast, thioredoxin variants poised at a higher redox potential were found to accumulate predominantly in the reduced form, which can serve as a catalyst for disulfide bond isomerization.
DsbC is also maintained in a reduced state in the cytoplasm. In vitro data strongly argue that reduced DsbC is an effective catalyst of disulfide isomerization (31). In the periplasm, the folding of proteins with multiple disulfide bonds such as mouse urokinase, bovine pancreatic trypsin inhibitor, and human tPA depends heavily on the presence of reduced DsbC (21, 30). In a similar fashion, the expression of DsbC in the cytoplasm greatly increased the active yield of complex proteins. However, coexpression of DsbA, which is a poor catalyst of disulfide isomerization, had the opposite effect, giving lower yields of active vtPA or full length tPA relative to FA113 alone.
Surprisingly, in certain of our strains, higher yields of active protein could be obtained in the cytoplasm than when the same protein was expressed in the periplasmic space, where disulfide bond formation normally takes place (Fig. 5). This effect was not attributable to differences in expression or inefficient export into the periplasm. There are at least two reasons why, under certain conditions, the cytoplasm of the trxB gor supp strain may be, ironically, a more favorable compartment for oxidative folding of proteins with complex disulfide patterns. First, the kinetics of protein oxidation in the cytoplasm are rather slow. As shown in Fig. 3, the half-life for the oxidation of alkaline phosphatase in the cytoplasm is well over 1 min. In contrast, alkaline phosphatase in the periplasm is nearly fully oxidized within <40 seconds (ref. 1; L. Debarbieux and J.B., unpublished work). A slower oxidation rate is likely to be more favorable because, in that case, disulfide bond formation is more likely to be determined by the conformational preferences of the polypeptide chain, which should result in the alignment of the proper cysteine residues. Second, the oxidation of proteins in the periplasm by DsbA may be detrimental for the folding of those proteins with multiple disulfides. DsbA is a very efficient enzyme; however, it tends to place disulfide bonds in polypeptides randomly, with little regard for the native conformation. Random oxidation results in the formation of scrambled disulfides which can be difficult to rearrange. Jonda et al. (33) have shown that the Grx-like TrxA variant is more efficient than DsbA in promoting folding because oxidation occurs more slowly, favoring formation of correct disulfide bonds by allowing intramolecular isomerization. Likewise, addition of reduced glutathione to the growth medium, rendering the periplasmic space less oxidizing, increases the yield of eukaryotic disulfide-bonded proteins coexpressed in the periplasm of E. coli with DsbA or rat PDI (34, 35). The implication is that somewhat more reducing conditions facilitate the folding of eukaryotic proteins containing multiple disulfide bonds.
Our results demonstrate that FA113 (and possibly FA112) engineered to coexpress cytoplasmic DsbC or the Grx-like TrxA variant should be very useful for the cytoplasmic expression of normally secreted eukaryotic proteins with multiple disulfide bonds. Although some exported proteins can be expressed in the bacterial periplasm at very high levels (36), often high level secretion, particularly of heterologous proteins, can interfere with the normal function of the Sec pathway causing cell toxicity. This was the case when the dsbC gene was expressed from a strong promoter (Fig. 2). Expression in the cytoplasm, together with either signal sequenceless DsbC or Grx-like TrxA, circumvents this problem. Thus, not only can complex disulfide bonds be formed more readily in the cytoplasm, greater cell yields can be achieved as well.
The rapid growth of the trxB gor supp strain indicates that bacteria can tolerate large perturbations in their cytoplasmic thiol-disulfide redox potential. This implies that the vast majority of native cytoplasmic proteins in FA113 are unable to form aberrant disulfides, even under oxidizing conditions. This is not as surprising as it first seems. The kinetics of disulfide bond formation in our strains are slow enough so that many native proteins will have folded into their tertiary structures, rendering their cysteines inaccessible to the catalysts. The model proteins we have used cannot fold into their tertiary structures without the involvement of disulfide bond formation and, therefore, may have their cysteines accessible to the catalysts.
Finally, we consider explanations for the poor growth of the trxB gor and trxB gshA strains and the nature of the suppressor mutations. Perhaps the most likely explanation is that the absence of the two major disulfide reducing pathways in the cytoplasm renders inactive the catalytic cycle of an essential enzyme, such as ribonucleotide reductase, that requires these reductants. The suppressor mutations may result in the turning on of expression of another disulfide reductant that can substitute, e.g., in the ribonucleotide reductase catalytic cycle but that does not alter the redox state of the thioredoxins or affect the oxidation state of heterologous proteins. Alternatively, the slow growth of the trxB gor strain may indicate that aberrant disulfide bonds are formed in an essential cytoplasmic protein that, as a result, becomes inactivated. Clearly identifying the nature of the suppressor mutation in FA112 and FA113 would aid in explaining the nature of the growth defect and its suppression. Unfortunately, fast-growing derivatives of trxB gor and trxB gshA strains arise in media without an exogenous reductant at a very high frequency. For this reason, it is not possible to select for the transfer of the suppressor allele from FA112 or FA113 into the respective parental strains, a prerequisite for genetic analysis of the suppressor.
Acknowledgments
We are grateful to Ekkehard Mössner and Rudi Glockshuber for providing some of the TrxA mutants and to Ji Qiu for plasmid construction and for carrying out preliminary experiments. This work was supported by National Institutes of Health Grant 5RO1 GM55090-02 to G.G. and J.B.; P.H.B. was supported in part by a National Institutes of Health Biotechnology Training Grant.
Abbreviation
- PDI
protein disulfide isomerase
References
- 1.Bardwell J C, McGovern K, Beckwith J. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 2.Kamitani S, Akiyama Y, Ito K. EMBO J. 1992;11:57–62. doi: 10.1002/j.1460-2075.1992.tb05027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peek J A, Taylor R K. Proc Natl Acad Sci USA. 1992;89:6210–6214. doi: 10.1073/pnas.89.13.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomb J F. Proc Natl Acad Sci USA. 1992;89:10252–10256. doi: 10.1073/pnas.89.21.10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J, Webb H, Hirst T R. Mol Microbiol. 1992;6:1949–1958. doi: 10.1111/j.1365-2958.1992.tb01368.x. [DOI] [PubMed] [Google Scholar]
- 6.Locker J K, Griffiths G. J Cell Biol. 1999;144:267–279. doi: 10.1083/jcb.144.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aslund F, Zheng M, Beckwith J, Storz G. Proc Natl Acad Sci USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson A S, King J. Nat Struct Biol. 1997;4:450–455. doi: 10.1038/nsb0697-450. [DOI] [PubMed] [Google Scholar]
- 9.Kang J G, Paget M S, Seok Y J, Hahn M Y, Bae J B, Hahn J S, Kleanthous C, Buttner M J, Roe J H. EMBO J. 1999;18:4292–4298. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rietsch A, Beckwith J. Annu Rev Genet. 1998;32:163–184. doi: 10.1146/annurev.genet.32.1.163. [DOI] [PubMed] [Google Scholar]
- 11.Aslund F, Beckwith J. J Bacteriol. 1999;181:1375–1379. doi: 10.1128/jb.181.5.1375-1379.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derman A I, Prinz W A, Belin D, Beckwith J. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 13.Stewart E J, Aslund F, Beckwith J. EMBO J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prinz W A, Aslund F, Holmgren A, Beckwith J. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 15.Boyd D, Manoil C, Beckwith J. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman L M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link A J, Phillips D, Church G M. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derman A I, Puziss J W, Bassford P J, Jr, Beckwith J. EMBO J. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mossner E, Huber-Wunderlich M, Rietsch A, Beckwith J, Glockshuber R, Aslund F. J Biol Chem. 1999;274:25254–25259. doi: 10.1074/jbc.274.36.25254. [DOI] [PubMed] [Google Scholar]
- 20.Obukowicz M G, Gustafson M E, Junger K D, Leimgruber R M, Wittwer A J, Wun T C, Warren T G, Bishop B F, Mathis K J, McPherson D T, et al. Biochemistry. 1990;29:9737–9745. doi: 10.1021/bi00493a033. [DOI] [PubMed] [Google Scholar]
- 21.Qiu J, Swartz J R, Georgiou G. Appl Environ Microbiol. 1998;64:4891–4896. doi: 10.1128/aem.64.12.4891-4896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber H E, Russel M, Model P, Richardson C C. J Biol Chem. 1986;261:15006–15012. [PubMed] [Google Scholar]
- 23.Mossner E, Huber-Wunderlich M, Glockshuber R. Protein Sci. 1998;7:1233–1244. doi: 10.1002/pro.5560070519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Sutter K, Hostens K, Vandekerckhove J, Fiers W. Gene. 1994;141:163–170. doi: 10.1016/0378-1119(94)90566-5. [DOI] [PubMed] [Google Scholar]
- 25.Waldenstrom M, Holmgren E, Attersand A, Kalderen C, Lowenadler B, Raden B, Hansson L, Pohl G. Gene. 1991;99:243–248. doi: 10.1016/0378-1119(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 26.Joly J C, Swartz J R. Biochemistry. 1997;36:10067–10072. doi: 10.1021/bi9707739. [DOI] [PubMed] [Google Scholar]
- 27.Zander T, Phadke N D, Bardwell J C. Methods Enzymol. 1998;290:59–74. doi: 10.1016/s0076-6879(98)90007-6. [DOI] [PubMed] [Google Scholar]
- 28.Grauschopf U, Winther J R, Korber P, Zander T, Dallinger P, Bardwell J C. Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 29.Aslund F, Berndt K D, Holmgren A. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 30.Rietsch A, Bessette P, Georgiou G, Beckwith J. J Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zapun A, Missiakas D, Raina S, Creighton T E. Biochemistry. 1995;34:5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
- 32.Lundstrom J, Krause G, Holmgren A. J Biol Chem. 1992;267:9047–9052. [PubMed] [Google Scholar]
- 33.Jonda S, Huber-Wunderlich M, Glockshuber R, Mossner E. EMBO J. 1999;18:3271–3281. doi: 10.1093/emboj/18.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wunderlich M, Glockshuber R. J Biol Chem. 1993;268:24547–24550. [PubMed] [Google Scholar]
- 35.Ostermeier M, De Sutter K, Georgiou G. J Biol Chem. 1996;271:10616–10622. doi: 10.1074/jbc.271.18.10616. [DOI] [PubMed] [Google Scholar]
- 36.Joly J C, Leung W S, Swartz J R. Proc Natl Acad Sci USA. 1998;95:2773–2777. doi: 10.1073/pnas.95.6.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]