Abstract
The drought stress tolerance of two Solanum tuberosum subsp. andigena landraces, one hybrid (adg×tbr) and Atlantic (S. tuberosum subsp. tuberosum) has been evaluated. Photosynthesis in the Andigena landraces during prolonged drought was maintained significantly longer than in the Tuberosum (Atlantic) line. Among the Andigena landraces, ‘Sullu’ (SUL) was more drought resistant than ‘Negra Ojosa’ (NOJ). Microarray analysis and metabolite data from leaf samples taken at the point of maximum stress suggested higher mitochondrial metabolic activity in SUL than in NOJ. A greater induction of chloroplast-localized antioxidant and chaperone genes in SUL compared with NOJ was evident. ABA-responsive TFs were more induced in NOJ compared with SUL, including WRKY1, mediating a response in SA signalling that may give rise to increased ROS. NOJ may be experiencing higher ROS levels than SUL. Metabolite profiles of NOJ were characterized by compounds indicative of stress, for example, proline, trehalose, and GABA, which accumulated to a higher degree than in SUL. The differences between the Andigena lines were not explained by protective roles of compatible solutes; hexoses and complex sugars were similar in both landraces. Instead, lower levels of ROS accumulation, greater mitochondrial activity and active chloroplast defences contributed to a lower stress load in SUL than in NOJ during drought.
Keywords: Chaperones, drought, flavonoids, metabolites, oligonucleotide-based microarray, osmolytes, potato, ROS, subspecies Andigena
Introduction
Plants have evolved different ways to respond to drought stress, with either avoidance or the development of adaptation mechanisms that provide tolerance to variable, species and ecotype or line-specific degrees. Once adapted, plants use mechanisms that lower tissue dehydration, either maintaining water potential, or tolerating lower tissue water potential. This can be accomplished by minimizing water loss, which is principally achieved by closing stomata, the accumulation of osmotically active metabolites, water retention by proteins or altered resistance to water flow. Stress responses are based on altered hormone metabolism and subsequent alterations in transcript expression and new protein activities. During drought stress, abscisic acid (ABA) biosynthesis increases (Radin, 1981; Robertson et al., 1985), which initiates a series of signalling events that induce or enhance plant tolerance to the stress. ABA-dependent and ABA-independent pathways have been defined (Shinozaki and Yamaguchi-Shinozaki, 1997; Zhu et al., 2002). ABA increases the amount or activity of transcription factors, such as MYBs, DREBs, and bZIPs that facilitate the expression of other genes (Yamaguchi-Shinozaki and Shinozaki, 2006). However, ABA levels are high only for a short time in acute stress treatments, lasting usually for a few hours after the perception of the stress. ABA is metabolized rapidly later (Kiyosue et al., 1994; Verslues and Bray, 2006). Over time, the induction of ABA-responsive control circuits leads to the engagement of ‘downstream’ genes and proteins, among them heat shock proteins (Rizhsky et al., 2002; Wang et al., 2004), antioxidant proteins (Pastori and Foyer, 2002; Walz et al., 2002), and osmolyte-producing metabolic enzymes (Satoh et al., 2002; Yoshiba et al., 1997).
Our work focused on potato, the fourth most important food crop in the world, with an annual production over 300 million tons (FAO/ESS, 2005). Potatoes regularly suffer drought stress in most of their growing regions. The most commonly consumed potato, S. tuberosum subsp. tuberosum Hawkes (hereafter referred to as Tuberosum) is highly susceptible to drought stress (Weisz et al., 1994). This domesticated tetraploid subspecies originated from Andean stock (Hawkes, 1990). S. tuberosum subsp. andigena Hawkes (hereafter referred to as Andigena) is a tetraploid Andean potato cultivated throughout the Andes from Venezuela to Northern Argentina (Huaman and Ross, 1985). It is cultivated at altitudes as high as to 3500 m above sea level (Terrazas et al., 1996) and is therefore adapted to harsh climate conditions. Its similar tetraploid genome allows for hybrid generation with Tuberosum (Kumar and Kang, 2006; Tai and Tarn, 1980). Therefore, Andigena is an ideal candidate to study genes related to drought tolerance that in the future could be introgressed into Tuberosum. Improvement of productivity and quality of cultivated potato using conventional breeding and biotechnology are the major objectives of CIP (Centro Internacional de la Papa).
Previous work with Andigena accessions revealed that accessions that adapted best to drought stress showed higher, constitutive expression levels of genes encoding enzymes of the flavonoid pathway (Watkinson et al., 2006). Another report with Sullu and SA2563 (an Andigena clone), both tolerant to drought stress, showed that both lines induced the expression of antioxidants, flavonoid genes, alternative oxidase pathway, HSPs and LEAs, among other genes (Schafleitner et al., 2007b). The physiological and phenotypic responses to drought of three Andigena genotypes, an Andigena×Tuberosum hybrid, and the cultivar Atlantic (Tuberosum), are evaluated here. In addition to physiological characterizations and metabolite profiling, gene expression of two of the Andean genotypes, Sullu (SUL) and Negra Ojosa (NOJ) were evaluated, using custom oligonucleotide-microarrays with 44 000 probes. Gene expression analysis suggested that drought resistance was correlated with up-regulation of plastid heat shock protein (HSP) and antioxidant genes, suggesting superior protection of the plastids against the reactive oxygen species (ROS). The response of known osmolytes, such as proline and trehalose, and the GABA pathway to drought appeared not to be correlated with relative resistance and/or the ability to recover, but rather with the degree of stress experienced by different genotypes.
Materials and methods
Plant material and drought stress application
Three Andigena genotypes, Leona, NOJ, and SUL, and two bred varieties, Costanera (Andigena×Tuberosum) and Atlantic (Tuberosum) (Fig. 1; see Supplementary Table 1 at JXB online), were clonally propagated in vitro and rooted in rooting medium. Plants were transferred to 9 cm square pots containing Pro-Mix BX (Premier Horticulture, Quakertown, PA, USA) in the greenhouse and grown under natural light (July, Blacksburg, VA, latitude 37.229 N longitude –80.414 W) at 20–25 °C for 2 weeks. Tip cuttings were made, dipped in Rootone® F brand powder (Bayer Cropscience, Research Triangle Park, NC, USA), transferred to new 9 cm square pots with a mix of 2 parts Pro-Mix BX and 1 part sand, maintained at high humidity for 2 d in the dark and then grown under the same conditions. After 4 weeks, each plant was transferred to 28×19 cm containers containing the same potting mix (2 Pro Mix BX:1 sand v:v) and continuing the natural light greenhouse conditions for six additional weeks. Plants were fertilized weekly with a half-strength solution of Peters professional soluble plant food (Scotts-Sierra Horticultural Products, Marysville, OH, USA). At this time (September) water was withheld from the plants being drought-stressed. The plants were monitored by taking photosynthetic measurements (Li-Cor 6400, Lincoln, NE, USA) 5 h after sunrise. Control plants were watered every 10 d. Photosynthesis remained stable under this watering regime and averaged between 8 and 12 μmol CO2 m−2 s−1 throughout the experiment. Samples were harvested when photosynthesis was reduced to 50–60% in the most susceptible genotype (18 d after water stress imposition) and to 25% or less in the other genotypes. The experiment was continued after the 18 d time point, using the genotypes in which photosynthesis had been inhibited by less than 25%. When photosynthesis was reduced by 70% (25 d after water stress imposition) all remaining plants were harvested. After sampling, the plants were rewatered and photosynthesis measurements taken 24 h later. Photosynthesis was monitored every 3–4 d on the 4th fully expanded leaf up to the point when a genotype's level was reduced by 50–60%; after that, photosynthesis was monitored every other day. The 4th fully expanded leaf was harvested at three time points (18 d and 25 d after withholding water, and at the 24 h recovery point). The leaves were flash-frozen in liquid nitrogen and stored at –80 °C. Half of each sample was used for RNA isolation and the other half was used for metabolite analysis. Tubers were also harvested at each point, counted, weighed, and flash-frozen in liquid nitrogen. At each point, three treated and three control plants of each genotype were harvested. Relative water content (RWC) was measured as follows: the 3rd leaf of each plant was weighed (fresh weight, FW) and then placed in a 50 ml polypropylene Falcon tube containing distilled water and stored at 4 °C overnight. The next day, excess water was removed by blotting each leaf in a paper towel prior to weighing (turgid weight, TW) and then dried in an oven overnight at 90 °C. After drying they were reweighed (dry weight, DW). RWC was calculated with the following formula: RWC%= [(FW–DW)/(TW–DW)]×100.
Fig. 1.
Negra Ojosa, Leona, Sullu, and Costanera plants after 18 d of stress. The lines are identified from left to right. After 17 d of stress. Atlantic had already wilted and harvested, which is why it is not included in the picture.
RNA isolation
RNA was isolated from approximately 2 g of leaf tissue using a phenol-based method retrieved from the TIGR website: http://www.tigr.org/tdb/potato/microarray_SOPs.shtml.
Microarray hybridizations
RNA isolated from leaves at the point of maximum stress (25 d of withholding water) from three treated plants and three control plants was used in the synthesis of cRNA according to the Agilent protocol (http://www.chem.agilent.com/temp/rad2AB07/00001389.PDF). The cRNA was hybridized against custom printed in situ synthesized 60-mer oligonucleotide microarrays containing 42 034 unique potato features (Agilent Technologies, Santa Clara, CA). The oligo microarray was developed as part of the Potato Oligo Chip Initiative (POCI) consortium and has been described in detail by Kloosterman et al. (unpublished data). All the reagents were purchased from Agilent and hybridization followed the Agilent protocol: http://www.chem.agilent.com/temp/rad2AB07/00001389.PDF.
Identification of Arabidopsis orthologues
The potato oligo (60-mers) sequences represented on the array were compared against the potato cDNA database made up of sequences obtained from the Sol Genomics Network (SGN; http://www.sgn.cornell.edu/) and TIGR (STGI; http://www.tigr.org/tdb/potato/) using standalone NCBI-BLAST v2.2.14 (Altschul et al., 1997). The top potato cDNA sequence hit of at least a 50 base-pair match to an oligo was considered as the representative gene for that oligo. Sequences corresponding to the spotted oligonucleotides mapped were used to identify putative orthologues in Arabidopsis thaliana (AGI; TIGR6 release) using BLASTX. AGI hits with e-values of at least 1e-10 were considered. In cases where multiple oligos mapped to the same AGI number but had different directions of expression, only the one with the lowest e-value was considered. All bioinformatics analyses were carried out on a MacBook with custom PERL scripts.
Statistical analysis and data mining
The microarray analysis platform TM4 was used (Saeed et al., 2003). The following normalization steps were carried out on Median Intensitiy Values (MIV) using the following components of the MIDAS pipeline: total intensity normalization, Lowess normalization, standard deviation regularization, and low intensity filtering. The output of the MIDAS pipeline was used to identify differentially expressed genes using a one class t test in the Multi Experiment Viewer (MEV) module of TM4. Microarray analyses used in this study follow those described in Sioson et al. (2006). After TM4 microarray analysis, a gene was considered to be induced or repressed if the log2 (fold change) after TM4 analysis was found to be statistically significant with a P-value lower than 0.05. The physiology data were analysed in R (http://www.R-project.org) using an ANOVA model where the main and interaction effects of genotype and treatment on tuber number, average tuber weight, tuber yield, and root weight were analysed. Differences indicated by the ANOVAs were further analysed using Tukey's honestly significant difference (HSD) test.
Gene clustering and heat maps
The differentially expressed genes were clustered using pairwise single-linkage hierarchical clustering based on Pearson correlation: Cluster 3.0 (de Hoon et al., 2004). Heat maps were generated using Java Treeview (Saldanha, 2004).
Quantitative real-time PCR
Two step real-time PCR was performed. Total RNA was DNase-treated with the DNAfree kit (Ambion, Austin, TX, USA). After cleaning, 4 μg of RNA were reverse transcribed using the cDNA archive kit (Applied Biosystems, Foster City, CA, USA) and oligo dT18V primers. Real time PCR was performed with 5 μl of cDNA (from a 20 ng μl−1 dilution) using SYBR® Green PCR Master Mix (Applied Biosystems) in a 25 μl reaction volume on an ABI Prism 7700 Sequence Detection System (Applied Biosystems), with 0.5 μM primer final concentration and the following cycling steps: initial denaturation for 10 min at 94 °C, followed by 34 cycles at 15 s at 94 °C, 30 s at 56 °C, and 30 s at 72 °C, and a 20 minute gradient from 60 °C to 90°C to obtain a melting curve. The data were collected at the extension step (72 °C). Primers to obtain larger amplicons (700–900 bp) were designed from S. tuberosum sequences found in the TIGR potato EST database. These PCR reaction products were electrophoresed and isolated from agarose. Ten-fold dilutions of the purified PCR products in a range between 10 pg to 0.1 fg were used to generate a standard curve.
A 183 bp amplicon of adenosine kinase served as an internal control since its expression level does not change in potato during drought stress (Watkinson et al., 2006). Real-time primers were designed from cloned sequences of candidate genes (see Supplementary Table 2 at JXB online). All primer pairs were tested for dimer formation before using them with the actual samples. At least three technical repeats per biological repeat were carried out. Deviations from threshold values were less than a 0.5 cycle for technical replicates and less than 1 cycle for biological replicates.
Metabolite profiling
Polar phase extractions from 10–15 mg of dried potato leaves were derivatized (Fiehn et al., 2000; Roessner et al., 2000). One to 2 μl of sample were injected with an 8:1 split ratio and analysed on an HP5890 gas chromatograph equipped with a HP5973 mass selective detector (Agilent Inc, Palo Alto, CA, USA). Gas chromatography was carried out using a 30 m SPB-50 column with a 0.25 mm ID and 0.25 μm film thickness (Supelco, Belfonte, CA, USA). The injection temperature was 230 °C and the interface was 250 °C. The carrier gas was helium set at a constant flow rate of 1 ml min−1. The temperature program was: 70 °C for 5 min, 5 °C min−1 to 310 °C, 310 °C for 10 min. Spectra were evaluated according Lozovaya et al. (2006).
Data sets contained five replicates per sample and were statistically analysed by t test and one-way ANOVA using the algorithm incorporated into Microsoft Excel 2002 (Microsoft Corporation, Seattle, WA, USA). Differences were scored as significant at the P <0.05 level.
Principal component analysis
Principal component analysis (PCA) was used to represent the contribution of metabolites that discriminate between the two Andigena potato lines. PCA transforms large sets of related variables into a smaller set of variables, termed principal components, that reveal the degree of variation or correlation (Joliffe, 1986). PCA was performed using the XLSTAT-Pro (v.7.5.3) program (Addinsoft, NY, USA). The eigenvalues that reflect the quality of the projection from the N-dimensional initial table to a lower number of dimensions were analysed. Each eigenvalue corresponds to a factor (PC), and each factor to one dimension (axis) such that each object is characterized by its proximity to a particular axis (Taylor et al., 2002). A factor is a linear combination of initial variables, representing the weight of the initial variability, and all factors are uncorrelated (r=0). The contribution of each variable to a particular PC can then be calculated. Ideally, the first two or three PCs will correspond to the highest percentage of the variance between samples, ensuring that maps based on these first two or three factors provide a high quality projection of the initial multi-dimensional table.
Results
Photosynthesis in the Andigena landraces studied is more tolerant to drought stress compared to the Tuberosum bred variety
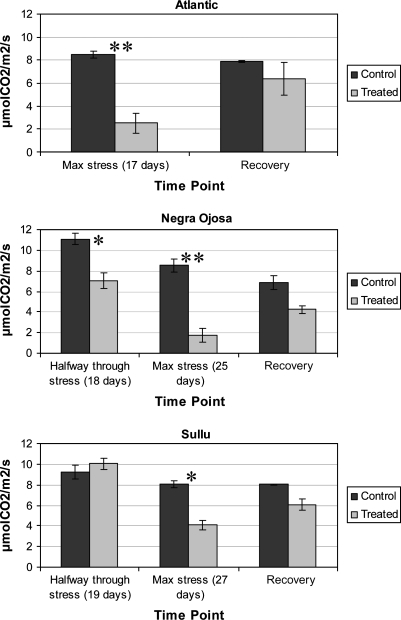
S. tuberosum subsp. tuberosum (Atlantic) was used to compare the level of drought stress in the plants analysed. After 17 d of drought stress, photosynthesis in Atlantic was reduced by almost 80% and Costanera (which comes from a Tuberosum and Andigena cross) showed photosynthetic reduction (20% of controls) after 22 d. Atlantic leaves were bigger than those of SUL and NOJ leaves, which have a similar leaf size (Fig. 1), while Costanera leaves were not as big as those of Atlantic, but were still twice as large as those of SUL and NOJ. However, NOJ was more affected by drought compared to SUL, showing 60% net photosynthesis compared with the non-stressed controls. In SUL, drought-stressed plants exhibited photosynthetic rates close to those of the watered, control plants (Fig. 2). By day 25, photosynthesis in NOJ had been reduced to 20% of the control while, in SUL, net photosynthesis was close to 60% of the control.
Fig. 2.
Effects of prolonged drought stress and subsequent recovery on photosynthesis in Atlantic, NOJ and SUL. Net photosynthesis during drought stress is shown at an intermediate stress stage, at the point of maximum stress imposed and 24 h after watering (recovery). All measurements were performed 5 h after sunrise. The asterisks indicate statistically significant differences between the control and treated samples, with Tukey test (* P-value <0.05, ** P-value <0.01).
One day after rewatering the plants, Atlantic and SUL recovered photosynthesis to 80% of the control, while NOJ only recovered to 23% of the control rate. Therefore, even though NOJ could stand water deficits longer than Atlantic, it could not recover from the level of stress in contrast to SUL. Leona, the other Andigena genotype showed photosynthetic behaviour similar to that of NOJ (photosynthetic reduction by 80% after 25 d of drought), but it recovered to 70% after rewatering (data not shown). The relative leaf water content (RWC) of the Andigena landraces at maximum drought stress was between 70–80%. Control plants had a leaf RWC around 90%.
Drought stress also affected root mass, tuber number, and average tuber weight, however, the variability was high and, after ANOVA analysis, there were not statistically significant differences (P <0.05) for these traits (see Supplementary Table 3 at JXB online).
Choice of genotypes for gene expression and metabolite analyses
Among the three Andigena landraces studied, NOJ and SUL exhibited the extremes of drought stress tolerance with respect to photosynthetic responses (SUL being the most tolerant and NOJ being the most susceptible). They also differed in their capacity to recover and, therefore, gene expression analysis (microarrays) was performed on these two clones.
Table 1 shows a summary of the transcriptomic responses at the point of maximum stress in SUL and NOJ (see Materials and methods and the legend to Table 2 for a description of the statistical method employed). The focus here is on some of the most striking examples of differences in gene expression between the two genotypes. Much activity was observed among genes encoding glycolytic and mitchondrial function. TFs, transporters, protection and repair genes, and antioxidant genes were also responsive.
Table 1.
Number of genes significantly (P >0.05) differentially expressed in SUL and NOJ compared with their respective controls
| Induced | Repressed | No change | |
| SUL | 4680 | 2036 | 35318 |
| NOJ | 2553 | 2055 | 37426 |
| Shareda | 704 | 309 | 31828 |
Statistical analysis was carried out as described in the Materials and methods.
Genes with the same response in SUL and NOJ.
Table 2.
Effect of prolonged drought stress on metabolite levels in the leaves of SUL and NOJ (μg g−1 DW)
| SUL control | SUL treated | NOJ control | NOJ treated | |
| Organic acids | ||||
| p-Hydroxybenzoic acid | 116.5±2.4 | 144.3±18.3 | 79.5±3.1 | 62.2±1.5 |
| α-Ketoglutaric acid | 738.7±37.5 | 702.3±11.6 | 805.0±1.4 | 711.3±51.5 |
| Ascorbic acid | 227.9±8.3 | 167.1±5.1 | 156.8±8.2 | 115.4±2.0 |
| Caffeic acid | 869.3±5.9 | 1814.4*±123.0 | 314.9±3.5 | 356.4±8.7 |
| Citric acid | 10681.8±141.1 | 17042.1±267.4 | 4717.0±203.9 | 5172.9±153.9 |
| Fumaric acid | 367.1±9.4 | 281.7±13.3 | 148.5±6.0 | 112.9±7.7 |
| Isocitric acid | 2130.0±26.2 | 2209.5±25.6 | nd | 1385.0*±39.9 |
| Malic acid | 19630.0±24.2 | 18977.6±103.5 | 13013.8±124.6 | 9043.4*±49.0 |
| Malonic acid | nd | nd | 90.6±1.5 | 1119.9*±33.7 |
| Quinic acid | 519.4±3.7 | 414.7±13.6 | 832.4±29.3 | 402.7*±15.1 |
| Succinic acid | 1150.7±22.4 | 1017.3±14.1 | 944.0±36.9 | 741.5±5.4 |
| Threonic acid | 718.0±17.3 | 585.9±17.2 | 397.2±10.6 | 350.4±7.4 |
| Total | 37149.5±153.7 | 43356.9±315.2 | 21499.9±244.1 | 19573.9±178.7 |
| Amino acids | ||||
| GABA | 391.8±5.7 | 557.6*±11.6 | 532.9±24.4 | 1098.4*±24.6 |
| Alanine | 60.3±1.1 | 60.2±0.6 | 70.5±4.2 | 74.5±2.1 |
| Aspartic acid | 746.9±17.6 | nd | nd | nd |
| Glutamic acid | 271.9±5.1 | 339.3±38.8 | 402.1±4.4 | nd |
| Glycine | 393.4±4.4 | 464.8±13.0 | 96.9±0.1 | 89.9±0.5 |
| Proline | nd | 295.1*±7.8 | nd | 613.9*±18.2 |
| Pyroglutamic acid | 132.3±2.6 | 180.6±1.5 | 47.7±2.6 | 78.7±2.4 |
| Serine | 226.5±7.9 | nd | 143.4±3.3 | nd |
| Threonine | 49.3±1.5 | 57.5±1.9 | 53.4±1.5 | 51.7±1.1 |
| Total | 2272.4±21.5 | 1955.1±43.3 | 1346.9±25.5 | 2007.0±30.8 |
| Sugars | ||||
| Arabinose | 310.5±6.9 | 320.8±5.4 | 115.8±12.2 | 82.0±1.7 |
| Fructose | 16968.1±279.7 | 18172.0±752.0 | 13299.7±20.4 | 13169.8±208.7 |
| Galactose | 4060.0±216.2 | 6585.0±332.2 | 6724.8±55.5 | 8264.2*±254.6 |
| Glucose | 13993.8±73.5 | 16203.2±704.0 | 12912.3±173.1 | 15240.2*±206.9 |
| Glucose-6-phosphate | 545.2±22.6 | 236.1*±3.2 | 599.5±23.1 | 396.3*±10.5 |
| Isomaltose | 366.9±11.8 | 603.2*±14.7 | 238.2±3.8 | 1101.2*±21.3 |
| Maltose | 538.2±18.3 | 632.8±2.6 | 818.9±87.5 | 514.8*±32.1 |
| Melezitose | 319.3±17.5 | 170.2*±2.6 | 253.2±3.1 | 125.1±10.9 |
| Rhamnose | 91.7±0.8 | 107.3±3.5 | 88.3±0.7 | 91.7±2.8 |
| Ribose | 405.5±10.5 | 969.8*±3.2 | 182.8±1.9 | 270.8*±5.9 |
| Sorbose | 7962.0±87.0 | 9463.3±608.1 | 4175.9±460.7 | 3711.2±9.3 |
| Sucrose | 14282.2±673.7 | 14441.9±283.2 | 14960.6±810.4 | 14624.9±37.9 |
| Trehalose | 626.4±20.2 | 303.7*±43.1 | 625.0±19.2 | 688.6±34.2 |
| Xylose | 1539.9±51.9 | 600.8*±23.9 | 710.6±9.4 | 714.8±40.4 |
| Total | 62009.6±772.2 | 68810.1±1274.4 | 55705.6±954.6 | 58995.5±396.6 |
| Sugar alcohols | ||||
| Galactinol | 2676.9±50.1 | 4632.1*±143.7 | 4870.4±77.6 | 5322.5±30.6 |
| Glycerol | 1016.6±25.5 | 953.4±40.1 | 668.1±16.6 | 664.1±29.1 |
| Inositol | 2646.3±22.1 | 2484.5±10.5 | 3859.0±88.9 | 3399.3±17.5 |
| Total | 6339.9±60.4 | 8070.0±149.6 | 9397.5±119.2 | 9385.8±45.7 |
Two comparisons were made. (i) Values obtained for control and treated samples for each genotype were compared with each other. Significant differences between the genotypes are marked in bold. (ii) Treated and control samples of each genotype were compared. Significant differences are indicated with an asterisk. nd, not determined. The table shows the average value plus the standard error. The leaves were harvested after drought stress application (25 d for NOJ, and 27 d for SUL). Data sets contained five replicates per sample (three samples) and were statistically analysed by t test and one-way ANOVA using the algorithm incorporated into Microsoft Excel 2002 (Microsoft Corporation, Seattle, WA, USA). Differences were determined to be statistically significant at P <0.05. Principal Component Analysis (PCA) was performed using XLSTAT-Pro (v.7.5.3) program (Addinsoft, NY, USA).
Effects of drought stress on genes encoding glycolytic and mitchondrial function
Since the physiological phenotype used to categorize the potato genotypes as relatively resistant or susceptible was photosynthetic rate at the time of maximum stress, together with the ability to recover upon subsequent rewatering, it was of interest to determine how gene expression associated with chloroplast function responded at the point of maximum stress. However, most of the genes involved in chloroplast function, have a similar response between SUL and NOJ. Therefore, given the close coupling of chloroplast and mitochondrial activities in leaves under stress (Noctor et al., 2007), the expression of genes associated with mitochondrial function was examined (Fig. 3).
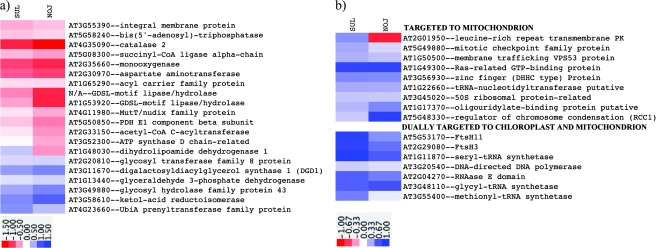
Fig. 3.
Effect of prolonged drought stress (‘maximum stress’) on the expression of genes associated with mitochondrial function. TM4 was used to analyse the microarray data. Normalization was done using MIV and MIDAS (total intensity normalization, Lowess normalization, standard deviation regularization, and low intensity filtering). Then a one-class t test in the Multi Experiment Viewer (MEV) module of TM4 was used to find differentially expressed genes. A gene was considered to be induced or repressed if the log2 (fold change) was found to be statistically significant with a P-value lower than 0.05. A total of three slides were analysed per genotype (each slide contained a biological replicate of a control and a treated plant). (a) Mitochondrial metabolism and (b) the regulation of gene expression in mitochondria and dually (chloroplast and mitochondria) targeted gene products. The colour scale for each heat map is located below each heat map.
Several genes associated with mitochondrial function were more negatively affected in NOJ than in SUL (Fig. 3a,b). Genes encoding components of the mitochondrial pyruvate dehydrogenase complex (PDH) were differentially affected in the genotypes. A homologue of At5g50850, the E1 component was significantly more repressed in NOJ than in SUL, and an At1g48030 homologue, dihydrolipoamide dehydrogenase, was induced in SUL but repressed in NOJ. At3g52300, the ATP synthase D chain, was also more repressed in NOJ than in SUL, as was an At4g11980 homologue, encoding a MutT/nudix family protein with similarity to a S. tuberosum ADP-sugar bisphosphatase. Two genes encoding members of the GDSL-motif lipase/hydrolase family of proteins, At1G53920, and a second GDSL motif gene with no known homologue in Arabidopsis, were repressed to a greater extent in NOJ than in SUL.
Increases in gene expression in SUL, and decreases in NOJ, were observed in UbiE/COQ5 methyltransferase (which has a low similarity to phosphatidylethanolamine N-methyltransferase), and pyruvate kinase, associated with glycolysis. A gene encoding β-carotene hydroxylase in the xanthophyll and ABA biosynthetic pathways was induced to a greater degree in SUL than in NOJ, as was a member of the FtsH protease gene family. FtsH11, which is targeted to both plastids and mitochondria (Fig. 3b), has been associated with thermotolerance in Arabidopsis (Chen et al., 2006).
Differential expression of transcription factors in SUL and NOJ
Since SUL was more tolerant compared to NOJ, the effects of drought on the expression of transcription factor genes was examined to determine whether any of the responses detected in SUL were related to known drought-stress responsive genes, or related to tolerance in other stresses. Figure 4 shows the expression of 39 transcription factors (TFs) that were significantly (P >0.05) induced or repressed in SUL and NOJ. Figure 4a, shows 22 TFs that were more induced in SUL compared to NOJ and, if repressed, repressed to a lesser extent than in NOJ. Among these 22 TFs, four TFs are induced in SUL while being repressed in NOJ; of these GAI (At1g14920), a DELLA protein, which is a gibberellic acid (GA) negative regulator, is also responsive to salt stress, ABA and ethylene (Achard et al., 2006). The other three genes that are induced in SUL and repressed in NOJ are a bZIP (At5g04840), a zinc finger (At1g16640), and a high mobility protein (At1g20636). There are five TFs that are induced to a higher level (2-fold or more) in SUL than in NOJ, and three of them are auxin responsive TFs (Fig. 4a).
Fig. 4.
Effect of prolonged drought stress (‘maximum stress’) on the expression of transcription factor genes in SUL and NOJ. Transcription factor genes that show a higher induction in SUL and, if repressed, are repressed to a lesser extent compared with NOJ are grouped in heat map (a). NOJ genes more induced or less repressed compared with SUL are shown in heat map (b). Data analysis as in Fig. 3.
Figure 4b shows TFs more highly induced in NOJ compared to SUL, and, if repressed, with the repression being less pronounced than in SUL. Among 17 TFs, only one gene is induced in NOJ but repressed in SUL: the jumonji domain-containing At1g78280. Also, six TFs are induced >2-fold in NOJ compared to SUL, including an ABA-responsive MYB (At3g09600). Another ABA-responsive MYB (AT1G74840) is 1.4 times more induced in NOJ than in SUL. The WRKY1 TF (At2g04880), was induced >2-fold in NOJ compared to SUL. This WRKY, known to be biotic stress induced, is involved in salicylic acid signalling (Duan et al., 2007), while five other WRKY TFs were repressed in both Andigena lines (Fig. 4a).
Differential expression of transporter genes in SUL and NOJ
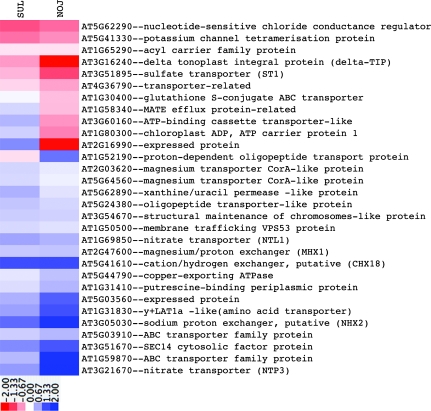
Given the material used in our studies, it is expected that the functional category ‘transport’ would provide information indicating how the two lines integrated stress at the level of the leaves. Transcripts in this category (Fig. 5) showed generally the same trend in response to water deficit in both genotypes, although the magnitude of the change was in some cases substantially different and NOJ showed more pronounced absolute changes. Among the down-regulated transcripts most affected appeared potassium and chloride conductance, sulphate transport, and an unspecified transporter in the major facilitator superfamily. In contrast, transcripts that controlled nitrate, amino acid, oligopeptide, phosphoglyceride movement, several antiporters in the proton/monovalent cation category, and uncharacterized ABC transporters were induced. Only a few of the transcripts in this category showed line-specific differences. Suppressed in NOJ, the more stress-sensitive line, were a chloroplast ADP/ATP carrier, an ABC transporter, a glutathione S-conjugate ABC transporter, and a transcript encoding an uncharacterized (trans)-membrane function, whose Arabidopsis counterpart, At2g16990, is annotated as an endomembrane tetracycline transporter, which is induced by osmotic and cold stresses. In SUL, these functions are either unaffected or strongly induced. In contrast, a transcript encoding a proton-dependent oligopeptide transporter is induced in NOJ but minimally suppressed in SUL. The Arabidopsis orthologue, At1g52190, is down-regulated during senescence and under most abiotic and biotic stress treatments.
Fig. 5.
Differential expression of transporter genes in SUL and NOJ. Responsive genes in SUL or NOJ encoding transport proteins are shown. Data analysis as in Fig. 3.
Differential expression of chaperone genes at maximum drought stress in SUL and NOJ
HSPs have been shown to be induced under different abiotic stress conditions, conferring thermotolerance (Hong and Vierling, 2001). It was of interest, therefore, to determine which HSPs were induced in the two lines. Figure 6a shows chaperone genes that were expressed differently in SUL and NOJ. Most of the HSPs (10 out of 15) are induced to a higher degree in NOJ. The heat shock factor (similar to HSFA5, At4g13980) is induced in NOJ while suppressed in SUL. The genes induced in NOJ include two HSP100s, one whose gene product localizes to the chloroplast and one to the cytosol. In SUL, most of the genes are not induced as much as in NOJ, but three genes whose product localizes to the chloroplast have a higher induction compared to NOJ: an sHSP, an HSP70, and HSP40/DnaJ.
Fig. 6.
Differential expression of HSP genes (a) and antioxidant related genes (b). The subcellular location (Cyt, cytoplasm; Chl, chloroplast; Mito, mitochondria; ER, endoplasmic reticulum; ES, endomembrane system; Nu, nucleus; Mem, membrane) and the family to which each gene belongs (Type) are indicated for each gene. Both heat maps (a and b) have the same colour scale. Data analysis as in Fig. 3.
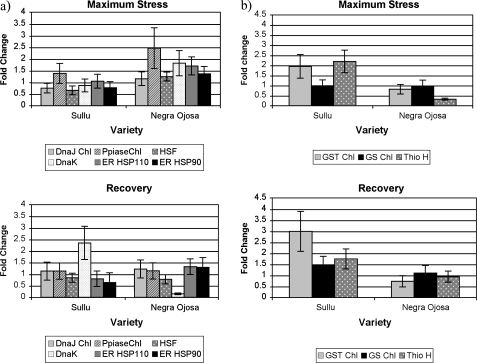
Real-time PCR confirmed the results found with microarrays (Fig. 7a). Other chaperone genes not included or not detected in the microarray hybridizations were also analysed: a homologue of BIP3 (luminal binding protein 3) and a homologue of GRP94 (ER resident HSP90). The general response during maximum stress of NOJ is higher than that of SUL. Most of the chaperone genes (two located in the chloroplast, two in the ER, one nuclear, and one in the cytosol) have a higher fold change in NOJ compared with SUL. No differences were observed between control samples in the two landraces (see Supplementary Table 4 at JXB online). During recovery, most of the genes analysed did not show any change, with the exception of HSP70, a homologue of the cytosolic HSC70-3 of tobacco, which is highly induced in SUL and, therefore, might have helped in the recovery of this plant after watering.
Fig. 7.
Real-time PCR results for selected chaperone genes (a) and antioxidant related genes (b) at maximum stress and recovery. Real-time PCR was carried out on cDNA from leaf samples from SUL and NOJ at maximum stress and 24 h after watering (recovery). HSP40 Chl and Ppiase Chl products are located in the chloroplast. GST, glutathione-S-transferase; GS, glutathione synthetase; Thio H; thioredoxin H.
Differential expression of antioxidant genes in SUL and NOJ
The expression of genes related to oxidative stress was investigated, since high levels of drought stress also causes oxidative stress (Selote et al., 2004).
Most of the genes directly related to antioxidant responses (Mittler et al., 2004) are induced in SUL, while they are repressed or induced to a lesser degree in NOJ (Fig. 6b). Moreover, most of the induced genes in SUL encode proteins that are involved in glutathione synthesis (one glutathione synthetase, homologue to At5g27380) or glutathione transport (one glutathione-S-transferase, homologue to At3g03190, and a glutathione transporter, homologue to At1g30400). Also, two thioredoxins that are highly induced in SUL are repressed or weakly induced in NOJ. One is a homologue to an Arabidopsis thioredoxin H At5g39950 (TrxH) and the other one to thioredoxin 3 in Plantago major (common plantain).
Real-time PCR was performed on three antioxidant genes: thioredoxin H (At5g39950), glutathione-S-transferase (At3g03190), and glutathione synthetase (At5g27380), for the points of maximum stress and recovery. In both cases, these genes are induced in SUL, compared to NOJ, where they are repressed or unchanged (Fig. 7b). This confirms the microarray results, and also adds the information that antioxidant genes are induced during recovery in SUL, especially glutathione-S-transferase. The homologue of this gene in Arabidopsis (AT3G03190) has been shown to be responsive to oxidative stress (Richards et al., 1998).
One gene associated with flavonoid synthesis was more induced in SUL compared to NOJ, a homologue to chalcone synthase (Fig. 6b). The homologue of this gene in Arabidopsis (At5g13930) localizes to the ER and the nucleus.
Metabolite responses during drought stress
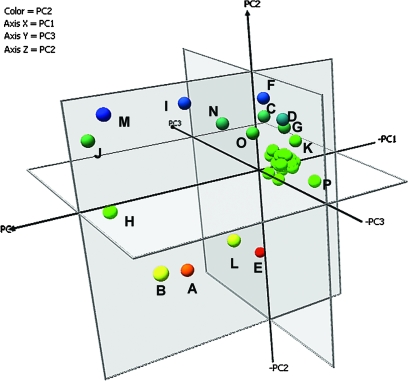
Metabolite profiling may be expected to provide additional information complementing and, to some degree, explaining transcript behaviour. The focus here was on the 38 most abundant metabolites in major carbon, amino acid, and sugar alcohol pathways. The PCA analysis (Fig. 8) showed that the first three PCs represented 98.87% of the initial variability contained in the original data. The three-dimensional plot of the PCA for the first three PCs allowed for a more detailed separation of the metabolites. Metabolites that are spatially close to each other are significantly positively correlated (r=1). Those appearing in an orthogonal orientation are not correlated (r=0). Those components on opposite sides of the centre are significantly negatively correlated (r= –1).
Fig. 8.
PCA of metabolites correlated with genotype (axis PC1) and with genotype×environment differences between lines (axis PC2 and PC3). Metabolites coloured in blue are most highly correlated with genotype×drought responses; in red and yellow are correlated with genotypic differences; and in green are not significantly correlated with genotype or genotype×drought. A, citrate; B, malate; C, malonate; D, quinate; E, glycine; F, proline; G, serine; H, fructose; I, galactose; J, glucose; K, melezitose; L, sorbose; M, sucrose; N, galactinol. O, inositol; P, ethanolamine.
SUL contained much higher levels of organic acids than NOJ in control and stress states. NOJ, in contrast, contained higher amounts of sugar alcohols. Individual organic acids that contributed to the differences in total organic acids between the genotypes under both conditions were five TCA cycle intermediates: succinic, citric, fumaric, isocitric, and malic acids. Of these five organic acids, only citric acid showed higher levels in drought-stressed plants. The glyoxylate cycle, utilizing carbon skeletons from the breakdown of fatty acids via β-oxidation, may also be operating at a higher level in SUL regardless of stress, since citrate, isocitrate, malate, and succinate levels were all higher in SUL than in NOJ under both conditions (Cornah and Smith, 2002). Other organic acids that had higher levels in SUL than in NOJ were caffeic, p-hydroxybenzoic, and threonic acids. Of these three, only caffeic acid showed higher levels in drought-stressed than in well-watered plants (Table 2).
Total amino acid amounts were comparable in both lines. However, in NOJ, proline and GABA were in the majority, while SUL maintained a more balanced composition of amino acids. Glycine and glutamic acid levels were both higher in SUL under drought stress than under well-watered conditions. In NOJ, glutamic acid was not detectable under drought conditions. Glycine levels were four times higher in drought-stressed SUL than in NOJ under the same conditions. Serine and aspartic acid levels declined to non-detectable levels in both lines under drought conditions. Phenylalanine and tyrosine, the amino acid precursors for the phenylpropanoid pathway, were present only in spurious amounts in all samples.
Total sugar levels were slightly higher in SUL than in NOJ under both conditions, although differences in behaviour under drought conditions were observed in both cases (Fig. 8; Table 2). The complexity and amount of various sugars increased in SUL under drought. Fructose, ribose, sorbose, and xylose levels were higher in SUL under well-watered conditions. Fructose, glucose, galactose, isomaltose, ribose, and sorbose levels in drought-stressed SUL were higher than in their corresponding controls. By contrast, in NOJ, ribose and sorbose levels were the same under both conditions, and galactose was higher than SUL in both cases, with an increase occurring under drought conditions. Isomaltose levels increased in NOJ under drought to twice the level observed in SUL under the same conditions. Trehalose levels under drought were also over twice as high in NOJ. This was due to a decline in levels of this sugar in drought conditions in SUL, while levels in stressed plants in NOJ were comparable to those observed at the non-stressed level.
Total sugar alcohol levels were higher in NOJ under both conditions. Galactinol levels increased in both genotypes, although the increase was greater in SUL than in NOJ. Raffinose could not be detected in either case in control or stressed samples, despite being in the same biosynthetic pathway as galactinol (Cook et al., 2004).
PC analysis of metabolite data
A table of the squared cosines allowed linkage of the PC2 and PC3 axes with the experimental conditions (drought versus control), while axis PC1 identified genotypic differences. PCA analysis of the contribution of each variable to the first three PCs identified nine metabolites as significant contributors to the variance (Fig. 8; Table 3).
Table 3.
Axis components of PCA analysis of metabolites from SUL and NOJ
| PC1 | PC2 | PC3 |
| Fructose | Citrate | Citrate |
| Glucose | Malate | Malate |
| Malate | Galactose | Glucose |
| Sucrose | Sucrose | Galactose |
| Citrate | Glucose | Sucrose |
| Sorbose | ||
| Galactinol | ||
| Inositol |
The list of metabolites for each PC identifies metabolites with highest membership in each PC that form the group of nine compounds (marked in bold) that are selected as significant by PCA. PC1 and PC2 explained >90% of the variability, with PC1 representing genotype difference and PC2 (and PC3) representing response differences.
Discussion
The Andigena landraces studied were better adapted to low water conditions compared to the Tuberosum genotype (Atlantic)
The Andigena landraces studied were better adapted to low water conditions compared to the Tuberosum genotype (Atlantic). Overall, the Andigena genotypes performed better under drought stress conditions compared to Atlantic. However, the Andigena lines showed a different phenotype (i.e. all showing smaller leaves; Fig. 1), and therefore a direct comparison with Tuberosum was not possible without determining stomatal numbers and densities. Nevertheless, there was a different response among the Andigena lines not dependent on their identical growth habits and leaf structure, since not all of them exhibited the same degree of drought resistance. Based on photosynthesis (Fig. 2; see Supplementary Table 3 at JXB online) and visual observation of morphological traits, it was possible to identify two extreme genotypes, the resistant SUL and the susceptible NOJ.
Mitochondrial responses
Samples were taken when photosynthesis in SUL was inhibited by 50% and by 80% in NOJ. Thus, it seems reasonable to assume that gene expression in the samples will be related to resistance mechanisms. Considering that SUL recovered completely within 24 h after rewatering, it was expected that resistance mechanisms are more successfully engaged in this line. One difference that appears to emerge from transcript as well as metabolite analysis points toward higher mitochondrial metabolic activity in SUL than in NOJ (Fig. 3a,b; Table 2). Signature functions pointing in this direction include the different expression, higher in SUL, of components of the PDH complex, the TCA cycle and mitochondrial ATP synthesis, at least at the point of maximal stress. By contrast, lipid breakdown may have been occurring at a higher rate in NOJ. Irrespectively, the induction of genes associated with signalling in the mitochondrion was of a similar extent in both genotypes.
The Arabidopsis orthologues of many drought-stress-responsive potato genes also respond to heat stress (Zimmermann et al., 2004; Busch et al., 2005). For example, of the 16 genes associated with chloroplast metabolism that responded to drought stress in our study (see Supplementary Fig. 1 at JXB online), 11 orthologues in Arabidopsis responded in like fashion to heat shock (Busch et al., 2005) in SUL and nine in NOJ, suggesting that either the plants were experiencing heat stress as a result of the closure of their stomata after 25 d of drought stress, or that the heat-responsive pathways also respond to drought.
The physiological differences between the genotypes may have been reflected in the differential response of the orthologue of the plastid- and mitochondrion-targeted FtsH11, which has been associated with thermal tolerance (Chen et al., 2006), where SUL showed a higher degree of induction than NOJ. The greater induction of chloroplast-localized antioxidants, including a gene in the xanthophyll biosynthetic pathway, and also chloroplast-localized chaperones in SUL, may be manifestations of a heat response pathway that enabled SUL to recover more effectively from drought stress.
Transcription factors induced in NOJ are involved in SA-responsive pathways
ABA-responsive TFs were induced in NOJ to a higher degree compared to SUL. The two Myb transcription factors, homologues of At1g74840 and At3g09600, apart from being regulated by ABA, are also induced by gibberellic acid (GA), jasmonic acid (JA), salicylic acid (SA), and salt (NaCl), but JA and SA induced this gene to a higher level compared to GA and ABA (Yanhui et al., 2006). The TF WRKY1 was induced in the experiment only in NOJ. WRKY 1 mediates responses in the SA signalling pathway (Duan et al., 2007). Thus, the degree of stress in NOJ might have activated biotic stress defence pathways that are controlled by SA. Moreover, cross-talk between abiotic and biotic stress-related genes has been shown (Cheong et al., 2002; Seki et al., 2002). ROS generation occurs during abiotic stress and biotic stress (Apel and Hirt, 2004) and SA can induce the formation of ROS molecules such as H2O2 (Kauss and Jeblick, 1995; Shirasu et al., 1997). ROS scavenging genes (antioxidants) were suppressed or not responsive in NOJ (Fig. 6b), probably resulting in ROS accumulation, that might have induced the expression of the TFs mentioned above.
Drought–mediated expression of HSP for plastid-localized functions may contribute to SUL tolerance to drought stress
As shown in Fig. 6a, two heat shock genes induced in SUL are part of the HSP70 chaperone complex (HSP70 and HSP40) whose products localize to the chloroplast. Moreover, a chloroplastic sHSP is also induced in SUL, and in vitro work has shown that sHSPs can help HSP70s in the repair of proteins (Lee and Vierling, 2000).
HSP genes targeted to the cytosol were induced in NOJ, but not in SUL. Two HSP40-DnaJ genes are induced to a higher degree in NOJ than in SUL. HSP40s play a role in mammalian and plant systems in the activation of the ATPase activity of HSP70/DnaK (Miernyk, 2001). One HSP70/DnaK gene, whose product also localizes to the cytosol, is also greatly induced in NOJ, suggesting the formation of the HSP70/HSP40 complex in the cytosol for protein refolding. Another HSP gene, a homologue of At1g74310, a cytosolic HSP100 gene was highly induced in NOJ, which is necessary for thermotolerance but dispensable for development (Hong and Vierling, 2001). This gene is induced almost 5-fold more in NOJ compared with SUL. Virus infection can induce the expression of HSP70s localized in the cytosol (Aparicio et al., 2005). Moreover, overexpression of unfolded proteins in the cytosol induced the expression of the same cytosolic HSP70s, suggesting that a mechanism similar to the ER unfolded protein response also occurs in the cytosol (Aparicio et al., 2005). This, together with the induction of a cytosolic HSP100, a protein needed for thermotolerance, suggest that there was a higher amount of unfolded proteins in NOJ, probably because of the higher degree of stress that NOJ was experiencing. A homologue of HSF5, cold and heat-responsive heat shock factor 5 (Miller and Mittler, 2006), was also induced in NOJ, but not in SUL.
Antioxidant gene expression under drought may help SUL tolerate drought stress
Most antioxidant genes that show a response in this experiment are induced in SUL, while they are not responsive or repressed in NOJ. Moreover, their gene products are localized to all subcellular compartments (with the exception of the mitochondrion, Fig. 6b) suggesting greater ROS responsiveness in SUL than in NOJ.
Many of the antioxidant genes induced in SUL are glutathione related (four out of eight genes induced). The GST gene induced in SUL is a homologue of the petunia GST An9 (79% similarity). In maize and petunia, this protein seems to be necessary for conjugation of anthocyanins before they can be transferred to the vacuole (Alfenito et al., 1998). So, in addition to its antioxidant function, GSTF11 might be involved in the transport of anthocyanins to the vacuole. It has previously been shown that genes associated with flavonoid metabolism are induced upon recovery from stress in drought-resistant accessions of Andigena potato as well as in loblolly pine (Watkinson et al., 2003, 2006). In SUL, but not in NOJ, chalcone synthase, the first enzyme in the flavonoid pathway, is induced during maximum drought stress, suggesting flavonoid biosynthesis. The fact that these genes are being induced at the time of maximum stress suggests that flavonoid production is not only crucial to the restoration of photosynthetic capability upon recovery from stress, as has previously been shown (Watkinson et al., 2003, 2006), but to protection during stress exposure itself.
NOJ experiences stress to a higher degree than SUL
Metabolite profiles of NOJ are characterized by compounds indicative of stress, such as proline, which accumulate to a higher degree than in SUL. This stress-dependent increase of proline is observed in many organisms. Its function, typically assumed to act as a compatible solute and protective osmolyte, however, is not clear (Deuschle et al., 2004). The increase of proline in NOJ, whose physiological phenotype indicated a higher stress level than SUL, seems to dispute the general view that proline accumulation is the sole contributer to stress tolerance. This accumulation of proline by the sensitive phenotype has also been observed in other Andean potato physiology studies (Schafleitner et al., 2007a). Studies in Arabidopsis mutants with altered low water potential response have shown that not only proline accumulates under stress but also other solutes (Verslues and Bray, 2004). Proline excess under non-stress conditions is toxic to plants (Hellmann et al., 2000; Nanjo et al., 2003). Studies in mutants defective in proDH suggested that catabolism of proline is as important as its accumulation after osmotic stresses to achieve recovery (Hellmann et al., 2000; Nanjo et al., 2003).
An analogous conclusion can be drawn from the results obtained for GABA, where, although levels rose under drought in both cases, the level attained in NOJ in drought-stressed samples was twice as high as that detected in SUL under the same conditions. The GABA metabolic pathway, which also includes glutamate, is known to respond to both abiotic and biotic stresses (Bouche and Fromm, 2004), although its function in plant cells is not yet clear. There is evidence, however, that suppression of the GABA shunt in Arabidopsis leads to increased susceptibility to oxidative stress (Bouche et al., 2003). Again, as for the proline result, the data presented here are compatible with the view that NOJ is suffering from a greater stress load than is SUL and, therefore, has had to marshal its defence processes to a greater degree.
The higher levels of TCA intermediates under both conditions in SUL suggests greater mitochondrial activity than in NOJ, which is directed towards generating more reductant and ATP, but possibly also into mitochondrial fatty acid formation (Wada et al., 1997) and for the provision of carbon skeletons for amino acid biosynthesis. This higher activity in the mitochondria is correlated with gene expression. Genes encoding subunits of pyruvate dehydrogenase were differentially expressed in the two genotypes, and the higher level of organic acids may also be an indication of gluconeogenic activity. Malonic acid can act as an inhibitor of the TCA cycle enzyme succinic dehydrogenase (Greene et al., 1993). The accumulation of malonic acid in NOJ may have led to the observed lower levels of TCA cycle intermediates in that genotype than in SUL.
In conclusion, the Andigena genotypes studied are more resistant to drought stress than the Tuberosum genotypes studied. This is associated with morphological differences. However, there is a diverse response to drought among Andigena genotypes, which is independent of morphology/habit. The higher drought stress resistance found in SUL is correlated with concerted induction of heat shock proteins and antioxidant genes encoding proteins located in the chloroplast and genes for anthocyanin synthesis and transport. The data are compatible with higher levels of fatty acid biosynthesis and mitochondrial activity in the more drought-resistant landrace. The response of well-studied osmolytes and of the GABA pathway to the stress was not correlated with resistance and ability to recover, but rather with the degree of stress experienced by the plant. The metabolite data provide validation for the transcript profile data. Future work will include a functional genomic examination of the effects of drought stress on tubers, in the same experimental system, and also detailed comparisons between drought stress responses in Andigena and Tuberosum genotypes.
Supplementary data
Supplementary tables and figures are available at JXB online.
Supplementary Table 1. Information about the potato genotypes.
Supplementary Table 2. Primers used for real-time PCR.
Supplementary Table 3. ANOVA results for evaluated traits.
Supplementary Table 4. RT PCR expression data normalized against adenosine kinase.
Supplementary Fig. 1. Effect of drought stress in the expression of genes associated with chloroplast function.
Supplementary Material
Acknowledgments
The work has been supported by NSF DBI 0223905 and IBN0219322 and by CIP, UIUC, and VT institutional grants. We are grateful to members of the POCI consortium for affording us access to the arrays and annotations.
Glossary
Abbreviations
- SUL
Sullu
- NOJ
Negra Ojosa
- ROS
reactive oxygen species
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R, Walbot V. Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. The Plant Cell. 1998;10:1135–1149. doi: 10.1105/tpc.10.7.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio F, Thomas CL, Lederer C, Niu Y, Wang D, Maule AJ. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiology. 2005;138:529–536. doi: 10.1104/pp.104.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Bouche N, Fait A, Bouchez D, Moller SG, Fromm H. Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proceedings of the National Academy of Sciences, USA. 2003;100:6843–6848. doi: 10.1073/pnas.1037532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Fromm H. GABA in plants: just a metabolite? Trends in Plant Science. 2004;9:110–115. doi: 10.1016/j.tplants.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. The Plant Journal. 2005;41:1–14. doi: 10.1111/j.1365-313X.2004.02272.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Burke JJ, Velten J, Xin Z. FtsH11 protease plays a critical role in Arabidopsis thermotolerance. The Plant Journal. 2006;48:73–84. doi: 10.1111/j.1365-313X.2006.02855.x. [DOI] [PubMed] [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiology. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornah J, Smith SM. Synthesis and function of glyoxylate cycle enzymes in plants. In: Baker A, Graham I, editors. Plant peroxisomes. The Netherlands: Kluwer; 2002. pp. 57–101. [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunze R, Frommer WB. The role of [Delta]1-pyrroline-5-carboxylate dehydrogenase in proline degradation. The Plant Cell. 2004;16:3413–3425. doi: 10.1105/tpc.104.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M-R, Nan J, Liang Y-H, Mao P, Lu L, Li L, Wei C, Lai L, Li Y, Su X-D. DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Research. 2007;35:1145–1154. doi: 10.1093/nar/gkm001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO/ESS. Major Food and Agricultural Commodities and Producers. 2005 [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nature Biotechnology. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Greene JG, Porter RH, Eller RV, Greenamyre JT. Inhibition of succinate dehydrogenase by malonic acid produces an ‘excitotoxic’ lesion in rat striatum. Journal of Neurochemistry. 1993;61:1151–1154. doi: 10.1111/j.1471-4159.1993.tb03634.x. [DOI] [PubMed] [Google Scholar]
- Hawkes JG. The potato: evolution, biodiversity and genetic resources. London, UK: Belhaven Press; 1990. [Google Scholar]
- Hellmann H, Funck D, Rentsch D, Frommer WB. Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiology. 2000;123:779–789. doi: 10.1104/pp.123.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E. Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. The Plant Journal. 2001;27:25–35. doi: 10.1046/j.1365-313x.2001.01066.x. [DOI] [PubMed] [Google Scholar]
- Huaman Z, Ross RW. Updated listing of potato species names, abbreviations and taxonomic status. American Potato Journal. 1985;62:629–641. [Google Scholar]
- Joliffe A. Prinicipal Components Analysis. New York: Springer; 1986. [Google Scholar]
- Kauss H, Jeblick W. Pretreatment of parsley suspension cultures with salicylic acid enhances spontaneous and elicited production of H2O2. Plant Physiology. 1995;108:1171–1178. doi: 10.1104/pp.108.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Molecular Biology. 1994;25:791–798. doi: 10.1007/BF00028874. [DOI] [PubMed] [Google Scholar]
- Kumar R, Kang GS. Usefulness of Andigena (Solanum tuberosum ssp andigena) genotypes as parents in breeding early bulking potato cultivars. Euphytica. 2006;150:107–115. [Google Scholar]
- Lee GJ, Vierling E. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiology. 2000;122:189–198. doi: 10.1104/pp.122.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovaya V, Ulanov A, Lygin A, Duncan D, Widholm J. Biochemical features of maize tissues with different capacities to regenerate plants. Planta. 2006;224:1385–1399. doi: 10.1007/s00425-006-0328-7. [DOI] [PubMed] [Google Scholar]
- Miernyk JA. The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones. 2001;6:209–218. doi: 10.1379/1466-1268(2001)006<0209:tjdpoa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Annals of Botany (London) 2006;98:279–288. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends in Plant Science. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Nanjo T, Fujita M, Seki M, Kato T, Tabata S, Shinozaki K. Toxicity of free proline revealed in an Arabidopsis T-DNA-tagged mutant deficient in proline dehydrogenase. Plant and Cell Physiology. 2003;44:541–548. doi: 10.1093/pcp/pcg066. [DOI] [PubMed] [Google Scholar]
- Noctor G, De Paepe R, Foyer CH. Mitochondrial redox biology and homeostasis in plants. Trends in Plant Science. 2007;12:125–134. doi: 10.1016/j.tplants.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Pastori GM, Foyer CH. Common components, networks, and pathways of cross-tolerance to stress. The central role of ‘redox’ and abscisic acid-mediated controls. Plant Physiology. 2002;129:460–468. doi: 10.1104/pp.011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radin JW. Water relations of cotton plants under nitrogen deficiency. III. Stomatal conductance, photosynthesis, and abscisic acid accumulation during drought. Plant Physiology. 1981;67:115–119. doi: 10.1104/pp.67.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC. Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiology. 1998;116:409–418. doi: 10.1104/pp.116.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiology. 2002;130:1143–1151. doi: 10.1104/pp.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JM, Pharis RP, Huang YY, Reid DM, Yeung EC. Drought-induced increases in abscisic acid levels in the root apex of sunflower. Plant Physiology. 1985;79:1086–1089. doi: 10.1104/pp.79.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. The Plant Journal. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview: extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Satoh R, Nakashima K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiology. 2002;130:709–719. doi: 10.1104/pp.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafleitner R, Gaudin A, Gutierrez Rosales RO, Alvarado Aliaga CA, Bonierbale M. Proline accumulation and real time PCR expression analysis of genes encoding enzymes of proline metabolism in relation to drought tolerance in Andean potato. Acta Physiologiae Plantarum. 2007a;29:19–26. [Google Scholar]
- Schafleitner R, Gutierrez Rosales RO, Gaudin A, Alvarado Aliaga CA, Nomberto Martinez G, Tincopa Marca LR, Avila Boliva L, Mendiburu Delgado F, Simon R, Bonierbale M. Capturing candidate drought tolerance traits in two native Andean potato clones by transcription profiling of field grown plants under water stress. Plant Physiology and Biochemistry. 2007b;45:673–690. doi: 10.1016/j.plaphy.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Selote DS, Bharti S, Khanna-Chopra R. Drought acclimation reduces O2* accumulation and lipid peroxidation in wheat seedlings. Biochemical and Biophysical Research Communications. 2004;314:724–729. doi: 10.1016/j.bbrc.2003.12.157. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiology. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defence mechanisms. The Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioson AA, Mane SP, Li P, Sha W, Heath LS, Bohnert HJ, Grene R. The statistics of identifying differentially expressed genes in Expresso and TM4: a comparison. BMC Bioinformatics. 2006;7:215. doi: 10.1186/1471-2105-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai GCC, Tarn TR. Multivariate analyses of potato hybrids. 2. Discrimination between Tuberosum–Andigena hybrid families and their relationship to their parents. Canadian Journal of Genetics and Cytology. 1980;22:279–286. [Google Scholar]
- Taylor J, King RD, Altmann T, Fiehn O. Application of metabolomics to plant genotype discrimination using statistics and machine learning. Bioinformatics. 2002;18:S241–S248. doi: 10.1093/bioinformatics/18.suppl_2.s241. [DOI] [PubMed] [Google Scholar]
- Terrazas F, Suarez V, Watson G, Thiele G, Walker T, Devaux A. Analysing potato productivity in farmers' fields in Bolivia. In: CIP, ed. CIP Program Report: 1996 CIP. [Google Scholar]
- Verslues PE, Bray EA. LWR1 and LWR2 are required for osmoregulation and osmotic adjustment in Arabidopsis. Plant Physiology. 2004;136:2831–2842. doi: 10.1104/pp.104.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Bray EA. Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. Journal of Experimental Botany. 2006;57:201–212. doi: 10.1093/jxb/erj026. [DOI] [PubMed] [Google Scholar]
- Wada H, Shintani D, Ohlrogge J. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proceedings of the National Academy of Sciences, USA. 1997;94:1591–1596. doi: 10.1073/pnas.94.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz C, Juenger M, Schad M, Kehr J. Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. The Plant Journal. 2002;31:189–197. doi: 10.1046/j.1365-313x.2002.01348.x. [DOI] [PubMed] [Google Scholar]
- Wang WX, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Watkinson JI, Sioson AA, Vasquez-Robinet C, et al. Photosynthetic acclimation is reflected in specific patterns of gene expression in drought-stressed loblolly pine. Plant Physiology. 2003;133:1702–1716. doi: 10.1104/pp.103.026914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson JI, Hendricks L, Sioson AA, Vasquez-Robinet C, Stromberg V, Heath LS, Schuler M, Bohnert HJ, Bonierbale M, Grene R. Accessions of Solanum tuberosum ssp. andigena show differences in photosynthetic recovery after drought stress as reflected in gene expression profiles. Plant Science. 2006;171:745–758. [Google Scholar]
- Weisz R, Kaminski J, Smilowitz Z. Water-deficit effects on potato leaf growth and transpiration: utilizing fraction extractable soil-water for comparison with other crops. American Potato Journal. 1994;71:829–840. [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of levels of proline as an osmolyte in plants under water stress. Plant and Cell Physiology. 1997;38:1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093. [DOI] [PubMed] [Google Scholar]
- Zhu J, Gong Z, Zhang C, Song CP, Damsz B, Inan G, Koiwa H, Zhu JK, Hasegawa PM, Bressan RA. OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. The Plant Cell. 2002;14:3009–3028. doi: 10.1105/tpc.006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–263. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.