Abstract
We report high resolution solution 19F NMR spectra of fluorine-labeled rhodopsin mutants in detergent micelles. Single cysteine substitution mutants in the cytoplasmic face of rhodopsin were labeled by attachment of the trifluoroethylthio (TET), CF3-CH2-S, group through a disulfide linkage. TET-labeled cysteine mutants at amino acid positions 67, 140, 245, 248, 311, and 316 in rhodopsin were thus prepared. Purified mutant rhodopsins (6–10 mg), in dodecylmaltoside, were analyzed at 20°C by solution 19F NMR spectroscopy. The spectra recorded in the dark showed the following chemical shifts relative to trifluoroacetate: Cys-67, 9.8 ppm; Cys-140, 10.6 ppm; Cys-245, 9.9 ppm; Cys-248, 9.5 ppm; Cys-311, 9.9 ppm; and Cys-316, 10.0 ppm. Thus, all mutants showed chemical shifts downfield that of free TET (6.5 ppm). On illumination to form metarhodopsin II, upfield changes in chemical shift were observed for 19F labels at positions 67 (−0.2 ppm) and 140 (−0.4 ppm) and downfield changes for positions 248 (+0.1 ppm) and 316 (+0.1 ppm) whereas little or no change was observed at positions 311 and 245. On decay of metarhodopsin II, the chemical shifts reverted largely to those originally observed in the dark. The results demonstrate the applicability of solution 19F NMR spectroscopy to studies of the tertiary structures in the cytoplasmic face of intact rhodopsin in the dark and on light activation.
Keywords: G-protein-coupled receptors, signal transduction, conformational change, site-directed 19F labeling, membrane proteins
Light-catalyzed isomerization of 11-cis-retinal in rhodopsin results in a conformational change in the cytoplasmic face. Precise description of this change in rhodopsin and the corresponding conformational changes in G-protein coupled receptors in general on ligand binding is a long range goal of studies on signal transduction. Considerable insights into the tertiary structure in the cytoplasmic face of rhodopsin in the dark and its change on light activation have been obtained recently (1–4). However, none of the approaches used to date can be expected to provide detailed resolution of the tertiary structures involved. Ideally, three-dimensional structures of rhodopsin in the two states are needed. However, neither rhodopsin nor any other G-protein coupled receptor has so far yielded crystals satisfactory for such analysis. Two-dimensional studies using electron diffraction techniques have proven more successful, and these account for the progress made to date in the study of rhodopsin (5). However, the level of resolution remains extremely low for the cytoplasmic and the intradiscal domains.
In recent years, NMR spectroscopy has emerged as an alternative powerful approach for studies of protein structure and dynamics (6). While the technique has proved much more fruitful in studies of water-soluble proteins, increasing applications are now being made to hydrophobic polypeptides and membrane proteins (examples in refs. 7–11). However, the latter, because of their size and the necessity of studying them in micellar systems, present difficulties for NMR studies. Therefore, solution NMR application has been mostly limited to studies of peptides that represent segments of the membrane proteins of interest (examples in refs. 8 and 12–14).
NMR experiments with membrane proteins are exceptionally demanding of the current gene expression systems in vitro in that they require many milligrams of material. The recent development of high level expression of the bovine opsin gene and its mutants in mammalian cell lines (15) now makes possible the application of NMR spectroscopy. Recently, efficient incorporation of 15N-lysine and 13C-glycine into rhodopsin using a defined growth medium was realized. This enabled application of magic angle spinning NMR to the observation of the 15N-resonance corresponding to the protonated retinylidene Schiff base nitrogen in rhodopsin (16). A similar chemical shift was observed for the protonated retinylidene Schiff base nitrogen in isotope labeled rhodopsin obtained from expression in the Baculovirus/Sf9 system (17).
Solution 19F NMR offers distinct advantages in the study of protein structure and conformational changes because of its high sensitivity and the lack of 19F background in proteins (18, 19). Thus, 19F NMR has found application in structural studies of a number of membrane proteins (20, 21). The retinal chromophore in rhodopsin was also studied by the use of 19F-substituted retinal isomers (22).
Here we report on our first experiments that demonstrate the applicability of solution 19F NMR spectroscopy to detergent-solubilized rhodopsin. A number of rhodopsin mutants containing single cysteine residues in different regions of the cytoplasmic face (Fig. 1) have been prepared, and a general method for the facile attachment of the trifluoroethylthio (TET) group by disulfide linkage to the cysteine groups in the mutants has been developed (Fig. 2). Thus, TET-labeled derivatives of single cysteines located at positions 67, 140, 245, 248, 311, and 316 in the cytoplasmic face were prepared. Solution 19F NMR spectra recorded in the dark showed resolved chemical shifts for the majority of the TET-labeled mutants. On illumination to form metarhodopsin II (Meta II), the TET derivatives at positions 248 and 316 showed downfield shifts and those at positions 67 and 140 upfield shifts whereas little or no change in chemical shift was observed at positions 311 and 245. Furthermore, on decay of Meta II, the opsins formed showed return of the chemical shifts largely to their original positions in the dark.†
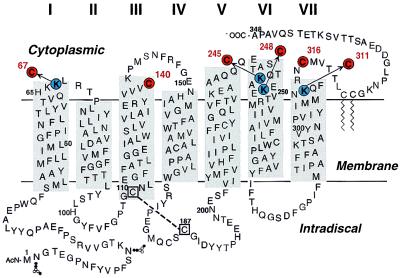
Figure 1.
Six single cysteine mutants of rhodopsin studied in this report, K67C, Cys-140, K245C, K248C, K311C, and Cys-316, are highlighted in a secondary structural model of bovine rhodopsin. For details of their preparation, see text.
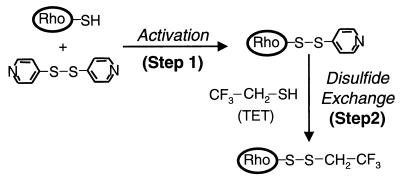
Figure 2.
A general method for attachment of an 19F label (TET) to the sulfhydryl groups of cysteine residues in the cytoplasmic face of rhodopsin.
Materials and Methods
Frozen retinas were purchased from J. A. Lawson Co. (Lincoln, NE). 11-cis-Retinal was a gift from Rosalie Crouch (University of South Carolina and the National Eye Institute of the National Institutes of Health U.S. Public Health Services). It was also prepared from all-trans-retinal after a published procedure (24). 4,4′-dithiodipyridine (4-PDS) was purchased from Sigma, dodecylmaltoside (DM) was from Anatrace (Maumee, OH), and dimyristoyl-phosphatidyl choline (DMPC) was from Avanti (Alabaster, AL). Antirhodopsin monoclonal antibody 1D4 (25) was purified from a myeloma cell line provided by R. S. Molday (University of British Columbia). It was coupled to cyanogen bromide-activated Sepharose 4B (Sigma) as described (25), at a level of ≈10 mg/ml of swollen Sepharose beads. The nonapeptide (Massachusetts Institute of Technology Biopolymers Laboratory) corresponding to the C-terminal sequence of rhodopsin, the antibody epitope, was used to elute rhodopsin mutants from 1D4-Sepharose. The sources of all reagents for cell culture have been described (15).
Buffers used were as follows: Buffer A, 137 mM NaCl, 2.7 mM KCl, 1.8 mM KH2PO4, 10 mM Na2HPO4 (pH 7.2); Buffer B, Buffer A + 1% DM; Buffer C, Buffer A + 0.05% DM; Buffer D, 2 mM NaH2PO4/Na2HPO4 (pH 6); Buffer E, Buffer D + 0.05% DM; Buffer F, Buffer D + 0.02% DM; Buffer G, Buffer D + 0.88% octyl glucoside (OG); Buffer H, Buffer E + 70 μM nonapeptide; Buffer I, Buffer F + 0.3 M α-methyl mannopyranoside; Buffer J, Buffer G + 0.3 M α-methyl mannopyranoside; Buffer K, 50 mM Tris (pH 8), 0.15 M NaCl; Buffer L, 20 mM Pipes, 1 mM MgCl2, 1 mM CaCl2, 0.1 mM EDTA, 0.15 M NaCl; Buffer M, Buffer K + 1% DM; Buffer N, Buffer L + 4% OG; Buffer O, Buffer L + 0.05% DM; Buffer P, Buffer L + 0.88% OG; Buffer Q, 20 mM NaH2PO4/Na2HPO4 (pH 6) in 99.9% D2O (Cambridge Isotopes, Andover, MA).
UV-visible Absorption and Fluorescence Spectroscopy.
UV-visible absorption spectra were recorded by using a spectrophotometer (Perkin–Elmer λ6). The molar extinction value used for rhodopsin was 40,600 M−1⋅cm−1 (26). Illumination of rhodopsin samples to form Meta II and the rates of their decay were measured by fluorescence spectroscopy as described (27).
19F NMR Spectroscopy.
19F NMR spectra were recorded on a Varian INOVA 500 spectrometer (frequency 470.668 MHz) at the instrumentation facility of our department of chemistry. Data acquisition and analysis was carried out by using vnmr 6.1 software (Varian). The sample was locked on deuterium. The relaxation delay was 0.5 sec. Line broadening was 20 Hz. Samples were referenced to an internal standard, trifluoroacetate (TFA). Acquisition time, number of scans averaged, and specific parameters were as indicated in the text or legends.
Conditions Used for 19F NMR Spectroscopy.
DM has proved to be the most suitable detergent in all of the structure–function work carried out so far with rhodopsin. Therefore, the present NMR spectroscopy of TET-labeled cysteine mutant was carried out in this detergent. However, because of much higher concentration of the detergent (between 4 and 7%; see below) during preparation of the mutants, the effect of detergent concentration on 19F NMR spectra was investigated as described in Results.
For spectra of the mutants after illumination, the detergent solubilized 19F labeled mutants were irradiated with a Dolan Jenner Fibre Lite P-2000 illuminator for 30 sec with a 495-nm cut-off and a heat-absorption filter before data acquisition. Meta II (λmax 380 nm) formed normally; however, the rate of its decay was increased relative to the rates previously recorded at 0.05% DM (1, 3). The T1/2 was ≈10 min at room temperature (RT) versus ≈13 min at 0.05% DM. [A similar effect has been observed previously in going from 0.02% DM concentration to 0.1% concentration (28)].
Construction of Plasmids Containing Synthetic Bovine Opsin Mutant Gene With Single Cysteine Codons.
Synthetic bovine opsin mutant genes containing single cysteine substitution codons were first created in the vector pMT4 (29). The following two single cysteine mutants have been described previously: K67C (3) and K311C (1). For preparation of the mutant genes containing single cysteine codons at positions K245C and K248C, the previously prepared plasmids (30) were digested with MluI/NotI, to excise the small fragments containing C322S and C323S substitutions. The resulting large fragments were ligated to the small MluI/NotI fragments from a mutant gene containing C140S/C316S replacement codons but that retained Cys-322 and Cys-323, the palmitoylation sites.
Next, the mutant opsin genes were excised from pMT4 as EcoRI/NotI DNA fragments, and these were inserted into the expression plasmid pACHrhoC as described (15). The vector pACHEnc was kindly provided by A. Shafferman (Israel Institute for Biological Research).
Preparation of HEK293S Stable Cell Lines Containing the Single Cysteine Opsin Mutant Genes.
HEK293S cells were transfected by the method of Chen and Okayama (31) as modified by O’Mahoney and Adams (32). Stable cell lines expressing the opsin genes at high levels were selected by using elevated concentrations of geneticin (2–3 mg/ml) as described (15).
Growth of Cells in Suspension and Reconstitution of the Opsins with 11-cis-Retinal.
Cells were grown as described by Reeves et al. (15) with minor modifications. Suspension cultures were set up by using 2-liter spinner flasks containing 500 ml of DMEM supplemented with 10% FBS, 0.1% Pluronic F-68, and 50 μg/ml heparin. Spinner flasks were inoculated by using four 15-cm tissue-culture dishes confluent with cells (6–9 × 107) per 500 ml of DMEM and were incubated at 37°C in a humidified incubator. After 6 days, the culture medium was further supplemented with 6 ml of 20% glucose and 4 ml of 8% NaHCO3. Cells were harvested on day 8. Cell pellets from a 500 ml culture were resuspended in 20 ml of buffer A containing benzamidine (0.005%) and phenylmethylsulfonylfluoride (0.1 mM). 11-cis-retinal was added to give 5 μM concentration, and the suspension was incubated for at least 1.5 hours. The concentration of reconstituted mutant rhodopsin was estimated by UV-visible absorbance difference spectrum analysis of an aliquot solubilized in buffer B. Addition of 11-cis-retinal was repeated until no further increase at 500 nm was observed by difference spectroscopy. Cell suspensions from different batches (the total volume depending on the expression level of the mutant) were combined to give at least 10 mg of the rhodopsin mutant, and the cells were harvested by centrifugation.
Immunoaffinity Purification of Rhodopsin Cysteine Mutants.
The cells harvested above were solubilized in 200–400 ml of buffer B (1 hour) in the dark at 4°C, and, after centrifugation, the supernatant was added to 1D4-Sepharose beads (10–15 ml, binding capacity 1 mg of rhodopsin per ml of settled beads). About 10% excess of 1D4-Sepharose over rhodopsin content was used. After end-over-end agitation for at least 6 hours at 4°C, the suspension was packed into a column (2.7 cm in diameter × 2–3 cm). The packed beads were washed at RT with at least 50 column volumes of buffer C followed by 10 bed volumes of buffer E. The flow rate was 0.5–1 ml/min.
TET-Labeling of Rhodopsin Cysteine Mutants Bound to 1D4-Sepharose.
The efficiency of the following labeling procedure was first checked on a small scale experiment as described in ref. 3. The total beads containing ≈10 mg of bound rhodopsin mutant after washing as above were resuspended in 40 ml of buffer E, and 4-PDS was added from a 1 M stock solution in ethanol to give a final concentration of 1 mM. After nutation for 5 min at RT, excess reagent was removed by multiple washing with 40 ml of buffer E under slight Argon pressure. A total of at least 35 times the column volume of buffer E was used. Complete removal of 4-PDS was tested spectrophotometrically (3). The beads were then resuspended in 30 ml of buffer E, and TET (3 μl of 11.2 M) was added to give a final concentration of ≈1 mM. After end-over-end agitation for several hours at RT, excess reagent was removed as described above for 4-PDS removal. Absence of TET in the effluent was checked spectrophotometrically by reaction with 4-PDS (3).
Elution of TET-Labeled Rhodopsin Mutants from 1D4-Sepharose.
After TET-labeling as above, the rhodopsin mutants were eluted from 1D4-Sepharose by using buffer H at a flow rate of 0.3–0.35 ml/min, the effluent being monitored by UV-visible absorption spectroscopy. Complete elution of the rhodopsin mutants usually required 50–70 ml of buffer H.
Concentration and Exchange to Buffer Q of Rhodopsin Mutants for NMR Analysis.
Fractions from 1D4-immunoaffinity chromatography containing >0.2 μM rhodopsin (A280/A500, 1.6–1.8) were pooled and concentrated using the membrane filter Centricon 30 (Amicon). About 6–10 mg amounts of rhodopsin mutants were concentrated to final volumes of 0.05–0.1 ml, and the solvent was exchanged to buffer Q by the addition of 1 ml buffer Q, followed by membrane filtration to a volume below 0.1 ml as above. This operation was repeated three times. The final volume was then adjusted to 0.3 ml with buffer Q. TFA was added from a 50 mM stock solution (in D2O) to give a final concentration of 0.2 mM. The samples were transferred to Shigemi NMR tubes.
Concentrations of the Detergents and Phospholipid in Rhodopsin Mutants Solutions Used for NMR Analysis.
In the standard method for preparation of the mutants, the DM concentration in buffer E was 0.05%. After concentration of the cysteine mutants eluted in buffer H, the concentration of DM in the samples to be used for NMR spectroscopy rose to 4–7%. The effect of the variations in concentrations of DM on chemical shift was tested for TET-Cys-140, for which the purification procedure was modified as follows: Cys-140, labeled with TET, was prepared as described in detail below. It was purified by absorption to Con A Sepharose as described (33) except that column washing was with buffer O, followed by washing with 10 bed volumes of buffer F and elution in buffer I. Con A Sepharose has a higher binding capacity (4–5 mg of rhodopsin per ml of settled beads), such that 7 mg of the rhodopsin was eluted in a volume of 1.8 ml. This was concentrated as above to 500 μl with a resulting DM concentration of 0.07% in buffer Q. The concentration was adjusted to 0.1%, and the NMR spectrum was recorded. Then, 20% DM in buffer Q was added to give a final DM concentration of 10%, and the NMR spectrum was again recorded.
For preparation of TET-Cys-140 in OG and in OG-DMPC, TET-Cys-140 was purified by using Con A Sepharose as above except that buffers O, F, and I were replaced by buffers P, G, and J, respectively. Solutions of TET-Cys-140 in 1% OG and 10% OG were obtained by addition of the appropriate volumes of 50% OG in buffer Q. In a separate experiment, DMPC was added from a suspension of 20% DMPC in 1% OG, buffer Q, to a TET-Cys-140 solution containing 1% OG.
Preparation of TET-Labeled Cys-316 and TET-Labeled Cys-140 Derivatives of Wild-Type (WT) Rhodopsin.
TET-labeling of both Cys-316 and Cys-140 in WT rhodopsin.
Pelleted rod outer segment membranes were suspended in buffer D (32 ml) to a final concentration of 0.375 mg/ml rhodopsin. 4-PDS (320 μl of 100 mM) was added to a final concentration of 1 mM. After end-over-end agitation for 3 hours at RT in the dark, the rod outer segment membranes were centrifuged in a 45 Ti rotor (Sorvall Ultracentrifuge) at 35,000 rpm for 30 min at 4°C. The pellet was resuspended in buffer D by passage through a #23 needle to a concentration of 1 mg/ml rhodopsin in a final volume of 12 ml. Liquid TET (1 μl) was added to give a final concentration of 1 mM, and the solution was nutated end-over-end for at least 5 hours in the dark at RT. The suspension was then centrifuged for 30 min at 35,000 rpm as above. Solubilization and purification of TET-labeled rhodopsin on 1D4-Sepharose was as described above for single cysteine mutants.
Selective TET-labeling of Cys-316 in WT rhodopsin.
Rod outer segment membranes were suspended at 0.375 mg/ml concentration at pH 6 (buffer D), and the entire procedure described above was followed, except that the reaction time with 4-PDS was 5 min at 20°C and the final concentration of 4-PDS was 0.1 mM.
Selective TET-labeling at Cys-140 in WT rhodopsin.
Pelleted rod outer segment membranes were suspended in buffer K at rhodopsin concentrations as above. N-ethylmaleimide, 100 fold excess over rhodopsin, was added from a stock solution of 1 M N-ethylmaleimide. The membrane suspension was incubated at RT for 5 min and then was pelleted. The pellet was resuspended in buffer D as above. The treatment with 4-PDS (3 hours in the dark) and subsequent procedure for purification was as described above.
Results
High Resolution Solution 19F NMR Spectra in the Dark of TET-Labeled Cysteine Mutants of Rhodopsin.
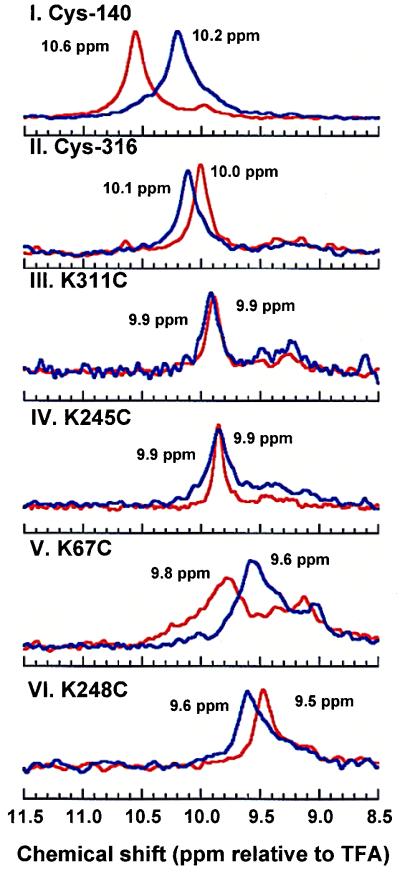
One-dimensional 19F NMR spectra, recorded in the dark, of TET-derivatives of the six cysteine mutants (Fig. 1) are assembled in Fig. 3A. The chemical shifts observed for the individual mutants are indicated in the figure, the spectra being arranged in the order of the magnitude of the chemical shifts. These are indicated relative to TFA, which was present at 0.2 mM concentration as internal standard in all of the samples. As seen, all of the signals were downfield from that of free TET, which was observed at 6.5 ppm. Thus, distinct chemical shifts were observed for the mutants Cys-140, Cys-316, K67C, and K248C, with Cys-140 (Fig. 3A, I) showing the most downfield and K248C (Fig. 3A, VI) the least downfield. The mutants K311C and K245C showed very similar chemical shifts. The integration intensity of the signals relative to that of the internal standard TFA indicated between 80 and 100% labeling efficiency.
Figure 3.
(A) Solution 19F NMR spectra in the dark of the single cysteine mutants (Fig. 1). The cysteine residues were derivatized with the TET group as in Fig. 2. All samples contained 0.2 mM TFA, an internal reference, which was set at 0 ppm. Between 400 and 4000 scans were averaged. I, Cys-140 (and small amount of Cys-316); II, Cys-316, (and small amount of Cys-140); III, K311C; IV, K245C; V, K67C; VI, K248C; VII, side products of the labeling procedure (Fig. 2), oxidized TET (TET-TET) and the mixed disulfide (TET-PDS). (B) Dark 19F NMR spectra of TET-labeled Cys-140 rhodopsin in different concentrations of DM, of OG, and in the presence of DMPC. I, DM, 0.1 and 10%; II, OG, 1 and 10%; III, OG 1% and OG (1%) + DMPC (0.15%).
Whereas all of the labeled mutants show major well resolved peaks, secondary peaks are also seen (Fig. 3A). The spectra for the TET-labeled Cys-140 and Cys-316 (Fig. 3A, I and II) show second minor peaks, which are attributable to contamination from the second cysteine (Cys-316 in Cys-140 and vice versa) in WT rhodopsin. In the spectra of the labeled mutants K311C, K245C, and, particularly, K67C, peaks additional to the main chemical shifts were observed at ≈9.2 ppm and 9.5 ppm at varying intensities. To determine whether these peaks are attributable to 19F- contaminants introduced during the labeling procedure, the reactants, 4-PDS and TET (Fig. 2), were mixed at different molar ratios in 1% DM. In Fig. 3A, VII is included the spectrum for one such mixture. Two signals can be seen, at 9.2 ppm and at 9.5 ppm. At a high concentration of 4-PDS in the mixture, which would be excepted to favor the formation of the mixed disulfide (thiopyridinyl-TET), the signal at 9.5 ppm predominated. On the other hand, with increasing concentration of TET, where TET would be expected to form homodimers (TET-TET), the intensity of the signal at 9.2 ppm increased. The two signals additional to the main peaks at 9.2 ppm and 9.5 ppm are therefore assigned, respectively, to the TET homodimer and to thiopyridinyl-TET. The similar position of these signals to the secondary peaks seen in the spectra of mutants K311C, K245C, and K67C suggests that the secondary peaks probably correspond to contamination of the preparations by oxidized TET and the mixed disulfide.
Solution 19F NMR Spectra of TET-Cys-140 at Different Concentrations of DM and of OG and in the Presence of the Phospholipid DMPC.
The possible effect on chemical shifts of variation in DM concentration were studied by using 0.1% (≈10× above critical micelle concentration) and 10% DM. The results are shown in Fig. 3B, I. A shift (−0.08 ppm) upfield at the higher concentration from 10.70 ppm (0.1% DM) to 10.62 ppm (10% DM) was observed.
As an example for the use of a different detergent, the chemical shifts in TET-Cys-140 rhodopsin were also recorded in the detergent OG at two concentrations (Fig. 3B, II). There was a significant effect on the chemical shift in OG relative to that in DM. Thus, an upfield shift to 10.31 ppm (1% OG) and 10.25 ppm (10% OG) were observed.
Effect of the addition of DMPC, a phospholipid used most often in reconstitution studies of rhodopsin and bacteriorhodopsin (35), to OG was also tested (Fig. 3B, III). The signal in the presence of 0.15% DMPC in 1% OG was at 10.45 ppm as compared with 10.33 ppm in its absence.
Comparisons of 19F NMR Spectra of TET-Cysteine Mutants in the Dark and After Illumination to Form Meta II.
The spectra obtained for all six mutants in the dark (red lines) and after illumination (blue lines) are shown in Fig. 4. TET-Cys-140 (Fig. 4, I) and TET-Cys-67 (Fig. 4, V) showed upfield shifts, −0.4 ppm and −0.2 ppm, respectively, whereas TET-Cys-316 (Fig. 4, II) and TET-Cys-248 (Fig. 4, VI) each showed a downfield shift of 0.1 ppm. It is noteworthy that TET-Cys-245 (Fig. 4, IV) and TET-Cys-311 (Fig. 4, III) showed no chemical shift change on illumination.
Figure 4.
Changes in 19F NMR spectra of TET-labeled cysteine mutants on illumination: red lines, dark spectra; blue lines, spectra after illumination. Each spectrum was taken within 2 min acquisition time (average of 160 scans).
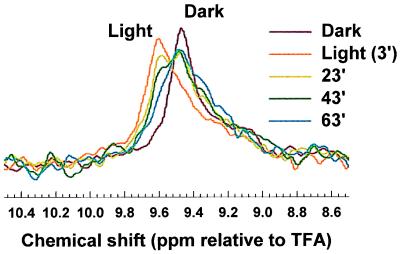
Changes in 19F NMR Spectrum of TET-Labeled K248C During Meta II Decay.
In Fig. 5 are shown the spectra for the mutant K248C, in the dark, 3 min after illumination and at three subsequent time intervals. Three features of the spectra are noteworthy: First, the amplitude of the spectral line on illumination is less than theoretically expected. This is attributable to the elapse of time during the recording of the spectrum. Even at 1 min after illumination, significant decay of Meta II would have occurred. Second, during Meta II decay, the spectral lines show shifts toward those that were originally observed in the dark. Finally, the intensity of this chemical shift increases with time, but the full amplitude of the dark chemical shift is not regained. Similar behavior after Meta II decay was found for the other mutants (data not shown). The rate at which these shifts progress toward the shift observed in the dark parallel the known rate of decay of Meta II. However, the full amplitude of the dark chemical shift is not regained, a result completely consistent with previous observations of alternative competing structures in opsin (34).
Figure 5.
19F NMR spectral changes in the TET-labeled cysteine mutant K248C: in the dark, 3 min after illumination, and at different subsequent time intervals. Acquisition parameters were as in Fig. 4.
Discussion
No information exists at this time on the three-dimensional structure of rhodopsin by x-ray analysis. NMR spectroscopy offers a viable alternative approach to the structural problem. Recently, by using 15N-lysine-labeled rhodopsin, magic angle spinning NMR of the protonated Schiff base at lysine 296 has been reported from two laboratories (16, 17). We now have reported on the first application of solution NMR to the study of rhodopsin. The 19F NMR spectra reported herein were recorded in DM which has proved to be the most satisfactory detergent for work with rhodopsin. Indeed, DM has been used in essentially all of the structure–function studies reported on rhodopsin as well as in studies of its interactions in vitro with transducin and rhodopsin kinase. Previously, solution NMR has been applied to studies of rhodopsin structure by using peptide segments corresponding in sequences to the cytoplasmic loops (14). Clearly, there are reservations in extrapolation of the results obtained from peptide studies to derivation of the structure of rhodopsin itself. Also, work with peptide segments precludes information on the conformational change on light activation and structural studies of subsequent protein–protein interactions.
In the present work, the influence of the detergent concentration and of the nature of the detergent on NMR spectra were examined. The chemical shifts of a representative 19F-labeled rhodopsin in different concentrations of DM were compared with the NMR spectra recorded in different concentrations of OG, a detergent frequently used for solubilization of membrane proteins, as well as in detergent–phospholipid mixed micelles. The observed effects of the nature and concentration of detergents and of added phospholipid are all consistent with the previous results on stability and structure of rhodopsin and bacteriorhodopsin in different detergents (34, 35).
Our overriding interest lies in understanding the nature of the conformational change in the cytoplasmic domain of rhodopsin on light activation. Therefore, in the present initial study of the cytoplasmic domain, rhodopsin mutants containing single reactive cysteines were engineered in different regions of this domain, and each derivatized with the 19F label TET. The cysteine mutants showed well resolved distinctly different chemical shifts, except for two mutants in which the shifts were similar. Further, in 19F NMR spectra of the different mutants, there appeared to be significant linewidth differences between labeling positions (Fig. 3A). These could be attributable to variability in the mobility and accessibility of the probe, or different degrees of static heterogeneity at the various sites.
Although single major peaks were observed in all six mutants, additional minor peaks (at 9.15–9.25 ppm and at 9.4–9.5 ppm) were also observed in the mutants. The latter peaks are probably attributable to interactions between the reagents (4-PDS and TET) used during the labeling procedure. They are close to those (9.2 ppm and 9.5 ppm) observed in mixtures of pure 4-PDS and TET in 1% DM (Fig. 3A, VII). The slight variation of ≈0.1 ppm could arise from differences in the detergent concentration (4–7%) used in the rhodopsin preparations.
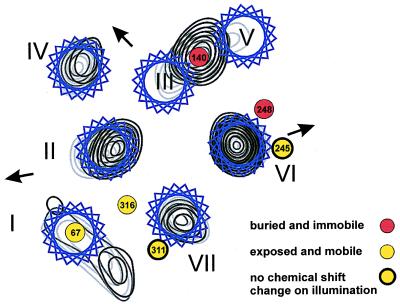
On illumination to form Meta II, upfield changes in chemical shifts were observed for 19F labels at positions 67 and 140 and downfield changes for positions 248 and 316 whereas little or no change was observed at positions 245 and 311. Quantitative understanding of these features lies far into the future. However, qualitatively, the shifts are entirely consistent with the conclusion from previous biochemical studies, namely the presence of a tertiary structure in the cytoplasmic face and its change on light activation (3). Also, the changes in chemical shifts observed on light activation fit quite well with the conclusions drawn on light-induced conformational changes from recent EPR studies of a large number of spin-labeled single cysteine mutants (2, 4, 36, 37). These EPR data allowed the proposal of a model for the superposition of residues onto electron density maps that indicate the packing of the helices and their light-induced movements (Fig. 6). In this model, the exposed residues 67 and 316 are not in helical regions (refs. 2 and 4; unpublished spin-spin interaction and disulfide bond formation data by J.K.-S., C. Altenbach, J. Hwa, W. L. Hubbell, and H.G.K., using double cysteine mutants including the Cys-67, Cys-316 pair) whereas exposed residues 245 and 311 face the outside of the helical bundle. It is significant that the latter two residues are the only ones that failed to show a 19F chemical shift change. This is consistent with their exposed location on a stable secondary structure element. In contrast, buried residues 140 and 248 do show 19F chemical shift changes, presumably because of the relative movement of helix VI with respect to helix III (Fig. 6), which has been demonstrated as an important feature of light activation (38, 39). Similarly, the model proposes that the sequence connecting helices I and II changes its position with respect to that of 316 above helix VII (J.K.-S., C. Altenbach, J. Hwa, W. L. Hubbell, and H.G.K., unpublished work cited above). Thus, 19F labels at positions 67 and 316 also show changes in chemical shift on light activation.
Figure 6.
Superimposition of the residues studied by 19F NMR on the arrangement of the 7-transmembrane helical bundle based on cryoelectron microscopy studies (5). The electron density contour sections at 13, 15, and 17 Å (gray to black) from the center of the membrane are shown. The contour sections were from a three-dimensional map at an effective resolution 7.5 Å in the membrane plane and 16.5 Å normal to the plane. Residue positions are mapped onto the electron contour sections on the basis of site-directed spin labeling studies (2, 4, 36, 37). The figure has been adapted from Fig. 5 in ref. 4.
Changes in the NMR spectra of the mutant K248C were studied on the decay of its Meta II intermediate. The chemical shift gradually returned to that originally observed in the dark spectrum (Fig. 5). The rate of this change matched the rate of Meta II decay as previously determined by the standard fluorescence increase assay (27). Thus, NMR spectroscopy provides an alternative method for following Meta II decay. Also in agreement with the previous results, the dark state opsin structure was not quantitatively regained because of the occurrence of alternative multiple structural changes in the opsin (34).
Although further work on NMR can follow a wide variety of lines, the following directions for immediate work should be mentioned. The fact that high-resolution NMR spectra were obtained offers the promise that other nuclei such as 15N, 13C, and 1H will also yield satisfactory signals. This will allow the application of more conventional NMR structure determination experiments the prospects for which have recently been improved for proteins of large size (40). For this, isotope incorporation will be required, and this has already been demonstrated for rhodopsin expressed in HEK293S cells (16) and in the baculovirus/Sf9 system (17).
Further, the question of distance measurements between 19F labels needs to be addressed immediately. For this, the preparation of mutants containing cysteine pairs is already underway. The choice of these is guided by the previous extensive studies on proximity relationships deduced from spontaneous disulfide bond formation and EPR spectroscopy (refs. 38, 41, and 42; K. Cai and J.K.-S., unpublished results of this laboratory). In addition to studies of dipole–dipole interactions between two 19F nuclei in solution, solid state rotational echo double resonance measurements should be used to determine specific distances between two different nuclei. Such pairs of different nuclei will be introduced by differential labeling of double cysteine mutants for 19F-13C rotational echo double resonance experiments as well as phosphorylation of single cysteine mutants for 19F-31P rotational echo double resonance measurements.
Acknowledgments
We thank Prof. J. Falke at the University of Colorado for critically reading the manuscript and helpful suggestions. We have benefited greatly from discussions with Prof. U. L. RajBhandary, all of our colleagues at the Massachusetts Institute of Technology, and Prof. S. Smith, Drs. M. Eilers, and W. Ying at the State University of New York at Stony Brook. We also thank the staff of the Instrumentation Facility in the Department of Chemistry (Massachusetts Institute of Technology) for technical advice on NMR experiments. Ms. Judy Carlin’s sustained assistance during the manuscript preparation is gratefully acknowledged. Research reported here was supported by National Institutes of Health Grants GM28289 and EY11716 (H.G.K.). J.K-S. is the recipient of a Howard Hughes Medical Institute Predoctoral Fellowship. E.V.G. is the recipient of an ARVO/Japan National Society for the Prevention of Blindness Research Fellowship. M.C.L. is the recipient of a Natural Sciences and Engineering Research Council of Canada fellowship.
Abbreviations
- WT
wild-type
- Meta II
metarhodopsin II
- 4-PDS
4,4′-dithiodipyridine
- TET
trifluoroethylthio
- DM
dodecylmaltoside
- OG
octyl glucoside
- DMPC
dimyristoyl-phosphatidyl choline
- TFA
trifluoroacetate
- RT
room temperature
- A280
absorbance at λ280 nm
- T1/2
half-life.
Footnotes
This is paper 37 in the series “Structure and Function in Rhodopsin.” Paper 36 is ref. 23.
References
- 1.Cai K, Klein-Seetharaman K, Farrens D, Zhang C, Altenbach C, Hubbell W L, Khorana H G. Biochemistry. 1999;38:7925–7930. doi: 10.1021/bi9900119. [DOI] [PubMed] [Google Scholar]
- 2.Altenbach C, Cai K, Khorana H G, Hubbell H G. Biochemistry. 1999;38:7931–7937. doi: 10.1021/bi9900121. [DOI] [PubMed] [Google Scholar]
- 3.Klein-Seetharaman J, Hwa J, Cai K, Altenbach C, Hubbell W L, Khorana H G. Biochemistry. 1999;38:7938–7944. doi: 10.1021/bi990013t. [DOI] [PubMed] [Google Scholar]
- 4.Altenbach C, Klein-Seetharaman J, Hwa J, Khorana H G, Hubbell W L. Biochemistry. 1999;38:7945–7949. doi: 10.1021/bi990014l. [DOI] [PubMed] [Google Scholar]
- 5.Unger V M, Hargrave P A, Baldwin J M, Schertler G F X. Nature (London) 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 6.Wüthrich K. NMR in Structural Biology. Singapore: World Scientific; 1995. [Google Scholar]
- 7.Girvin M E, Rastogi V K, Abildgaard F, Markley J L, Fillingame R H. Biochemistry. 1998;37:8817–8824. doi: 10.1021/bi980511m. [DOI] [PubMed] [Google Scholar]
- 8.MacKenzie K R, Prestegard J H, Engelman D M. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 9.Williams K A, Farrow N A, Deber C M, Kay L E. Biochemistry. 1996;35:5145–5157. doi: 10.1021/bi952897w. [DOI] [PubMed] [Google Scholar]
- 10.Almeida F C L, Opella S J. J Mol Biol. 1997;270:481–495. doi: 10.1006/jmbi.1997.1114. [DOI] [PubMed] [Google Scholar]
- 11.Patzelt H, Ulrich A S, Egbringhoff H, Düx P, Ashurst J, Simon B, Oschkinat H, Oesterhelt D. J Biomol NMR. 1997;10:95–106. [Google Scholar]
- 12.Rohl C A, Boeckman F A, Baker C, Scheuer T, Catterall W A, Klevit R E. Biochemistry. 1999;38:855–861. doi: 10.1021/bi9823380. [DOI] [PubMed] [Google Scholar]
- 13.Pellegrini M, Bisello A, Rosenblatt M, Chorev M, Mierke D F. Biochemistry. 1998;37:12737–12743. doi: 10.1021/bi981265h. [DOI] [PubMed] [Google Scholar]
- 14.Yeagle P L, Alderfer J L, Albert A D. Biochemistry. 1997;36:9649–9654. doi: 10.1021/bi970908a. [DOI] [PubMed] [Google Scholar]
- 15.Reeves P J, Thurmond R L, Khorana H G. Proc Natl Acad Sci USA. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eilers M, Reeves P J, Ying W, Khorana H G, Smith S O. Proc Natl Acad Sci USA. 1999;96:487–492. doi: 10.1073/pnas.96.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Creemers A F L, Klaassen C H W, Bovee-Geurts P H M, Kelle R, Kragl U, Raap J, de Grip W J, Lugtenburg J, de Groot H J M. Biochemistry. 1999;38:7195–7199. doi: 10.1021/bi9830157. [DOI] [PubMed] [Google Scholar]
- 18.Danielson M A, Falke J J. Annu Rev Biophys Biomol Struct. 1996;25:163–195. doi: 10.1146/annurev.bb.25.060196.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerig J T. Methods Enzymol. 1989;177:3–23. doi: 10.1016/0076-6879(89)77003-8. [DOI] [PubMed] [Google Scholar]
- 20.Falke J J, Luck L A, Scherrer J. Biophys J. 1992;62:82–86. doi: 10.1016/S0006-3495(92)81787-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rule G S, Pratt E A, Simplaceanu V, Ho C. Biochemistry. 1987;26:549–556. doi: 10.1021/bi00376a029. [DOI] [PubMed] [Google Scholar]
- 22.Colmenares L U, Niemczura W P, Asato A E, Liu R S H. J Phys Chem. 1996;100:9175–9180. [Google Scholar]
- 23.Cai K, Klein-Seetharaman J, Hwa J, Hubbell W L, Khorana H G. Biochemistry. 1999;38:12893–12898. doi: 10.1021/bi9912443. [DOI] [PubMed] [Google Scholar]
- 24.Knowles A, Priestley A. Vision Res. 1978;18:115–116. doi: 10.1016/0042-6989(78)90086-x. [DOI] [PubMed] [Google Scholar]
- 25.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wald G, Brown P K. J Gen Physiol. 1953;37:189–200. doi: 10.1085/jgp.37.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farrens D L, Khorana H G. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 28.Thurmond R L, Creuzenet C, Reeves P, Khorana H G. Proc Natl Acad Sci USA. 1996;94:1715–1720. doi: 10.1073/pnas.94.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franke R R, Sakmar T P, Oprian D D, Khorana H G. J Biol Chem. 1988;263:2119–2122. [PubMed] [Google Scholar]
- 30.Yang K, Farrens D L, Hubbell W L, Khorana H G. Biochemistry. 1996;35:12464–12469. doi: 10.1021/bi960848t. [DOI] [PubMed] [Google Scholar]
- 31.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Mahoney J V, Adams T E. DNA Cell Biol. 1994;13:1227–1232. doi: 10.1089/dna.1994.13.1227. [DOI] [PubMed] [Google Scholar]
- 33.De Grip W J. Methods Enzymol. 1982;81:197–207. doi: 10.1016/s0076-6879(82)81032-x. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto T, Khorana H G. Proc Natl Acad Sci USA. 1995;92:249–253. doi: 10.1073/pnas.92.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reeves P J, Hwa J, Khorana H G. Proc Natl Acad Sci USA. 1999;96:1927–1931. doi: 10.1073/pnas.96.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farahbakhsh Z T, Ridge K D, Khorana H G, Hubbell W L. Biochemistry. 1995;34:8812–8819. doi: 10.1021/bi00027a033. [DOI] [PubMed] [Google Scholar]
- 37.Altenbach C, Yang K, Farrens D L, Farahbakhsh Z T, Khorana H G, Hubbell W L. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 38.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 39.Sheikh S P, Zvyaga T A, Lichtarge O, Sakmar T P, Bourne H R. Nature (London) 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 40.Pervushin K, Riek R, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang K, Farrens D L, Altenbach C, Farahbakhsh Z T, Hubbell W L, Khorana H G. Biochemistry. 1996;35:14040–14046. doi: 10.1021/bi962113u. [DOI] [PubMed] [Google Scholar]
- 42.Cai K, Langen R, Hubbell W L, Khorana H G. Proc Natl Acad Sci USA. 1997;94:14267–14272. doi: 10.1073/pnas.94.26.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]