Abstract
Stabilization of p53 in response to DNA damage is caused by its dissociation from Mdm2, a protein that targets p53 for degradation in the proteasome. Dissociation of p53 from Mdm2 could be caused by DNA damage-induced p53 posttranslational modifications. The ATM and ATR kinases, whose activation in response to ionizing radiation (IR) and UV light, respectively, is required for p53 stabilization, directly phosphorylate p53 on Ser-15. However, phosphorylation of Ser-15 is critical for the apoptotic activity of p53 and not for p53 stabilization. Thus, whether any p53 modifications, and which, underlie disruption of the p53–Mdm2 complex after DNA damage remains to be determined. We analyzed the IR- and UV light-induced stabilization of p53 proteins with substitutions of Ser known to be posttranslationally modified after DNA damage. Substitution of Ser-20 was sufficient to abrogate p53 stabilization in response to both IR and UV light. Furthermore, both IR and UV light induced phosphorylation of p53 on Ser-20, which involved the majority of nuclear p53 protein and weakened the interaction of p53 with Mdm2 in vitro. ATM and ATR cannot phosphorylate p53 on Ser-20. We therefore propose that ATM and ATR activate an, as yet unidentified, kinase that stabilizes p53 by phosphorylating it on Ser-20.
The integrity of the genome is guarded by cellular checkpoints (1–3). One of the major checkpoints in mammalian cells is the p53 tumor suppressor protein, a sequence-specific DNA-binding transcription factor that induces cell cycle arrest or apoptosis in response to DNA damage (4–7). Because of its function as a “guardian of the genome,” p53 suppresses the occurrence of developmental defects and cancer (8, 9). p53 also affects therapy of tumors that retain wild-type p53 function (10), such as most childhood cancers and about half of all cancers in adults (11), because many cancer therapeutics are DNA-damaging agents that kill tumor cells by inducing p53-dependent apoptosis. Accordingly, understanding how p53 is activated by DNA damage could impact the prevention and therapy of developmental defects and cancer.
DNA damage regulates both the protein levels of p53 (4, 12) and its affinity for specific DNA sequences (13–15). The increase in DNA-binding affinity involves dephosphorylation of Ser-376 of p53 and subsequent binding of 14-3-3 protein family members to p53 (16). The increase in p53 levels is caused by decreased degradation of p53 (4, 12, 17) and to a lesser extent, from increased translation efficiency of the p53 mRNA (18). Degradation of p53 involves Mdm2, an intracellular protein that binds to p53 (19) and exports it out of the nucleus (20, 21). In the cytoplasm, Mdm2 targets p53 for ubiquitin-dependent proteolysis (22–24). In response to DNA damage, p53 is stabilized because of inhibition of Mdm2-dependent p53 degradation (25). Thus, modified p53 proteins that fail to interact with Mdm2 are expressed at high levels and are not further stabilized after DNA damage (26, 27).
Inhibition of Mdm2-dependent p53 degradation after DNA damage is from dissociation of p53 from Mdm2 (25). DNA damage induces multiple p53 posttranslational modifications, including phosphorylation of Ser residues 6, 9, 15, 20, 33, 37, and 392, dephosphorylation of Ser-376, and acetylation of Lys residues 320, 373, and 382 (16, 25, 28–36), thereby raising the possibility that one or more of these modifications disrupt the interaction of p53 with Mdm2. Yet, identifying the critical modification has not been straightforward. In vitro, there has been conflicting information as to which posttranslational modifications weaken the interaction of p53 with Mdm2; potential culprits include phosphorylation of Ser-15 (25), phosphorylation of Thr-18 (37), and phosphorylation of Ser-20 (38). In vivo, stabilization of p53 in response to ionizing radiation (IR) and UV light is dependent on the ATM (ataxia–telangiectasia-mutated) and ATR (ATM + Rad3-related) kinases, respectively (39–43). ATM and ATR directly phosphorylate p53 on Ser-15 in response to DNA damage (43–46); however, replacement of Ser-15 with Ala compromises the apoptotic activity of p53 (47) and not its stabilization after DNA damage (26, 27). We describe here a series of genetic and biochemical experiments that point to phosphorylation of Ser-20 as the critical posttranslational modification leading to dissociation of p53 from Mdm2 in vitro and p53 stabilization in response to IR and UV light in vivo.
Materials and Methods
Analysis of p53 Protein Levels in Response to DNA Damage.
All plasmids used to express p53 in mammalian cells were derived from pSV2hp53wtB (48). U2-OS osteosarcoma cells were transfected by calcium phosphate precipitation with 0.5 μg of hemagglutinin (HA)-tagged p53 expression plasmid and 29 μg of pBC12/PLseap carrier plasmid (48). The cells were exposed to 50 J/m2 UV light or 9 Gy IR 36 and 48 h after transfection, respectively. Whole-cell extracts were prepared 16 h after exposure to UV light or 2 h after exposure to IR by lysis in buffer consisting of 50 mM Tris⋅HCl (pH 8), 120 mM NaCl, 0.5% Nonidet P-40, 1 mM DTT, 0.4 μg/ml Pefabloc SC (Pentapharm), 2 μg/ml pepstatin, 0.2 μM wortmannin, 0.1 μM staurosporine, 15 mM NaF, and 1 mM sodium vanadate. p53 protein levels were assayed by immunoblotting with anti-HA Y11 antibody (Santa Cruz Biotechnology). p53-nullizygous mouse embryo fibroblasts (MEFs) were transfected as described above with 5 μg of p53 expression plasmid, 1 μg of plasmid pSV7neo, and 24 μg of pBC12/PLseap carrier plasmid (48). Transfected cells were selected with G418, pooled, and exposed to DNA-damaging agents as described above or treated with 3 μM lactacystin (Calbiochem) for 3 h. p53 protein levels were assayed by immunoblotting with antibody DO1 (Calbiochem).
Phosphospecific Antibodies.
AbS20p, a polyclonal antibody (clone 430) that recognizes p53 phosphorylated on Ser-20, was obtained from New England Biolabs. DO1 is a monoclonal antibody that, under high-salt conditions, recognizes p53 only when Ser-20 is not phosphorylated. The specificity of these antibodies was established by incubating 6 μg of biotinylated peptides corresponding to residues 7–29 of human p53 either not phosphorylated or phosphorylated on Ser-15, Thr-18, or Ser-20 attached to avidin-agarose beads (Pierce) with 200 ng of antibody AbS20p or DO1 in a buffer consisting of 20 mM Tris⋅HCl (pH 8), 200 mM (for AbS20p) or 800 mM (for DO1) NaCl, 1 mM EDTA, and 0.05% Tween-20. Antibodies bound to the beads were detected by immunoblotting.
Phosphorylation State of p53.
Nuclear extracts from untreated cells or cells exposed to 9 Gy IR or 50 J/m2 UV light were prepared as described (16). Endogenous p53 protein in these extracts was immunoprecipitated with antibodies DO1 and AbS20p as described (16). The immunoprecipitation (IP) buffer for antibody DO1 was 20 mM Tris⋅HCl (pH 8)/800 mM NaCl/0.05% Tween-20, and the beads were washed with 1× IP buffer. The IP buffer for antibody AbS20p was 25 mM Hepes (pH 7.4)/100 mM NaCl/5 mM MgCl2/100 mM EDTA/0.2 mg/ml BSA/0.1% Tween-20, and the beads were washed with 2× IP buffer. The presence of p53 in the immunoprecipitates was monitored by immunoblotting with antibody DO7 as described (16). Where indicated, the extracts were incubated with 1,000 units of λ protein phosphatase (New England Biolabs) for 1 h at 30°C before IP with antibody DO1.
Binding of p53 Peptides to Mdm2.
To determine whether phosphorylation of Ser-20 compromises the interaction of p53 with Mdm2, 1 μg of biotinylated p53 peptides corresponding to residues 7–29 of human p53 either not phosphorylated or phosphorylated on Ser-15, Thr-18, or Ser-20 were coupled to avidin-agarose beads (Pierce) and incubated with in vitro-translated 35S-labeled full-length or N-terminally truncated Mdm2 protein in buffer consisting of 50 mM Hepes (pH 7.4), 200 mM NaCl, 10 mM MgCl2, 200 mM EDTA, 0.4 mg/ml BSA, and 0.2% Tween-20. The beads were washed three times in the same buffer, and bound Mdm2 was resolved by denaturing gel electrophoresis and detected by autoradiography.
Results
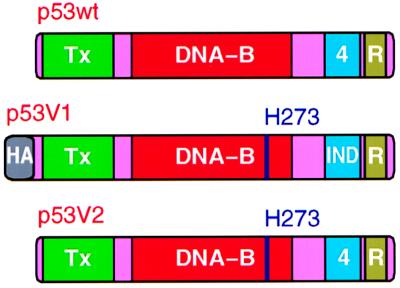
To identify residues that are critical for p53 stabilization in response to DNA damage, we examined the DNA damage response of p53 proteins with amino acid substitutions targeting putative regulatory residues. Two versions of p53 proteins were constructed for these experiments (Fig. 1). Version 1 allows the experiments to be performed in cells that have endogenous wild-type p53 and has three other modifications in addition to the substitutions of putative regulatory residues. First, an N-terminal HA tag was inserted to distinguish exogenous from endogenous p53. Second, an Arg-273 → His substitution compromises sequence-specific DNA-binding activity without altering the native p53 protein structure (48, 49). This ensures that the transfected p53 proteins are passive reporters of p53 stabilization and do not themselves modify the DNA damage response by transactivating Mdm2 or inducing cell cycle arrest. Third, the tetramerization domain was modified by introducing seven amino-acid substitutions (Leu-330 → Phe, Met-340 → Phe, Ala-347 → Ile, Leu-348 → Met, Ala-353 → Leu, Gln-354 → Leu, and Ala-355 → Asp). The p53 subunits bearing this modified tetramerization domain, hereafter referred to as IND (independent), still form homotetramers, but fail to heterooligomerize with endogenous p53 (50). Thus, the IND tetramerization domain allows the exogenous p53 subunits to be regulated independently of the endogenous wild-type subunits. Version 2 has only the Arg-273 → His substitution and is suitable for analysis only in cells that lack endogenous p53.
Figure 1.
Diagrams of wild-type p53 (p53wt) and of the variant p53 proteins (p53V1 and p53V2) that were used to study p53 stabilization after DNA damage. Tx, transactivation domain; DNA-B, sequence-specific DNA-binding domain; 4, native tetramerization domain; R, C-terminal regulatory region for the DNA-binding activity of p53; HA, hemagglutinin tag; IND, modified (independent) tetramerization domain that does not form heterotetramers with the native p53 tetramerization domain; H273, histidine substitution for Arg-273. Substitutions of Ser residues with Ala or Asp were performed in the context of p53V1 and p53V2.
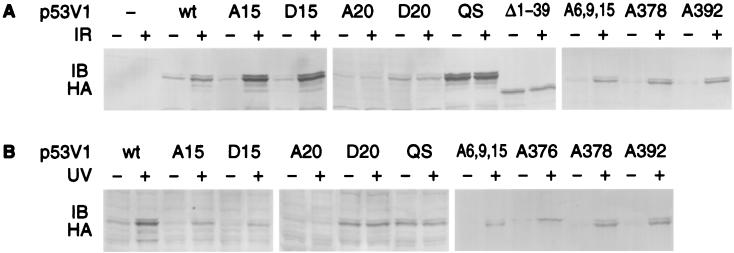
Levels of p53V1 proteins in transiently transfected U2-OS osteosarcoma cells were examined 2 h after exposure to 9 Gy IR and 16 h after exposure to 50 J/m2 UV light. These are the time points and radiation doses at which the levels of endogenous wild-type p53 protein peaked in response to DNA damage in U2-OS cells (data not shown). DNA damage led to increased levels of p53V1wt, which has no Ser or Thr substitutions. Stabilization was Mdm2 dependent, as confirmed by the absence of increased levels of mutant p53 proteins that do not associate with Mdm2 either because of a double substitution of Leu-22 and Trp-23 with Gln and Ser, respectively, or because of deletion of the N-terminal 39 residues (Fig. 2). Analysis of the p53 mutants with Ser/Thr substitutions revealed no effect from substitutions of serines 6, 9, 15, 33, 37, 376, 378, or 392 or Thr-18 on the increase in p53 protein levels in response to either IR or UV light. However, replacement of Ser-20 with Ala or Asp abrogated the increase in p53 protein levels after DNA damage (Fig. 2 and data not shown). The effect of the Ser-20 substitution was not limited to a specific time point, because similar results were obtained when the cells were examined over a 24-h period after exposure to DNA damage (data not shown).
Figure 2.
Substitution of Ser-20 abrogates p53 protein stabilization after DNA damage in U2-OS osteosarcoma cells. p53V1 proteins with Ser/Thr substitutions were expressed by transient transfection. The transfected cells were exposed to IR (A) or UV light (B) and p53 protein levels were determined by immunoblotting (IB) with an antibody that reacts with the HA tag in p53V1. Designations of the expressed p53 proteins correspond to the amino acid substitutions introduced in p53V1, indicating the position(s) of the replaced residue(s) and the type of residue introduced by using the single-letter code. p53V1wt has no Ser/Thr substitutions, p53V1QS has substitutions of Leu-22 → Gln and Trp-23 → Ser, and p53V1Δ1–39 has amino acids 1–39 deleted.
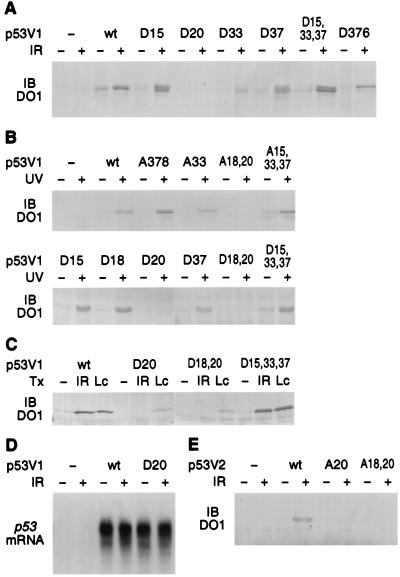
Stabilization of the mutant p53V1 proteins was subsequently examined in early-passage pools of stably transfected p53-nullizygous MEFs. We used the p53-specific antibody DO1 to monitor p53 protein levels, because DO1 is more sensitive than the anti-HA antibody and the MEFs did not have endogenous p53; however, similar results could be obtained with the anti-HA antibody. The protein levels of most p53 mutants increased after DNA damage; the only exception was the p53 mutants with substitutions targeting Ser-20, which were not stabilized in response to DNA damage (Fig. 3 A and B) even though they could be stabilized when the cells were treated with the proteasome inhibitor lactacystin (Fig. 3C). DNA damage and substitution of Ser-20 had no effect on expression of the p53 mutants at the mRNA level (Fig. 3D), thus ruling out the trivial possibility that differences in mRNA expression accounted for the differences in p53 protein levels. We also ruled out the possibility that the results were an artifact of the N-terminal HA tag or the IND tetramerization domain, which are present in p53V1, by repeating the key experiments in p53-nullizygous MEFs stably expressing p53V2 proteins, which lack the HA tag and have a wild-type tetramerization domain (Figs. 1 and 3E).
Figure 3.
Substitution of Ser-20 abrogates p53 protein stabilization after DNA damage in p53-nullizygous MEFs stably transfected with constructs expressing p53V1 (A–D) or p53V2 (E) proteins. The transfected cells were exposed to IR (A and C–E), UV light (B), or the proteasome inhibitor lactacystin (Lc) (C). p53 protein levels were determined by immunoblotting (IB) with antibody DO1 (A–C, E) and p53 mRNA levels by Northern blotting (D). The mutants are designated as in Fig. 2.
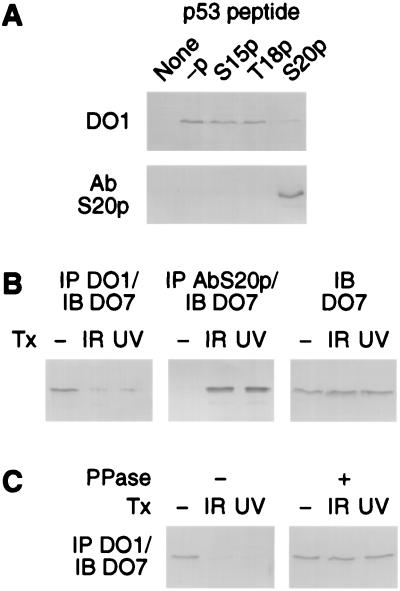
The identification of Ser-20 as the critical residue for p53 stabilization suggests that Ser-20 is phosphorylated in response to DNA damage, and this phosphorylation underlies the increase in p53 protein levels. Phosphorylation of p53 on Ser-20 has been reported in response to DNA damage in cell lines that express mutant p53, but not in cells that express wild-type p53 (36). This may be because wild-type p53 is expressed at lower levels than mutant p53 (36). Nevertheless, because the levels of mutant p53 proteins are unaffected in response to DNA damage (4, 36), it was important to establish whether endogenous wild-type p53 is phosphorylated on Ser-20 in a DNA damage-dependent manner. We used two antibodies to monitor Ser-20 phosphorylation: AbS20p, a polyclonal antibody specific for p53 phosphorylated on Ser-20, and DO1, a monoclonal antibody that is specific for p53 not phosphorylated on Ser-20 when the salt concentration in the IP buffer is 800 mM. The phosphospecificities of these antibodies were confirmed by using p53 peptides either not phosphorylated or phosphorylated on Ser-15, Thr-18, or Ser-20 (Fig. 4A). Phosphorylation of endogenous wild-type p53 on Ser-20 was monitored by immunoprecipitating nuclear extracts from untreated and irradiated U2-OS cells with antibodies AbS20p or DO1 and detecting the fraction of p53 in the precipitate by immunoblotting. The cell extracts were prepared 2 h after exposure of the cells to 9 Gy IR and 16 h after exposure to 50 J/m2 UV light, the same time points and doses used to study p53 stabilization. In the absence of DNA damage, endogenous p53 was precipitated by antibody DO1, but not by AbS20p; after exposure to either IR or UV light, the inverse pattern was observed (Fig. 4B). The switch in DO1 immunoreactivity after DNA damage could be reversed by treatment of the cell extracts with protein phosphatase, confirming that the modification affecting DO1 reactivity was phosphorylation (Fig. 4C). Thus, endogenous wild-type p53 becomes phosphorylated on Ser-20 after DNA damage. Furthermore, the very low reactivity of p53 to AbS20p in untreated cells and the low reactivity of p53 to DO1 in the cells exposed to DNA damage indicates that in untreated cells, the major fraction of nuclear p53 is not phosphorylated on Ser-20, but becomes phosphorylated on this residue after DNA damage.
Figure 4.
Phosphorylation of endogenous wild-type p53 on Ser-20 in response to DNA damage. (A) Specificity of antibodies DO1 and AbS20p for p53 not phosphorylated and phosphorylated on Ser-20, respectively. Biotinylated peptides corresponding to residues 7–29 of human p53 were coupled to avidin-agarose beads and incubated with antibodies DO1 or AbS20p. Antibodies bound to the beads were detected by fractionation on denaturing polyacrylamide gels and immunoblotting. -p, nonphosphorylated peptide; S15p, T18p, and S20p, peptides phosphorylated on Ser-15, Thr-18, and Ser-20, respectively. (B) Phosphorylation of p53 on Ser-20 in response to IR and UV light in U2-OS cells, as determined by immunoreactivity of p53 to antibodies DO1 and AbS20p. The fractions of p53 IP with antibody DO1 (not phosphorylated on Ser-20) and antibody AbS20p (phosphorylated on Ser-20) were determined by immunoblotting (IB) with antibody DO7. Because p53 protein levels increase in response to DNA damage, to facilitate comparisons between nonirradiated and irradiated cells, the amounts of cell extracts used were adjusted to have equal p53 protein levels in all reactions [as shown by immunoblotting (IB) with antibody DO7 control reactions not subjected to IP]. (C) Effect of protein phosphatase on the immunoreactivity of p53 to antibody DO1. Extracts from untreated or irradiated U2-OS cells were treated with protein phosphatase (PPase +) or mock-treated (PPase −) before IP with antibody DO1. The amounts of cell extracts used were adjusted to have equal p53 protein levels in all reactions.
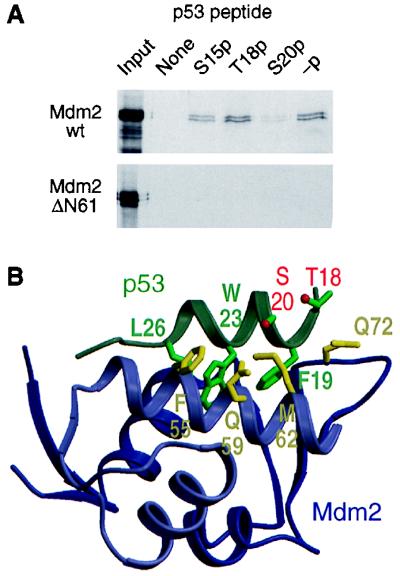
The genetic data presented above suggest that stabilization of p53 in response to DNA damage depends on phosphorylation of p53 on Ser-20. Furthermore, DNA damage leads to dissociation of p53 from Mdm2 (25). Because Ser-20 maps to the region of p53 that contacts Mdm2 (51), one would predict that phosphorylation of p53 on Ser-20 inhibits binding to Mdm2. Ser-15 and Thr-18 are also physically close to the p53–Mdm2 interface, but the genetic data suggest that their phosphorylation would not disrupt the interaction of p53 with Mdm2. One study has indeed reported that phosphorylation of Ser-20 weakens the interaction of p53 with Mdm2 (38), but in another study, phosphorylation of Thr-18, rather than Ser-20, affected binding of p53 to Mdm2 (37). In an attempt to address this discrepancy, we examined the effect that phosphorylation of p53 on Ser-15, Thr-18, or Ser-20 has on its interaction with Mdm2. Peptides that include the entire region of p53 that interacts with Mdm2 were synthesized with Ser-15, Thr-18, or Ser-20 phosphorylated and were assayed for binding to full-length Mdm2 or N-terminally truncated Mdm2 (ΔN61) lacking the domain that contacts p53, as a specificity control. Only phosphorylation of Ser-20 significantly compromised the interaction between p53 and Mdm2 (Fig. 5A). These results are consistent with the known three-dimensional structure of the p53–Mdm2 complex (51), in which the side chain of Ser-20 is within 1.5 Å of the hydrophobic side chain of Met-62 on Mdm2. The side chain of Thr-18 faces away from the p53–Mdm2 interface, and Ser-15 is outside the region of p53 that contacts Mdm2 (Fig. 5B).
Figure 5.
Effect of phosphorylation of Ser-20 on the interaction of p53 with Mdm2. (A) Capture of 35S-labeled in vitro-translated full-length or N-terminally truncated (ΔN61) Mdm2 by phosphorylated and nonphosphorylated p53 peptides corresponding to residues 7–29 of human p53; -p, nonphosphorylated peptide; S15p, T18p, and S20p, peptides phosphorylated on Ser-15, Thr-18, and Ser-20, respectively. (B) Three-dimensional structure of the p53–Mdm2 complex (PDB ID code 1YCR). The p53 peptide shown (residues 17–29) is the entire p53 region that associates with Mdm2. Selected amino acid side chains of p53 and Mdm2 are shown and labeled by using the codon number and single-letter residue code: F, Phe; L, Leu; M, Met; Q, Gln; S, Ser; T, Thr; and W, Trp. The oxygen atoms of the hydroxyl groups of Thr-18 and Ser-20 are colored red. The figure was prepared by using molscript (58) and raster3d (59).
Discussion
Ser-20 Phosphorylation and p53 Stabilization.
Activation of p53 in response to DNA damage involves an increase in p53 protein levels, which is caused by stabilization of the p53 protein (4, 12, 17). In turn, stabilization of p53 is caused by dissociation of p53 from Mdm2 (25), a protein that targets p53 for degradation (22–24). The involvement of Mdm2 in the regulation of p53 by DNA damage is well established, because modified p53 proteins that cannot associate with Mdm2 are not subject to Mdm2-dependent degradation and are not stabilized after DNA damage (22–24, 26, 27).
The mechanism by which DNA damage leads to dissociation of p53 from Mdm2 has been difficult to resolve, because regulation of p53 by DNA damage is very complex. There are 11 reported, and possibly even more, posttranslational modifications of p53 in response to DNA damage (16, 25, 28–36). Biochemical and genetic experiments have attempted to resolve which of these modifications are critical for p53 stabilization, but have yielded somewhat conflicting results. Biochemically, dissociation of p53 from Mdm2 has been attributed to p53 phosphorylation on Ser-15 (25), Thr-18 (37), or Ser-20 (38) by different groups, with no obvious explanation as to why different results were obtained. Analysis of mutant p53 proteins with substitutions targeting many of the Ser residues that are phosphorylated after DNA damage did not reveal major defects in p53 stabilization, leading to the conclusion that p53 phosphorylation does not regulate p53 stabilization (26, 27). However, this conclusion might have been premature. The genetic analysis of the mutant p53 proteins was somewhat incomplete, because one group did not study any p53 mutant with a Ser-20 substitution (27), whereas the second group reported that a p53 mutant with Ala substitutions of all N-terminal Ser, including Ser-20, had a partial stabilization defect. The latter group did not pursue their analysis by examining single-site mutants (26). Furthermore, neither of the two groups had available to them p53 proteins with modified tetramerization domains that allow the ectopic mutant p53 subunits to be regulated independently of endogenous wild-type p53.
On the basis of genetic and biochemical results, we propose that phosphorylation of p53 on Ser-20 leads to dissociation of p53 from Mdm2 and p53 stabilization in response to DNA damage. The most convincing evidence is that replacement of Ser-20 with Ala or Asp completely abrogated p53 stabilization after exposure to IR and UV light (Figs. 2 and 3). This result is quite striking in light of the observation that seven Ser residues in p53, including Ser-20, are phosphorylated in response to DNA damage, but substitutions targeting the other six Ser residues had no effect on p53 stabilization. In cells where endogenous wild-type p53 was stabilized in response to DNA damage, we could detect phosphorylation of p53 on Ser-20 after exposure to either IR or UV light (Fig. 4). Furthermore, the phosphorylation involved the major pool of nuclear p53 protein. Consistent with the genetic and biochemical analysis of p53 in cells, phosphorylation of p53 on Ser-20 weakened the interaction of p53 with Mdm2 in vitro, whereas phosphorylation of Ser-15 or Thr-18 did not have such an effect (Fig. 5). Although the results presented here argue that Ser-20 phosphorylation is required for p53 stabilization after DNA damage, we have not demonstrated that Ser-20 phosphorylation is sufficient for p53 stabilization; thus, our results are also consistent with a model whereby stabilization of p53 requires phosphorylation of p53 on Ser-20 and additional modification(s) targeting either p53 or even other proteins, such as Mdm2. We have excluded the possibility that the additional modification(s) target Ser residues 6, 9, 15, 33, 37, 376, 378, or 392 or Thr-18 of p53 (Figs. 2 and 3); however, there are still several known modifications of p53 in response to DNA damage (16, 25, 28–36), and potentially several, as-yet-unknown, modifications targeting p53 or Mdm2, one or more of which may cooperate with Ser-20 phosphorylation to induce p53 stabilization.
In addition to abrogating p53 stabilization after DNA damage, replacement of Ser-20 with Ala or Asp resulted in lower basal levels of p53 protein. This was particularly evident in the MEF (Fig. 3). A similar result was reported by Unger et al. (38) and was attributed to increased sensitivity to Mdm2-dependent degradation, because in cells that lack Mdm2, basal p53 protein levels were unaffected by replacement of Ser-20 with Ala. Increased degradation of p53 with Asp at position 20 further indicates that Asp is not functionally equivalent to phosphoserine in terms of its ability to weaken the interaction between p53 and Mdm2.
The proposed model linking Ser-20 phosphorylation and p53 stabilization in response to DNA damage does not contradict any previous reports, except for one study showing that the interaction of p53 with Mdm2 is disrupted by phosphorylation of Thr-18, and not by phosphorylation of Ser-20 (37). The reason for this discrepancy is unclear. We note, however, that we are not the only group reporting that phosphorylation of p53 on Ser-20 affects its interaction with Mdm2 (38).
DNA Damage-Signaling Pathways to p53.
Stabilization of p53 after exposure to IR requires ATM, a kinase implicated in DNA damage signaling (39–42). In response to UV light, stabilization of p53 is ATM independent and may require ATR, an ATM-related kinase (43). ATM and ATR phosphorylate p53 on Ser-15 in vitro and possibly in vivo (43–46); nevertheless, p53 stabilization cannot be mediated by direct phosphorylation of p53 on Ser-15 by ATM or ATR, because replacement of Ser-15 with Ala or Asp does not compromise p53 stabilization (refs. 26 and 27, and this study). Rather, p53 stabilization requires phosphorylation of Ser-20, and neither ATM nor ATR can phosphorylate p53 on Ser-20 (43–46). Because stabilization of p53 is dependent on ATM and ATR in response to IR and UV light, respectively, we propose that ATM and ATR activate other kinases that in turn directly phosphorylate p53 on Ser-20. Such a model is consistent with our current understanding of DNA damage signaling pathways in budding and fission yeast, where the ATM homologs Mec1 and Rad3 activate the downstream kinases Rad53 and Cds1, respectively, which in turn regulate the activities of transcription factors and mitotic regulators (52, 53). Regulation through a kinase cascade, rather than by Mec1 or Rad3 directly, allows amplification of the DNA damage signal and integration of signals from many checkpoint pathways (54). The human homolog of Rad53 and Cds1, referred to as Chk2 or hCdsl, has been cloned and is activated in an ATM-dependent manner in reponse to IR and in an ATM-independent manner in response to replication blocks, such as those caused by UV light (55–57). Chk2, like Cds1, regulates the activity of Cdc25C (55–57). It will be interesting to determine whether it also regulates the activity of transcription factors, such as p53.
Acknowledgments
We thank Daniel Scolnick, Frank Rauscher, III, and Giovanni Rovera for support and discussions. We also thank Andreas Nelsbach (New England Biolabs) for supplying us with antibody AbS20p before its commercial introduction. Financial support was provided by the American Cancer Society, the National Cancer Institute (CA76367), the W. W. Smith Charitable Trust (T.D.H.), and the Wistar Institute National Institutes of Health Training Grant T32 CA09171 (N.H.C.).
Abbreviations
- IR
ionizing radiation
- ATM kinase
ataxia–telangiectasia-mutated kinase
- ATR kinase
ATM + Rad3-related kinase
- IP
immunoprecipitation
- HA
hemagglutinin
- MEF
mouse embryo fibroblast
References
- 1.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 2.Carr A M. Microbiology. 1998;144:5–11. doi: 10.1099/00221287-144-1-5. [DOI] [PubMed] [Google Scholar]
- 3.Weinert T. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 4.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 5.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hooper M L, Wyllie A H. Nature (London) 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 7.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 8.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 9.Sah V P, Attardi L D, Mulligan G J, Williams B O, Bronson R T, Jacks T. Nat Genet. 1995;10:175–180. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- 10.Lowe S W, Bodis S, McClatchey A, Remington L, Ruley H E, Fisher D E, Housman D E, Jacks T. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 11.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 12.Maltzman W, Czyzyk L. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutzker S G, Levine A J. Nat Med. 1996;2:804–810. doi: 10.1038/nm0796-804. [DOI] [PubMed] [Google Scholar]
- 14.Chernov M V, Stark G R. Oncogene. 1997;14:2503–2510. doi: 10.1038/sj.onc.1201104. [DOI] [PubMed] [Google Scholar]
- 15.Haapajarvi T, Pitkanen K, Tsubari M, Laiho M. Mol Cell Biol. 1997;17:3074–3080. doi: 10.1128/mcb.17.6.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterman M J, Stavridi E S, Waterman J L, Halazonetis T D. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 17.Fritsche M, Haessler C, Brandner G. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]
- 18.Fu L, Benchimol S. EMBO J. 1997;16:4117–4125. doi: 10.1093/emboj/16.13.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 20.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lain S, Midgley C, Sparks A, Lane E B, Lane D P. Exp Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- 22.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 23.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 24.Midgley C A, Lane D P. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 25.Shieh S Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 26.Ashcroft M, Kubbutat M H, Vousden K H. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blattner C, Tobiasch E, Litfen M, Rahmsdorf H J, Herrlich P. Oncogene. 1999;18:1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 28.Knippschild U, Milne D M, Campbell L E, DeMaggio A J, Christenson E, Hoekstra M F, Meek D W. Oncogene. 1997;15:1727–1736. doi: 10.1038/sj.onc.1201541. [DOI] [PubMed] [Google Scholar]
- 29.Ko L J, Shieh S Y, Chen X, Jayaraman L, Tamai K, Taya Y, Prives C, Pan Z Q. Mol Cell Biol. 1997;17:7220–7229. doi: 10.1128/mcb.17.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blades J P, Hupp T. Oncogene. 1998;17:1045–1052. doi: 10.1038/sj.onc.1202014. [DOI] [PubMed] [Google Scholar]
- 32.Lu H, Taya Y, Ikeda M, Levine A J. Proc Natl Acad Sci USA. 1998;95:6399–6402. doi: 10.1073/pnas.95.11.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapoor M, Lozano G. Proc Natl Acad Sci USA. 1998;95:2834–2837. doi: 10.1073/pnas.95.6.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shieh S Y, Taya Y, Prives C. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bottger V, Bottger A, Garcia-Echeverria C, Ramos Y F, van der Eb A J, Jochemsen A G, Lane D P. Oncogene. 1999;18:189–199. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 38.Unger T, Juven-Gershon T, Moallem E, Berger M, Sionov R V, Lozano G, Oren M, Haupt Y. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 40.Khanna K K, Lavin M F. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- 41.Canman C E, Wolff A C, Chen C Y, Fornace A J, Jr, Kastan M B. Cancer Res. 1994;54:5054–5058. [PubMed] [Google Scholar]
- 42.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 43.Tibbetts R S, Brumbaugh K M, Williams J M, Sarkaria J N, Cliby W A, Shieh S Y, Taya Y, Prives C, Abraham R T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, et al. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 45.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 46.Khanna K K, Keating K E, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller S P, et al. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 47.Unger T, Sionov R V, Moallem E, Yee C L, Howley P M, Oren M, Haupt Y. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 48.Wieczorek A M, Waterman J L, Waterman M J, Halazonetis T D. Nat Med. 1996;2:1143–1146. doi: 10.1038/nm1096-1143. [DOI] [PubMed] [Google Scholar]
- 49.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 50.Stavridi E S, Chehab N H, Caruso L C, Halazonetis T D. Protein Sci. 1999;8:1773–1779. doi: 10.1110/ps.8.9.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 52.Huang M, Zhou Z, Elledge S J. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 53.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Nature (London) 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 55.Matsuoka S, Huang M, Elledge S J. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 56.Blasina A, de Weyer I V, Laus M C, Luyten W H, Parker A E, McGowan C H. Curr Biol. 1999;9:1–10. doi: 10.1016/s0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 57.Brown A L, Lee C H, Schwarz J K, Mitiku N, Piwnica-Worms H, Chung J H. Proc Natl Acad Sci USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 59.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]