Abstract
RNA recognition receptors are important for detection of and response to viral infections. RIG-I and MDA5 are cytoplasmic DEX(D/H) helicase proteins that can induce signaling in response to RNA ligands, including those from viral infections. LGP2, a homolog of RIG-I and MDA5 without the caspase recruitment domain required for signaling, plays an important role in modulating signaling by MDA5 and RIG-I, presumably through heterocomplex formation and/or by serving as a sink for RNAs. Here we demonstrate that LGP2 can be coexpressed with RIG-I to inhibit activation of the NF-κB reporter expression and that LGP2 protein produced in insect cells can bind both single- and double-stranded RNA (dsRNA), with higher affinity and cooperativity for dsRNA. Electron microscopy and image reconstruction were used to determine the shape of the LGP2 monomer in the absence of dsRNA and of the dimer complexed to a 27-bp dsRNA. LGP2 has striking structural similarity to the helicase domain of the superfamily 2 DNA helicase, Hef.
Innate immune responses provide the first line of defense against invading pathogens through the recognition of molecular patterns in pathogen-specific molecules (1). During RNA virus infection, double-stranded RNAs (dsRNA)3 generated during RNA replication are potent molecular patterns recognized by innate receptors (1–3). De novo-initiated but uncapped single-stranded RNAs (ssRNA) can also signal viral infections (4, 5).
Several innate immunity receptors can recognize RNA ligands (6). Within membranes, TLRs (Toll-like Receptors) 3, 7, and 8 can respond to single- and double-stranded RNAs (7). Within the cytoplasm, members of the superfamily 2 class of DEX(D/H) helicases and ATPases can sense RNAs to initiate antiviral responses. These include the RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation-associated gene 5) (8–12). The RIG-I protein can recognize both double-stranded RNAs and RNAs with a 5′-triphosphate (13–16). Ligand binding can induce RIG-I and MDA5 to release the caspase recruitment domains (CARDs) within these proteins (12), leading to binding of the mitochondrial adaptor IPS-1 (interferon-β promoter stimulator-1, also known as MAVS, VISA, and Cardif) that coordinates the activities of kinases to activate transcription factors required for cytokine and chemokine production (17, 18).
LGP2 (laboratory of genetics and physiology 2), originally identified as a highly expressed gene in mammary tumors, is another cytoplasmic DEX(D/H)-box helicase that can recognize RNA (19). Unlike RIG-I and MDA5, LGP2 lacks the CARDs (20). It has also been proposed to antagonize RIG-I- and MDA5-mediated signaling by forming heterodimeric complexes with these proteins (19, 21, 22). In Sendai virus-infected cells, LGP2 can coprecipitate with RIG-I in an interaction that requires the RIG-I CARD, suggesting a repression of signaling (19). In addition, LGP2 can interact with IPS-1 to block binding by IKKι that would otherwise activate IRF-3, a key transcription factor in interferon production (10, 21). Despite evidence that LGP2 may act as a negative regulator of innate signaling (20), its activity may be more complex because Lgp2-/- mice exhibit both a defect in IFN production in response to encephalomyocarditis virus infection and resistance to lethal doses of vesicular stomatitis virus (23).
The main goal of this study is to understand how LGP2 can respond to ligands. We characterized the structure and RNA binding by the recombinant LGP2 expressed in insect cells; imaged monomers, dimers, and oligomers of LGP2 using electron microscopy; and reconstructed three-dimensional images without presumed symmetry. The resulting model of the LGP2 monomer has obvious similarity to the crystal structure of the Hef helicase domain (24). The remaining electron density can accommodate the recently reported structure of the RIG-I regulatory domain (15, 16).
EXPERIMENTAL PROCEDURES
Cell-based Reporter Assays—HEK 293T cells were harvested from an actively growing culture and plated in CoStar white 96-well plates at 4.5 × 104 cells/well. Cells at ∼80% confluency were used for transfection with Lipofectamine 2000 (Invitrogen) mixed with plasmids pNiFty-Luc (30 ng, Invivogen) and phRL-TK (5 ng, Promega), which code for the firefly luciferase reporter under a promoter that contains NF-κB elements and the Renilla luciferase driven by the herpesvirus thymidine kinase promoter, respectively. Where appropriate, plasmids encoding Rig-I (pUNO-hRIG-I; Invivogen) and/or Lgp2 (pUNO-hLGP2; Invivogen) were added to the transfection mixture. Transfected cells were incubated for 24 h to allow expression from the plasmids. Ligand to induce RIG-I was transfected into the cells using Lipofectamine. The ligand shRNA was synthesized by in vitro transcription using a T7 RNA polymerase kit (Ambion, Inc.) and purified from a denaturing polyacrylamide gel as described by Kao et al. (25). After another 12-h incubation, the cells were harvested using the Dual-Glo Luciferase assay system (Promega). Luminescence was quantified using a FLUOstar OPTIMA Plate Reader (BMG LABTECH, Inc). All assays were performed with six independent replicates per experiment, and each result reported was consistent in at least two sets of assays.
LGP2 Expression and Purification—Sf9 (Spodoptera frugiperda 9) cells were cultured in HyQ® SFX-Insect™ medium (Hyclone) and grown at 27 °C in flasks.
Full-length Lgp2 was cloned into the baculovirus transfer vector pBAC-1 (Novagen, Inc.), and its sequence was verified by DNA sequencing. The expression constructs were cotransfected with linearized baculovirus DNA (BD Biosciences) into Sf9 cells, and the resultant recombinant virus was amplified and used for protein expression. At 72 h after infection with baculovirus encoding Lgp2, the cells were pelleted by centrifugation for 5 min at 680 × g and resuspended in ice-cold lysis buffer (200 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and 1% Nonidet P-40). Cell lysates were clarified by centrifugation at 30,600 × g for 15 min. HIS-Select (Sigma) nickel affinity resin was added to the clarified supernatant to bind the recombinant protein. The resin was washed four times with Buffer A (20 mm Tris-HCl, pH 7.5, and 150 mm NaCl) containing 10 mm imidazole. Bound protein was eluted with Buffer A containing 250 mm imidazole. Purified LGP2 was concentrated and further purified by gel filtration chromatography on a Superdex 200 column (HiLoad 16/60, GE Healthcare) eluted with Buffer A. Gel filtration chromatography used to study RNA binding used a Superdex 200 column (10/300 GL) calibrated with protein standards (Bio-Rad).
Gel-based RNA Protein-binding Assays—RNA RS46, which forms an intramolecular hairpin with a stem of 21 bp, was used for gel-based assays. RS46 was made by in vitro transcription using a T7 RNA polymerase kit (Ambion) and purified as a single band from a denaturing gel. The RNA was treated with alkaline phosphatase and radiolabeled with [32P]ATP using T4 polynucleotide kinase. Prior to use, RS46 was heated to 90 °C followed by slow cooling to room temperature to induce hairpin formation. The 27-nt single-stranded RNA (css27) was made by in vitro transcription using T7 RNA polymerase in the AmpliScribe kit (Ambion). The RNA was then purified from free NTPs, aborted oligonucleotides, and the digested DNA template by purification with a P-6 gel filtration column. An aliquot of the RNA was treated with alkaline phosphatase to remove the 5′-terminal phosphates and then adjusted to the same concentration as the untreated RNA.
UV-induced cross-linking reactions were performed with 1 μg of purified proteins and 40 fm RNA in a buffer containing 50 mm Tris-HCl, 4 mm MgCl2, and 25 mm NaCl (pH 8.1) by irradiation with UV light at 1200 mJ for 2 min. The cross-linking reactions were subjected to SDS-PAGE, and the bands were visualized using a PhosphorImager.
Electrophoretic mobility shift assays used labeled RNA (50 pm) incubated with a 125 nm final concentration of the desired protein in a 20-μl reaction containing 20 mm sodium glutamate, pH 8.2, 4 mm MgCl2, 12.5 mm dithiothreitol, and 0.5% (v/v) Triton X-100. After an incubation at 25 °C for 30 min, a 4-μl termination mix (containing 10 mm Tris, pH 8.2, 1 mm EDTA, 0.1% bromphenol blue, 0.1% xylene cyanol, and 30% glycerol) was added. The samples were electrophoresed on a native 10% polyacrylamide gel at 400 V for 1 h, after which the gel was wrapped in plastic and autoradiographed.
Fluorescence Anisotropy Assay—Cy3-labeled RNA probes for the binding assays were synthesized, purified by high pressure liquid chromatography, and quantified by the manufacturer (IDT Technologies). The probes were diluted to 200 nm in Buffer A. For the formation of dsRNA, the probes were heated at a 1:1 molar ratio with the complementary RNA in an 80 °C water bath, after which the bath was allowed to cool to room temperature. Fluorescence measurements were made at room temperature with a PerkinElmer Life Sciences luminescence spectrometer LS55 using cuvettes with an optical path length of 0.4 cm. Measurements were taken with an integration time of 1 s and a slit width of 5 nm. The excitation and emission wavelengths used were 490 and 520 nm, respectively. Anisotropy values were recorded 20 s after each protein addition to allow the sample to reach equilibrium. The total volume of protein added was <5% of the final volume. Represented anisotropy values are the average of 10 measurements. Binding data were analyzed by nonlinear least square fitting using KaleidaGraph software (Synergy Software, Reading, PA). The Hill equation, ΔA = BmaxXn/(Xn + K nd) was used to determine the dissociation constant (Kd). In this equation, ΔA is the anisotropy change caused by the ligand binding, Bmax is the maximum anisotropy change, X is the total concentration of the input protein, and the exponential term n is the Hill coefficient.
Electron Microscopy—LGP2 (∼0.6 ng/μl in Buffer A) was negatively stained on a freshly glow-discharged carbon-coated copper grid with a 1% (w/v) aqueous solution of uranyl acetate. The grid was viewed in a JEOL 1200 EX TEM at a calibrated magnification of ×56,200 and an operating voltage of 100 kV. The micrographs were digitized using an Epson Projection 3200 scanner at 1200 dpi corresponding to 4.1 Å/pixel at the specimen level. Three-dimensional reconstructions were calculated using the EMAN (version 1.8) software package (26) in procedures similar to those described by Sun et al. (27). More than 3,000 single particles of the LGP2 were selected using the boxer routine, filtered to remove high frequency noise, centered, and aligned. The particles were then used to generate class averages without imposed symmetry using EMAN′s “refine2d” routine. An initial three-dimensional model was calculated from a set of selected noise-free class averages. The reconstructions were iteratively refined until the structure was stable as judged by Fourier shell correlation. A molecular mass of 80 kDa was used for the surface-rendering threshold of the three-dimensional structure. Three-dimensional reconstructions were visualized using the Chimera software package (28).
To visualize oligomeric LGP2 complexed to dsRNA, it was mixed with a 4-fold molar excess with 40-bp poly(I-C) in Buffer A and incubated on ice for 10 min prior to fixing the sample on an electron microscopy grid and staining with uranyl acetate as described above. The sample was viewed in a JEOL 1200 EX electron microscope at an acceleration voltage of 100 kV and at a calibrated magnification of ×38,900. The micrographs were digitized at 1200 dpi, corresponding to 5.5 Å/pixel at the specimen level.
RESULTS
LGP2 Can Modulate RIG-I Activation of Gene Expression— To assess the biological activity of LGP2, we transiently transfected HEK 293T cells with plasmids expressing an innate immunity receptor along with two reporter constructs: a firefly luciferase reporter construct driven by a promoter containing the NF-κB binding element and a Renilla luciferase driven by the thymidine kinase promoter. The Renilla luciferase serves as a transfection control and a control for the general health of the cells (29). Results from the firefly luciferase were divided by the Renilla luciferase to generate the term Ratio-FF/R. In some of the samples, the Ratio-FF/R could be from ligand-induced versus uninduced cells. In each sample, at least three independent wells of cells were tested with each condition, and each set of results was repeated in at least two experiments and found to be consistent.
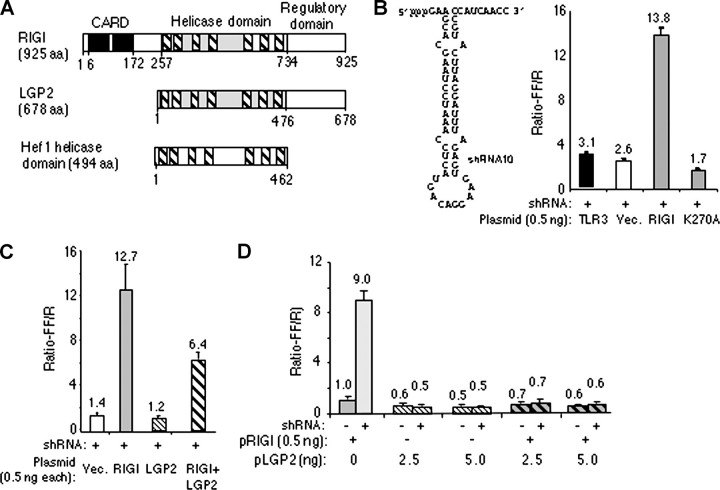
The plasmid expressing LGP2 was unable to activate the NF-κB promoter in the presence of several ligands, including ssRNA, hairpin RNA, or double-stranded RNA such as poly(I-C) (data not shown). However, the plasmid expressing RIG-I could activate the promoter by 8 of 14-fold when the cells were induced by transfecting poly(I-C) or a 60-nt hairpin RNA named shRNA10 (Fig. 1B and data not shown). No response was detected when shRNA10 or other ligands were added to the medium (data not shown), consistent with previous reports on the requirements of Rig-I induction (19). The activation was also specific to Rig-I because wild-type TLR3 and Rig-I with a mutation in the ATP-binding motif (K270A) had much lower responses to shRNA10 (Fig. 1B). Western blot analysis showed that K270A is expressed at a comparable level to wild-type RIG-I (data not shown).
FIGURE 1.
LGP2 can inhibit RIG-I activation of cell signaling. A, domain arrangements of RIG-I, LGP2, and the helicase domain of Hef. aa, amino acids. B, sequence and secondary structure of a RIG-I ligand, shRNA10, and its effect on Rig-I activation of reporter signaling from the NF-κB promoter. RIG-I was expressed from the plasmid pUNO-RIG-I (pRIG-I on the figure). The height of each bar represents the mean of the results of six independently transfected cultures of cells. The horizontal line above the bar represents the range for 1 S.E. The identity of the construct used is shown below each bar. The wild-type TLR3 construct used was described by Ranjith-Kumar et al. (29). K270A is a mutation in RIG-I that inactivates signaling (19). In this and other assays, the cell cultures were induced with shRNA10 transfected into the cells at 1 μm. Vec., vector. C and D, the effects of pUNO-LGP2 (pLGP2) on signaling by pUNO-RIG-I. C shows the results from an equal molar ratio of the two plasmids expressing RIG-I and LGP2. D shows the results from a 1:5 and 1:10 molar ratio of the plasmid expressing RIG-I to LGP2. The transfections of the ligand or buffer controls are shown as + and -, respectively. In parentheses are the numbers of nanograms of plasmid DNA transfected into cells.
Because RIG-I was active for inducing reporter activity, we examined the effect of the presence of LGP2 on RIG-I activity. At a 1:1 ratio of the two plasmids, an ∼50% reduction in RIG-I activity was observed (Fig. 1C). At a ×5 or 10 molar ratio of the plasmid encoding Lgp2 to Rig-I, RIG-I-dependent reporter activities were reduced to background (Fig. 1D). In these experiments, the same amounts of DNA were transfected; any differences were adjusted by the addition of the empty vector. The identical concentration of pUNO-LGP2 did not induce the NF-κB reporter (Fig. 1C). These results are consistent with those of Saito et al. (19), who demonstrated that LGP2 could inhibit RIG-I activation of the IFN-β promoter. Furthermore, these results demonstrate that Lgp2 cDNA used here is functional in cells and hence is appropriate for biochemical analysis.
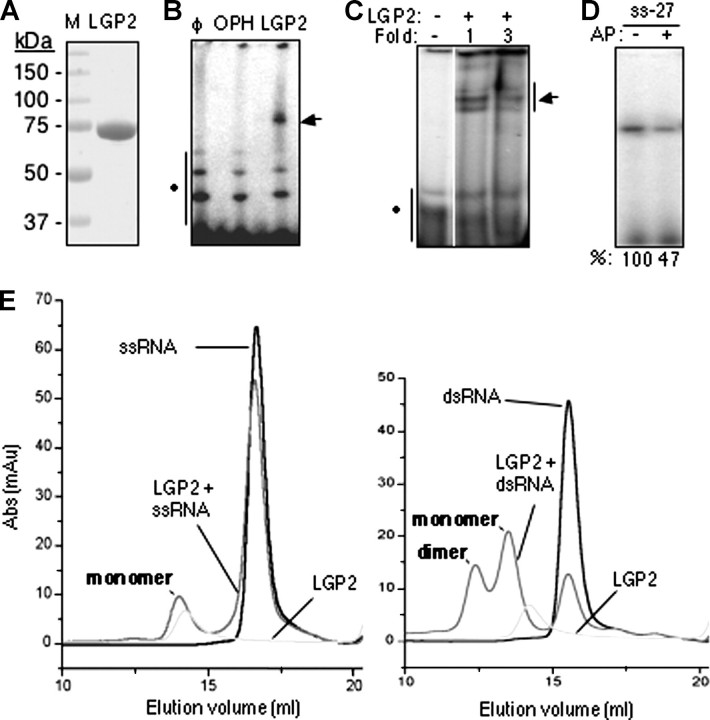
RNA Binding by Recombinant LGP2 Protein—Soluble LGP2 was produced using baculovirus-infected Sf9 cells and purified to a single band when visualized by staining with Coomassie Blue (Fig. 2A). LGP2 was eluted at ∼80 kDa in a gel filtration column when compared with calibrants, in excellent agreement with the mass expected of an LGP2 monomer.
FIGURE 2.
Purified LGP2 can bind RNA. A, SDS-PAGE analysis of purified recombinant LGP2 expressed from baculovirus-infected insect cells. The gel was stained with Coomassie Blue R250. B, RNA cross-linking of LGP2 to dsRNA of 46 nt named RS46. The symbol φ denotes a reaction containing only radiolabeled RS46. Lanes labeled with organophosphate hydrolase (OPH) and LGP2 contain reactions wherein purified recombinant LGP2 or organophosphate hydrolase was used in UV-induced cross-linking reactions to RS46. C, results from a gel mobility shift assay wherein 125 or 375 nm (final concentrations) purified LGP2 was incubated with 50 nm radiolabeled RS46. D, LGP2 binding to a 27-nt RNA with a 5′-triphosphate or the same RNA treated with alkaline phosphatase (AP). Cross-linked protein-RNA complexes were resolved on denaturing PAGE. The numbers at the bottom are from quantification of the radiolabel by phosphorimaging analysis. ss-27, 27-nt ssRNA. E, gel filtration chromatography shows that LGP2 preferentially binds dsRNAs and can form a 2:1 complex with dsRNA. The column was calibrated with molecular standards of 670-, 158-, 44-, 17-, and 13.5-kDa proteins. The elution volumes of monomeric and dimeric LGP2 are consistent with proteins of 85 and 150 kDa. The left panel was performed with an LGP2 and a 27-nt ssRNA. The right panel was performed with a 27-bp dsRNA. Abs, antibodies.
Several RNA binding assays were used to characterize the biochemical activity of LGP2. First, a UV-induced cross-linking assay was performed with a radiolabeled RNA named RS46 that contains a 21-bp perfect duplex capped by a GNRA hairpin. A shifted band was detected in denaturing PAGE with the reaction containing LGP2, whereas an equal concentration of organophosphate hydrolase, which does not bind RNA, served as a negative control (Fig. 2B). A similar result was observed by a nondenaturing electrophoretic mobility shift assay (Fig. 2C). These results qualitatively demonstrate that LGP2 can bind RNA.
RIG-I and the regulatory domain of the RIG-I protein were reported to preferentially bind RNAs with a 5′-triphosphate (15, 16). To examine whether LGP2 has a similar activity, we prepared a 27-nt RNA named css27 by in vitro transcription in the presence of internal radiolabels. An aliquot of css27 was then treated with alkaline phosphatase to remove the 5′ phosphates. Phosphatase treatment of css27 reduced cross-linking to LGP2 by about 2-fold (Fig. 2D). Thus, LGP2 exhibits preference for a ligand containing a 5′-triphosphate, although it can recognize RNA lacking a 5′-triphosphate.
Because LGP2 can bind to both ssRNA and dsRNA, we wanted to determine which form is preferred. To examine this and to determine whether RNA can induce LGP2 oligomerization, gel filtration chromatography was used. A chemically synthesized ss27 mixed with LGP2 resulted in a slight change in the LGP2 elution profile (Fig. 2E, left panel). However, dsRNA consisting of ss27 annealed to its complement css27 produced a larger shift in the LGP2 elution profile as well as a dramatic increase of a peak at ∼160 kDa that is indicative of an LGP2 dimer (Fig. 2E, right panel). These results using purified LGP2 are in agreement with observations that LGP2 present in a lysate can oligomerize in the presence of dsRNA (19). Furthermore, LGP2 preferentially binds to dsRNA rather than to ssRNA.
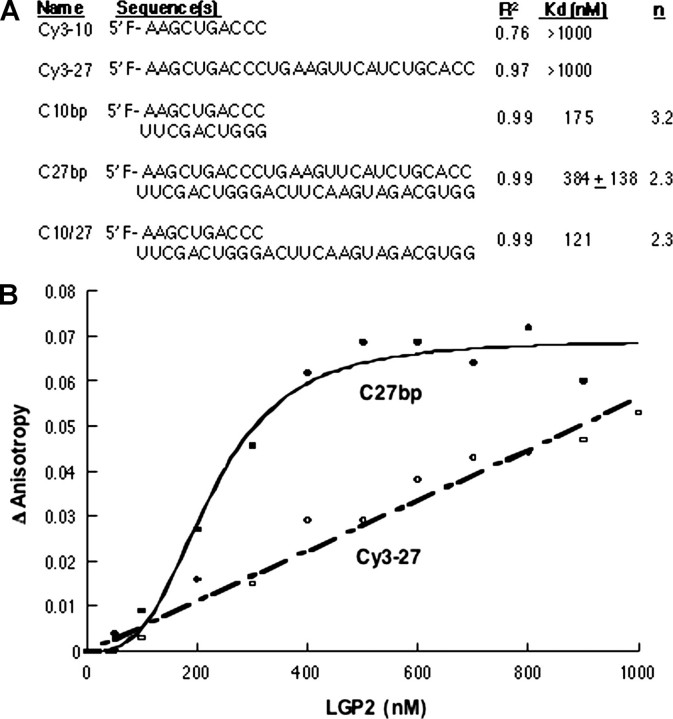
Quantitative Examination of RNA Binding by LGP2—A fluorescence anisotropy assay was used to determine the affinities of LGP2 binding to RNAs. A series of chemically synthesized RNAs of 10 or 27 nt with 5′-Cy3 fluorophores were used (Fig. 3A). A linear increase in the anisotropy of the single-stranded 10- and 27-nt RNAs was observed with increasing LGP2 concentrations (Fig. 3, A and B). However, the binding isotherms did not reach saturation with either ssRNAs, and the Kd values for either RNAs can only be estimated to be in excess of 1,000 nm, the highest concentration of LGP2 assayed (Fig. 3B).
FIGURE 3.
Fluorescence anisotropy to determine the binding affinity of LGP2 for RNA. A, a summary of the RNAs used to assess binding by LGP2 and parameters for their interactions with LGP2. The R2 column denotes the fit of the data to the Hill equation. The n column denotes the Hill coefficient for the binding isotherm. Where values are missing, the binding isotherm did not reach saturation under the conditions tested, and hence the parameters could not be determined. B, binding isotherms for LGP2 interaction with a ssRNA of 27 nt and a dsRNA of 27 bp.
The 10- and 27-nt ssRNAs were also annealed to complementary oligonucleotides to form dsRNA. Upon LGP2 addition, anisotropy values changed dramatically and reached saturation. The binding isotherms fitted to the Hill equation revealed that the Kd values were 175 and 384 + 138 nm for the 10- and 27-bp RNAs, respectively (Fig. 3, A and B). The value for the 27-bp RNA was derived from three independent experiments. We note that these binding assays were performed in a buffer that contained 150 mm monovalent salt to discourage nonspecific binding and could be better in a buffer with reduced salt. The Kd was 121 nm for LGP2 binding to a partial RNA duplex where the Cy3-labeled 10-nt RNA was annealed to C27 (generating a 10-bp duplex and a 17-nt overhang) (Fig. 3A). These results demonstrate that LGP2 clearly prefers binding to dsRNA over ssRNA and that RNAs that contain partially double-stranded regions are also recognized. Lastly, the Hill coefficients for LGP2 binding to dsRNAs and to the partial duplex were above 2.3, indicating positive cooperativity (Fig. 3A). Cooperative LGP2 binding to dsRNA may provide a basis for LGP2 oligomerization.
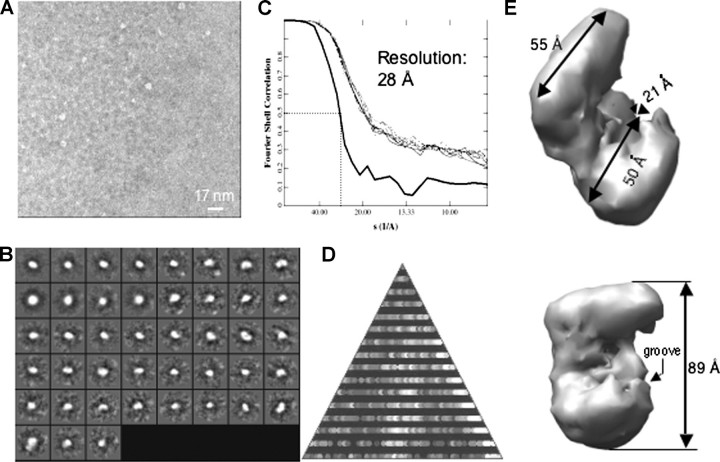
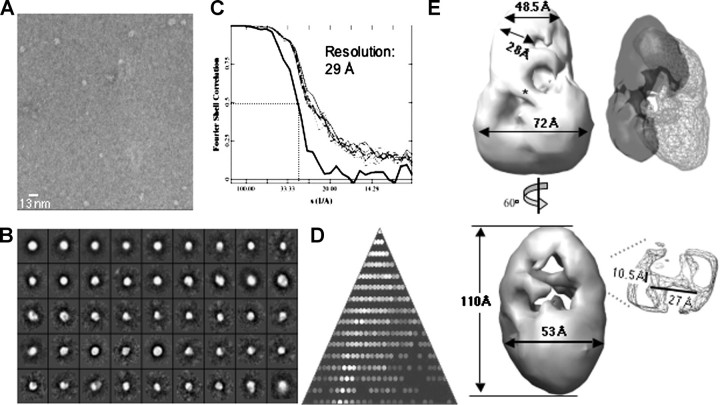
A Low-resolution Structure for LGP2—Image reconstruction of single particles from electron micrographs can be used to obtain low-resolution structures of proteins and protein complexes (30, 31). We performed this analysis without imposed symmetry for more than 3,000 individual particles of the monomeric LGP2 (Fig. 4, A and B). Refinement of the initial model for eight cycles resulted in a structure that converged to a resolution of ∼28 Å (Fig. 4C). As a check on the reconstruction, the particles used represented all of the expected views seen in the class averages (Fig. 4B). The overall shape of LGP2 resembles a partially opened clamp with a length of 89 Å (Fig. 4E). Notably, the lower portion of the clamp shown in Fig. 4E contains a groove (with a width of ∼21 Å) that may be involved in RNA binding.
FIGURE 4.
Reconstruction of the monomeric structure of LGP2. A, an electron micrograph of monomeric LGP2 is shown. The scale bar in the micrograph equals 17 nm. B, class averages of all the particles selected from the micrograph without assumption of symmetry are shown. C, Fourier shell correlation using the 0.5 σ criterion suggests a resolution of 28 Å. D, an asymmetric triangle shows a complete three-dimensional sampling of the particles used in the reconstruction. E, two views of the three-dimensional reconstruction of the monomeric LGP2 and the dimensions of the particles are shown.
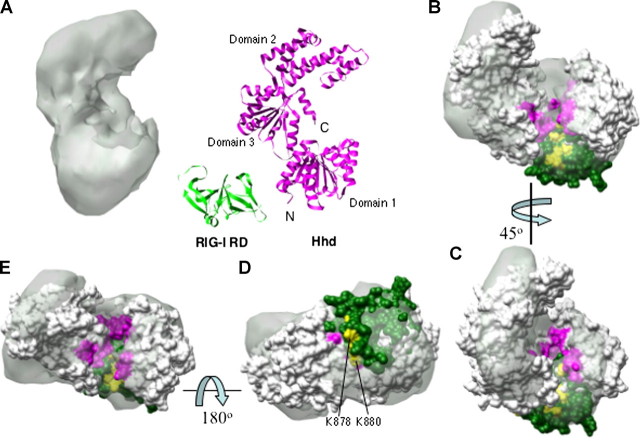
Given that LGP2 contains helicase-like motifs, we compared the electron density of LGP2 to the crystal structure of the Hhd (Hef helicase domain) (residues 1–494) of the Hef protein from the hyperthermophyllic archaeon Pyrococcus furiosus, another member of the superfamily 2 helicase (24). The crystal structure of Hhd consists of three subdomains (24, 32). Similar to other members of superfamily 2 helicase group, a parallel α-β domain contains the seven helicase motifs (24) and corresponds to domains 1 and 3 (Fig. 5A). A comparison of the helicase motifs in the Hhd and LGP2 is shown in supplemental Fig. 1. Like LGP2, the crystal structure of Hhd resembles a partially opened clamp with a concave groove associated with its lower portion (Fig. 5A). In Hhd, this groove contains the various DEAD-box helicase motifs that are involved in ATP recognition and hydrolysis and also RNA binding (33). Notably, the center of the LGP2 clamp is broader than the comparable region in Hhd (Fig. 5B), likely because of additional domains that are present in LGP2.
FIGURE 5.
A comparison of LGP2 structure to the Hef helicase domain. A, a comparison of the overall structures of the LGP2 and the Hef helicase domain (Protein Data Bank code 1WP9) and the C-terminal RIG-I regulatory domain (green) (Protein Data Bank code 2QFB). B–E, results from manual docking of Hhd and the RIG-I RD into the LGP2 electron shell. The orientations of the views relative to each other are indicated by the arrows. The helicase motifs of Hhd are colored in pink and fit nicely into the grooved part of the LGP2 reconstruction. The RD of RIG-I is in green, and the eight RNA-binding residues identified in Refs. 15 and 16 are in yellow. Two of the highlighted residues are in the surface away from the helicase motifs, as shown in D. It is possible that the observed effects of these two residues on RNA binding identified in Ref. 16 are due to an indirect effect of the presence of RNA.
LGP2, RIG-I, and MDA5 all share a C-terminal regulatory domain (RD) (also known as a C-terminal domain (16)) or the repressor domain (19) (Fig. 1A). The structure of the RIG-I RD (amino acids 802–925) was recently solved independently by x-ray crystallography (15) and NMR (16). The RD is a relatively flat structure with a concave and a convex side with dimensions of 45 × 35 × 30 Å (15). We manually docked the RD structure into the monomer of LGP2 with the concave surface to match the lower portion of LGP2. The RIG-I RD fits nicely to the larger of the unoccupied area next to the groove that is opposite the side of the C terminus of the Hhd (Fig. 5A). The other area does not contain sufficient density to accommodate the RD.
Three views of the location of the RD and Hhd within the LGP2 electron shell are shown in Fig. 5, B–E. To judge the fit, several residues in the RD that have been identified to be required to interact with RNA are highlighted in yellow: His830, Lys858, Lys861, Ile875, Lys878, Lys880, Lys888, and Lys907. Six of these residues are aligned with in the same groove that contains the Hhd helicase motifs (Fig. 5, B, C, and F). The remaining two (Lys880 and Lys878) are on the other side relative to the helicase motifs (Fig. 5D). The fit of the electron density and the alignment of the RNA-binding residues with the helicase motifs into one groove suggest that the LGP2 RD lies to the side of the parallel α-β domains.
Reconstruction of LGP2 Complexed with RNA—LGP2 complexed to a 27-bp dsRNA eluted as a dimer. To visualize the dimer, we collected the appropriate gel filtration fractions and immediately placed the samples onto electron microscopy grids to minimize dissociation. The negatively stained particles observed (Fig. 6A) are distinct in shape and size from the monomeric LGP2 particles seen in Fig. 4A. Processing of over 3,000 particles yielded the class averages, particle coverage, and a model calculated to have a resolution of 29 Å (Fig. 6, B–D). The overall structure had approximate bilateral symmetry (Fig. 6E). When bisected in the horizontal plane, both halves have a wider and narrower portion. However, the wider portion of the upper half is approximately perpendicular to the comparable portion in the lower half. Two projections of the dimer are shown in Fig. 6E.
FIGURE 6.
Reconstruction of the dimeric structure of LGP2. A, an electron micrograph of LGP2 in complex with 27-bp dsRNA is shown. The sample used was from a gel filtration column, an example of which is shown in Fig. 2E. B, class averages of all the particles selected from the micrograph without assumption of symmetry are shown. C, Fourier shell correlation using the 0.5 σ criterion suggests a resolution of 29 Å. D, an asymmetric triangle shows a complete three-dimensional sampling of the particles used in the reconstruction. E, a three-dimensional model of the LGP2 dimer with the relevant dimensions is shown. Docking of two monomeric LGP2 structures to fit into the dimeric LGP2 is shown (top right panel). One monomer is shown in charcoal and the other in outlined form. The best fit in the LGP2 electron shell is a head-to-tail arrangement of two dimers, although significant conformational changes are necessary for two dimers to fit into the electron shell of the dimer. Several contacting regions in the dimer are not observed in the monomer (the area identified by an asterisk). The dimensions of the putative dsRNA within the LGP2 dimer are shown in the bottom right panel.
When structures of two monomers of LGP2 helicases were fitted into the density of the dimer, the best fit was obtained with the monomers arranged in head-to-tail configuration and partially twisted around each other. Even with this arrangement of the two monomers, not all of the dimers' electron density can be accounted for. For example, the overall length of the monomer is 89 Å, in comparison to the dimer, which has a maximal length of ∼110 Å. In addition, several contacting regions in the dimer are not observed in the monomer (e.g. see the area identified by an asterisk on Fig. 6E). These two observations suggest that a significant change in the conformation of the monomer takes place upon formation of the dimer in association with dsRNA.
We also observed a rod-shaped density with an estimated diameter (∼10.5 Å) that is a good fit with dsRNA (Fig. 6E, lower right panel). This density was not observed in the reconstructions of the monomers and could correspond to the dsRNA. It is located at the lower portion of the clamp, the portion of the molecule that putatively contains the helicase motifs (Fig. 5B). The location of the dsRNA in the dimeric LGP2 structure and the interaction between the protein subunits need to await higher resolution analysis.
Oligomeric Structures of LGP2—LGP2 helicases larger than dimers were not observed with the 27-bp dsRNA in either the gel filtration profiles or the electron micrographs of LGP2. To determine whether a longer dsRNA could result in oligomer formation by LGP2, a 40-bp poly(I-C) was mixed with LGP2 at a 1:4 molar ratio and then processed for electron microscopy. Quantification of several areas of the grid that contained a total of over 500 particles showed that 11% of the particles exist in a higher oligomeric state (supplemental Fig. 2A). A representative gallery of oligomers is shown in Fig. 2B. The typical dimension of the oligomers is ∼270 Å in length and ∼70 Å in width. The width is consistent with that of an LGP2 dimer, but the length is significantly longer than that of the LGP2 monomer or dimer (supplemental Fig. 2). These longer particles were not observed with the 27-bp dsRNA or with poly(I-C) imaged in the absence of LGP2 (data not shown). We did not attempt to reconstruct the three-dimensional structure for these oligomers as they are heterogeneous, but their presence demonstrates that LGP2 can oligomerize when bound to longer dsRNA.
DISCUSSION
In this study we have 1) determined that LGP2 transiently expressed in HEK 293T cells with RIG-I inhibited RIG-I signaling; 2) characterized RNA binding by recombinant LGP2 protein; and 3) generated low-resolution images of monomers and dimers of LGP2 using electron microscopy, image reconstruction, and manual docking of the helicase domains from the Hef protein and the RD from RIG-I.
LGP2 is an efficient negative regulator of RIG-I signaling. A 1:1 ratio of pLGP2 to pRIG-I could reduce RIG-I signaling by about half. At a 5:1 ratio of pLGP2 to pRIG-I, RIG-I activation of signaling was reduced to background. Our results on the functional properties of LGP2 are consistent with the reports of Saito et al. (19), who showed that LGP2 could form a complex with RIG-I. The RIG-I RD was reported to efficiently suppress RIG-I activation of cell signaling (19). In our hands, the LGP2 RD (amino acids 476–678) could reduce RIG-I signaling in the absence of the remainder of the LGP2 protein. However, a 10-fold molar excess of the plasmid containing the LGP2 RD only reduced RIG-I signaling by about 40% (data not shown), much less than what was seen with the full-length LGP2. These results suggest that the helicase domain of LGP2 contributes to the inhibitory activity of LGP2.
Although LGP2 can bind either ssRNAs or dsRNAs, several differences are noteworthy. First, dsRNA is bound with higher affinity than ssRNA. Second, we observed cooperativity for LGP2 binding to dsRNA but not for ssRNA (Fig. 3B). Third, a 27-bp dsRNA can induce dimerization of LGP2, whereas a 27-nt ssRNA could not (Fig. 2E). These results can be compared with those from the RD of RIG-I, which binds triphosphorylated ssRNA with a Kd of ∼150 nm but which has significantly weaker binding when the RNA was treated with phosphatase or when the RNA was in double-stranded form (15). Lastly, although LGP2 does exhibit a preference for binding to a triphosphate-containing ssRNA (Fig. 2D), the difference we observed was more modest than reported for RIG-I (15, 16). We speculate that all of these differences between LGP2 and the RD of RIG-I may be due to the contributions of the helicase domain.
We have been unable to produce crystals of full-length LGP2. However, low-resolution structures of LGP2 and comparison to structural homologs do reveal useful insights. In fact, other superfamily 2 helicases, including the hepatitis C virus helicase, the DNA repair enzyme UvrB, and the Hef helicase, have two core parallel α-β domains that regulate ATP binding and hydrolysis and also ancillary domains that may confer additional properties, such as RNA binding (24, 32). In addition to RIG-I recognition of triphosphorylated RNA through its C-terminal domain (15, 16), the Escherichia coli DdpA and the Bacillus counterpart YxiN have a C-terminal domain that confers specific recognition for the 23 S rRNA (34).
The docking of the Hef helicase domain into the electron shell of LGP2 immediately suggests that LGP2 retains a structure comparable with the three domains of Hhd. Indeed, the helicase motifs are concentrated within a groove in the concave surface of the LGP2. The observation that the RIG-I RD can line up with the groove containing the helicase motifs further suggests that the two domains can cooperate in RNA binding (Fig. 5).
A small portion of the LGP2 electron density in the midsection of the clamp structure remains unaccounted for after the docking of Hhd and the RIG-I RD. We speculate that the 77 amino acids (residues 472–549) that connect the helicase domain to the RD will fit into this portion of the LGP2 molecule.
Lastly, docking of the electron shell of two monomers into the electron density of the dimer suggests that significant conformational changes in each monomer must take place in order for the stable dimer to form. It is increasingly apparent that all of the nucleic acid-binding receptors have sophisticated mechanisms to distinguish and to differentially respond to a variety of ligands. Furthermore, ligand binding can lead to an oligomerization state that is a central feature of the signaling mechanism (35). Our studies provide initial glimpses into the structural features and oligomerization of LGP2. Higher resolution structures will be necessary to confirm the arrangements of the various functional motifs in LGP2.
Supplementary Material
Acknowledgments
We thank S. Chinnaswamy for RS46 and the electrophoretic mobility shift assay results shown in Fig. 2C and L. Kao and S. Hoose for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 1RO1AI073335 from NIAID. This work was also supported by Centocor Inc. Grant CRA-Kao2007. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: dsRNA, double-stranded RNA; ssRNA, single-stranded RNA; CARD, caspase recruitment domain; NF-κB, nuclear factor κB; RD, regulatory domain; shRNA, short hairpin RNA; nt, nucleotide.
References
- 1.Meylan, E., and Tschopp, J. (2006) Mol. Cell 22 561-569 [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama, M., and Fujita, T. (2007) J. Biol. Chem. 282 15315-15318 [DOI] [PubMed] [Google Scholar]
- 3.Tenoever, B. R., and Maniatis, T. (2006) Immunity 24 510-512 [DOI] [PubMed] [Google Scholar]
- 4.Nallagatla, S. R., Hwang, J., Toroney, R., Zheng, X., Cameron, C. E., and Bevilacqua, P. C. (2007) Science 318 1455-1458 [DOI] [PubMed] [Google Scholar]
- 5.Saito, T., and Gale, M., Jr. (2007) Curr. Opin. Immunol. 19 17-23 [DOI] [PubMed] [Google Scholar]
- 6.Eisenacher, K., Steinberg, C., Reindl, W., and Krug, A. (2008) Immunobiology 212 701-714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underhill, D. M. (2004) Curr. Opin. Immunol. 16 483-487 [DOI] [PubMed] [Google Scholar]
- 8.Gaspari, A. A. (2006) J. Am. Acad. Dermatol. 54 S67-S80 [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi, T., Takaesu, G., and Yoshimura, A. (2006) Nat. Immunol. 7 123-124 [DOI] [PubMed] [Google Scholar]
- 10.Komuro, A., and Horvath, C. M. (2006) J. Virol. 80 12332-12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneyama, M., Kikuchi, M., Matsumoto, K., Imaizumi, T., Miyagishi, M., Taira, K., Foy, E., Loo, Y. M., Gale, M., Akira, S., Yonehara, S., Kato, A., and Fujita, T. (2005) J. Immunol. 175 2851-2858 [DOI] [PubMed] [Google Scholar]
- 12.Lee, M. S., and Kim, Y. J. (2007) Mol. Cells 23 1-10 [PubMed] [Google Scholar]
- 13.Pichlmair, A., Schulz, O., Tan, C. P., Naslund, T. I., Liljestrom, P., Weber, F., and Sousa, C. R. E. (2006) Science 314 997-1001 [DOI] [PubMed] [Google Scholar]
- 14.Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K. K., Schlee, M., Endres, S., and Hartmann, G. (2006) Science 314 994-997 [DOI] [PubMed] [Google Scholar]
- 15.Cui, S., Eisenacher, K., Kirchhofer, A., Brzozka, K., Lammens, A., Lammens, K., Fujita, T., Conzelmann, K. K., Krug, A., and Hopfner, K. P. (2008) Mol. Cell 29 169-179 [DOI] [PubMed] [Google Scholar]
- 16.Takahasi, K., Yoneyama, M., Nishihori, T., Hirai, R., Kumeta, H., Narita, R., Gale, M., Jr., Inagaki, F., and Fujita, T. (2008) Mol. Cell 29 428-440 [DOI] [PubMed] [Google Scholar]
- 17.Lee, M. S., and Min, Y. J. (2007) Annu. Rev. Biochem. 76 447-480 [DOI] [PubMed] [Google Scholar]
- 18.Johnson, C. L., and Gale, M. (2006) Trends Immunol. 27 1-4 [DOI] [PubMed] [Google Scholar]
- 19.Saito, T., Hirai, R., Loo, Y. M., Owen, D., Johnson, C. L., Sinha, S. C., Akira, S., Fujita, T., and Gale, M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 582-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothenfusser, S., Goutagny, N., DiPerna, G., Gong, M., Monks, B. G., Schoenemeyer, A., Yamamoto, M., Akira, S., and Fitzgerald, K. A. (2005) J. Immunol. 175 5260-5268 [DOI] [PubMed] [Google Scholar]
- 21.Vilasco, M., Larrea, E., Vitour, D., Dabo, S., Breiman, A., Regnault, B., Riezu, J. I., Eid, P., Prieto, J., and Meurs, E. F. (2006) Hepatology 44 1635-1647 [DOI] [PubMed] [Google Scholar]
- 22.Vitour, D., and Meurs, E. F. (2007) Science's STKE 2007 pe20. [DOI] [PubMed] [Google Scholar]
- 23.Venkataraman, T., Valdes, M., Elsby, R., Kakuta, S., Caceres, G., Saijo, S., Iwakura, Y., and Barber, G. N. (2007) J. Immunol. 178 6444-6455 [DOI] [PubMed] [Google Scholar]
- 24.Nishino, T., Komori, K., Tsuchiya, D., Ishino, Y., and Morikawa, K. (2005) Structure 13 143-153 [DOI] [PubMed] [Google Scholar]
- 25.Kao, C. C., Yang, X., Kline, A., Wang, Q. M., Barket, D., and Heinz, B. A. (2000) J. Virol. 74 11121-11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludtke, S. J., Baldwin, P. R., and Chiu, W. (1999) J. Struct. Biol. 128 82-97 [DOI] [PubMed] [Google Scholar]
- 27.Sun, J., DuFort, C., Daniel, M. C., Murali, A., Chen, C., Gopinath, K., Stein, B., De, M., Rotello, V. M., Holzenburg, A., Kao, C. C., and Dragnea, B. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1354-1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004) J. Comput. Chem. 25 1605-1612 [DOI] [PubMed] [Google Scholar]
- 29.Ranjith-Kumar, C. T., Miller, W., Sun, J., Xiong, J., Santos, J., Yarbrough, I., Lamb, R. J., Mills, J., Duffy, K. E., Hoose, S., Cunningham, M., Holzenburg, A., Mbow, M. L., Sarisky, R. T., and Kao, C. C. (2007) J. Biol. Chem. 282 17696-17705 [DOI] [PubMed] [Google Scholar]
- 30.Rixon, F. J., and Chiu, W. (2003) Adv. Protein Chem. 64 379-408 [DOI] [PubMed] [Google Scholar]
- 31.Zhou, Z. H., and Chiu, W. (2003) Adv. Protein Chem. 64 93-124 [DOI] [PubMed] [Google Scholar]
- 32.Caruthers, J. M., and McKay, D. B. (2002) Curr. Opin. Struct. Biol. 12 123-133 [DOI] [PubMed] [Google Scholar]
- 33.Cordin, O., Banroques, J., Tanner, N. K., and Linder, P. (2006) Gene (Amst.) 367 17-37 [DOI] [PubMed] [Google Scholar]
- 34.Kossen, K., Karginov, F. V., and Uhlenbeck, O. C. (2002) J. Mol. Biol. 324 625-636 [DOI] [PubMed] [Google Scholar]
- 35.Park, H. H., Logette, E., Raunser, S., Cuenin, S., Walz, T., Tschopp, J., and Wu, H. (2007) Cell 128 533-546 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.