Abstract
c-Abl is a ubiquitously expressed protein tyrosine kinase activated by DNA damage and implicated in two responses: cell cycle arrest and apoptosis. The downstream pathways by which c-Abl induces these responses remain unclear. We examined the effect of overexpression of c-Abl on the activation of mitogen-activated protein kinase pathways and found that overexpression of c-Abl selectively stimulated p38, while having no effect on c-Jun N-terminal kinase or on extracellular signal-regulated kinase. c-Abl-induced p38 activation was primarily mediated by mitogen-activated protein kinase kinase (MKK)6. A C-terminal truncation mutant of c-Abl showed no activity for stimulating p38 and MKK6, while a kinase-deficient c-Abl mutant still retained a residual activity. We tested different forms of c-Abl for their ability to induce apoptosis and found that apoptosis induction correlated with the activation of the MKK6-p38 kinase pathway. Importantly, dominant-negative MKK6, but not dominant-negative MKK3 or p38, blocked c-Abl-induced apoptosis. Because overexpression of p38 blocks cell cycle G1/S transition, we also tested whether the MKK6-p38 pathway is required for c-Abl-induced cell cycle arrest, and we found that neither MKK6 nor p38 dominant-negative mutants could relieve c-Abl-induced cell cycle arrest. Finally, DNA damage-induced MKK6 and p38 activation was diminished in c-Abl null fibroblasts. Our study suggests that c-Abl is required for DNA damage-induced MKK6 and p38 activation, and that activation of MKK6 by c-Abl is required for c-Abl-induced apoptosis but not c-Abl-induced cell cycle arrest.
c-Abl is a ubiquitously expressed protein tyrosine kinase that localizes in both the nucleus and the cytoplasm (1, 2). Although the transforming activity of Abl oncoproteins, v-Abl and Bcr-Abl, has been well established, the physiological function of c-Abl is not clear (3). The N-terminal region of c-Abl is similar to Src and includes Src homology regions 3 and 2 (SH3, SH2) and a kinase domain. However, unlike Src family kinases, c-Abl has a unique large C-terminal region. Multiple motifs have been defined on this C-terminal region, including nuclear localization signals (4), a nuclear exporting sequence (5), a DNA binding domain (6), an actin binding domain (2, 7), proline-rich regions serving as binding sites for proteins like Abi (8, 9), and a p53 binding region (10). Mutant mice null for c-Abl and mutant mice with the C terminus of c-Abl deleted exhibit the same pleiotropic defects, including high neonatal death rate, defects in T cell and B cell development, reduced fertility, and developmental abnormalities in the spleen and bone (11, 12).
c-Abl tyrosine kinase activity is tightly regulated in vivo. c-Abl kinase activity can be triggered by stimuli such as DNA-damaging agents (13, 14) and integrin-mediated cell adhesion (15). The activation of c-Abl by ionizing irradiation is mediated by ataxia–telangiectasia mutated (ATM), presumably by direct phosphorylation of Ser-465 in the c-Abl kinase domain by ATM (16, 17). An important effect of c-Abl activation is cell cycle arrest: overexpression of c-Abl blocks cell cycle G1/S transition (18), and cells with compromised c-Abl function have deregulated cell cycle (18, 19). c-Abl-induced cell cycle arrest requires p53, and possibly Rb, but not p21Cip (4, 10, 20). DNA damage also induces G1 cell cycle arrest, but whether c-Abl is required for DNA damage-induced G1 arrest is not clear (14, 16, 20). Another effect of c-Abl activation is induction of apoptosis. Overexpression of c-Abl induces apoptosis, and c-Abl is apparently required for DNA damage-induced apoptosis: MCF-7 cells harboring dominant-negative c-Abl and Abl−/− fibroblasts are relatively more resistant to DNA damage-induced apoptosis (21, 22).
The stress-activated kinase pathways have been implicated in mediating stress-induced apoptosis. Upstream activators of c-Jun N-terminal kinase (JNK) and p38, such as mitogen-activated protein kinase kinase kinase 1 (MEKK1) (23), mitogen-activated protein kinase kinase (MKK)6 (24, 25), apoptosis signal-regulating kinase 1 (ASK1) (26), and MAP three kinase 1 (MTK1) (27), induce apoptosis upon overexpression. Whether c-Abl induces stress-activated kinases or whether stress-activated kinases are important in c-Abl-induced cell cycle arrest and apoptosis is unclear. It was reported that overexpression of c-Abl could activate JNK and that cells deficient in c-Abl failed to activate JNK upon exposure to ionizing irradiation and alkylating DNA-damaging agents (13, 28); however, conflicting results from other labs render the issue controversial (14).
Although c-Abl has been implicated in DNA damage-induced apoptosis, the pathways downstream of c-Abl leading to apoptosis are not clear. In this study, we examined the role of the stress-activated kinase pathways in c-Abl-induced apoptosis and cell cycle arrest. Our study demonstrates that c-Abl is required for DNA damage-induced MKK6 and p38 activation, and that activation of MKK6 by c-Abl is required for c-Abl-induced apoptosis.
Materials and Methods
Cell Culture and Abs.
293 cells, COS7 cells, and NIH 3T3 cells were grown in DMEM supplemented with 10% FBS or bovine calf serum. Early passage abl+/+ and abl−/− mouse embryonic fibroblasts (MEFs) were generated from littermates of abl+/+ and abl−/− embryos and maintained in DMEM supplemented with 10% FBS. Anti-phospho-p38 and anti-phospho-MAPK Abs were from Promega; anti-phospho-JNK Abs were from New England Biolabs; anti-p38, anti-JNK, anti-extracellular signal-regulated kinase (ERK)2, and anti-Abl (K-12) Abs were from Santa Cruz Biotechnology; anti-Abl (8E9) Abs were from PharMingen; and anti-hemagglutinin epitope (HA) Abs were from Roche Molecular Biochemicals. Anti-MKK6 Abs were gifts from Eisuke Nishida (Kyoto University, Kyoto, Japan) (29).
Plasmids.
Murine type IV wild-type c-Abl, kinase deficient c-Abl, and c-Abl C-terminal truncation mutant (encoding amino acids 1–520) were all cloned into the mammalian expression vector pMT21. ERK2HA, p38HA, and JNKHA expression vectors and the glutathione S-transferase (GST)-ATF2 bacterial expression construct were gifts from Audrey Minden (Columbia University, New York) (30). MKK6HA, MKK3HA, MKK6(A), MKK3(A), MKK6(E), and p38(AF) mammalian expression vectors (all cloned in pCDNA3), and the GST-p38(M) bacteria expression vector were gifts from Jiahuai Han (The Scripps Research Institute, La Jolla, CA) (24). Dominant-negative JNKK1 was a gift from Anning Lin (University of Alabama, Birmingham, AL) (31). c-Jun mammalian expression construct was a gift from Tom Curran (St. Jude Children’s Research Hospital, Memphis, TN) (32). HPV16 E6 mammalian expression vector was a gift of Karl Munger (Harvard Medical School, Boston) (33). The GST-Jun-(1–109) bacterial expression vector was generated by cloning c-Jun cDNA in-frame into pGEX2TK (Amersham Pharmacia).
Immunoprecipitation and Immunoblot Analysis.
293 cells were transfected by the standard calcium phosphate method. COS7 cells were transfected by the DEAE-dextran method. Cells were lysed with RIPA buffer [10 mM sodium phosphate (pH 7.4)/100 mM NaCl/1% Triton X-100/0.5% sodium deoxycholate/0.1% SDS/10 mM NaF/10 mM β-glycerophosphate/protease inhibitors]. Immunoprecipitation and immunoblot analysis were performed as previously described (34).
In Vitro Kinase Assay.
Immunoprecipitation and in vitro kinase assays were performed as previously described (35, 36). GST-ATF2 was used as the substrate for p38, GST-Jun-(1–109) was used as the substrate for JNK, and GST-p38(M) was used as the substrate for MKK3 and MKK6.
Apoptosis Analysis.
NIH 3T3 cells were transfected with Lipofectamine reagents according the protocols provided by the manufacturer (GIBCO/BRL). Cells were harvested 42 hr after the start of transfection, and stained with rhodamine-conjugated anti-CD20 Abs (Dako). CD20-positive cells were sorted by fluorescence-activated cell sorting and centrifuged to glass slides with a Cytospin centrifuge. Cytospin preparations were subjected to terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) assay with the in Situ Cell Death Detection Kit (Roche Molecular Biochemicals). Slides were examined under a fluorescence microscope (Nikon). For each sample, >500 CD20-positive cells were scored for TUNEL staining.
Cell Cycle Arrest Assays.
NIH 3T3 cells were transfected with 0.2 μg of SV-β-galactosidase reporter construct, 1 μg of pMT21-Abl (or its derivatives), 1 μg of pCDNA (or other expression constructs), along with Lipofectamine reagents (GIBCO/BRL). Twenty-four hours after transfection, 5 bromo 2-deoxyuridine (BrdUrd) at 20 μM was added to the medium. Eighteen hours after the addition of BrdUrd, cells were fixed, and BrdUrd incorporation was detected with BrdUrd Labeling and Detection Kit I (Roche Molecular Biochemicals). After BrdUrd staining, cells were stained for β-galactosidase expression with rabbit polyclonal anti-β-galactosidase Abs (Cappel) and rhodamine-conjugated donkey anti-rabbit IgG Abs (Jackson ImmunoResearch). Coverslips were examined with an immunofluorescence microscope (Nikon). For each coverslip, >300 β-galactosidase-positive cells were scored for BrdUrd incorporation.
Results
Differential Activation of Mitogen-Activated Protein (MAP) Kinase Pathways by Overexpression of c-Abl.
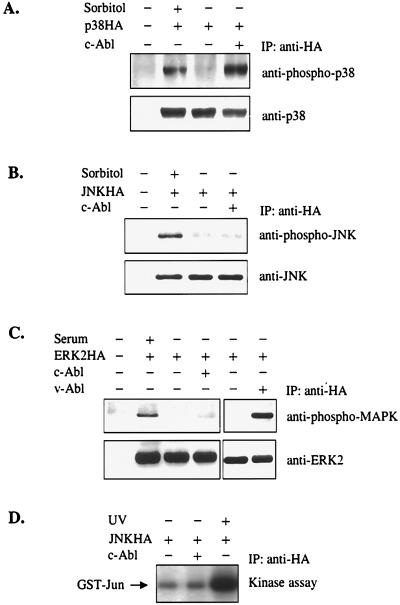
To determine the effect of the overexpression of c-Abl on the activation of MAP kinase pathways, a transient transfection strategy was employed. 293 cells were cotransfected with constructs expressing c-Abl and one of three HA-tagged MAP kinases: p38HA, JNKHA, or ERK2HA. The activation of MAP kinases was determined by immunoprecipitation and immunoblotting with phosphospecific Abs that recognize only the phosphorylated and active form of MAP kinases (Fig. 1). ERK2 was not activated by overexpression of c-Abl, although it was activated by serum treatment and by coexpression of v-Abl (Fig. 1C). This finding is consistent with the notion that v-Abl, but not c-Abl, is able to induce the activation of the Ras pathway. Furthermore, no activation of JNK by c-Abl was detected, although JNK was clearly activated by osmotic shock (Fig. 1B). Significantly, overexpression of c-Abl strongly activated p38 (Fig. 1A), contrasting with its small effect on JNK. To test for JNK activation in more detail, the same experiment was also performed in COS7 cells, which support higher levels of JNK and c-Abl expression. We consistently failed to detect the activation of JNK by c-Abl when we used anti-phospho-JNK Abs (data not shown). JNK activation in COS7 cells was further measured by an in vitro kinase assay, and no activation of JNK by c-Abl was detected (Fig. 1D). The JNK activity was also tested at different time points after transfection, and no JNK activation by c-Abl was detected (data not shown). Our data are consistent with a previous report (14) that overexpression of c-Abl does not seem to activate the JNK pathway.
Figure 1.
Overexpression of c-Abl selectively activates p38. 293 cells were transfected with 2 μg of p38HA (A), JNKHA (B), or ERK2HA (C) expression constructs, together with 6 μg of pMT21 or pMT21Abl. MAP kinases were immunoprecipitated (IP) with anti-HA Abs. The activation of MAP kinases was determined by immunoblot analysis with anti-phospho-p38 (A), anti-phospho-JNK (B), or anti-phospho-MAPK (C) Abs (Upper). Membranes were stripped and reblotted with anti-p38 (A), anti-JNK (B), or anti-ERK2 (C) Abs to show the equal loading (Lower). As a control for p38 and JNK activation, cells were also treated with 500 mM sorbitol for 10 min just before collecting cells (A and B). For detection of ERK2 activation, cells were starved with DMEM containing 0.2% FBS for 28 hr before cell collection. As positive controls, cells were either cotransfected with pGD-v-Abl or restimulated with 20% FBS for 10 min just before cell harvesting (C). (D) COS7 cells were cotransfected with indicated plasmids. JNK was immunoprecipitated and subjected to an in vitro kinase assay with GST-Jun-(1–109) as substrate. As a positive control, cells were treated with 40 J/m2 of UV light 1 hr before cell harvesting.
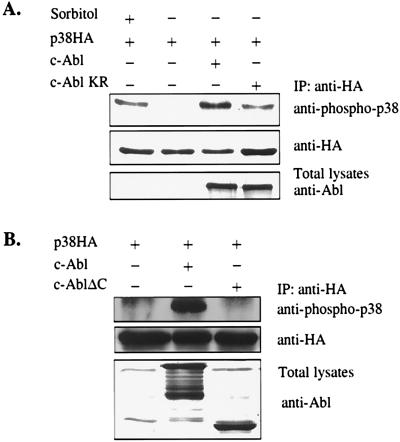
To determine the structural requirements of c-Abl for activating p38, a c-Abl kinase-deficient mutant and a c-Abl C-terminal truncation mutant were tested for their abilities to activate p38. The c-Abl C-terminal truncation mutant retains full kinase activity as judged by an in vitro kinase assay (data not shown). Surprisingly, the c-Abl kinase-deficient mutant retained some residual capacity for p38 activation (Fig. 2A), whereas the c-Abl C-terminal truncation mutant completely lacked this capability (Fig. 2B). These experiments suggest not only that c-Abl kinase activity is required for the full activation of p38, but also that the C-terminal region plays a critical role and is absolutely required for any detectable activation.
Figure 2.
The kinase activity and the C-terminal region of c-Abl are required for c-Abl-induced p38 activation. 293 cells were transfected with indicated plasmids. The experiments were performed as described in the legend of Fig. 1.
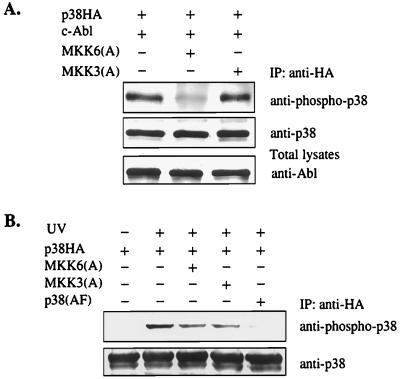
The Activation of p38 by c-Abl Is Primarily Mediated by MKK6.
p38 is specifically activated by two immediate upstream regulators, MKK3 and MKK6 (29, 37, 38). MKK3 and MKK6, like other MAP kinase family members, are activated by the dual phosphorylation on two Ser/Thr residues. Substitution of these phosphorylation sites with Ala or Glu results in a dominant-negative or a constitutively active form, respectively (24). To determine which of these two kinases is responsible for the c-Abl-induced activation of p38, MKK3(A) and MKK6(A) were included in the cotransfection assays. Strikingly, c-Abl-induced p38 MAP kinase activation was inhibited only by MKK6(A), not by MKK3(A) (Fig. 3A), suggesting that MKK6 mediates c-Abl-induced p38 activation. The effect of dominant-negative MKK6 and MKK3 on UV-induced p38 activation was also examined; both mutants partially inhibited UV-induced p38 activation, suggesting that both mutants are functional (Fig. 3B). p38 is activated by dual phosphorylation on Thr and Tyr, and a p38 mutant with these two residues changed to Ala and Phe, respectively, p38(AF), functions as a dominant negative for p38 (24). As expected, overexpression p38(AF) blocked UV-induced p38 activation (Fig. 3B).
Figure 3.
MKK6 is required for c-Abl-induced p38 activation. (A) MKK6 is required for c-Abl-induced p38 activation. (B) Both MKK6 and MKK3 are required for UV-induced p38 activation. 293 cells were transfected with indicated plasmids. The experiments were performed as described in the legend of Fig. 1. (B) Cells were treated with 40 J/m2 of UV light 1 hr before cell harvesting.
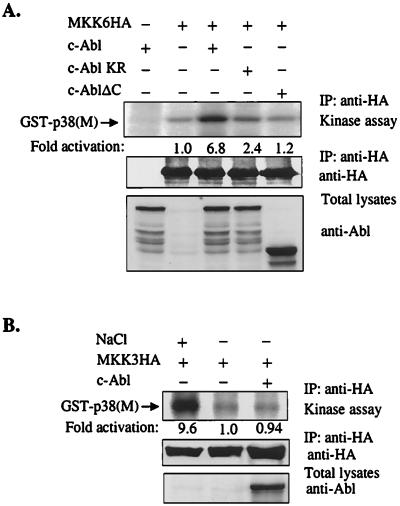
To directly assess the stimulation of MKK6 by c-Abl, cotransfection experiments were performed. HA-tagged MKK6 was immunoprecipitated with anti-HA Abs and subsequently examined for its ability to phosphorylate p38(M) by an in vitro kinase assay. c-Abl potently activated MKK6, while a c-Abl kinase-deficient mutant only slightly activated MKK6, and the c-Abl C-terminal truncation mutant failed to activate MKK6 (Fig. 4A). The abilities of different forms of c-Abl to activate MKK6 correlated with their abilities to activate p38. Using the same method, we also found that MKK3 was not activated by overexpression of c-Abl, although MKK3 can be potently activated by treatment with 700 mM NaCl (Fig. 4B). These data support the idea that MKK6, but not MKK3, mediates c-Abl-dependent p38 activation.
Figure 4.
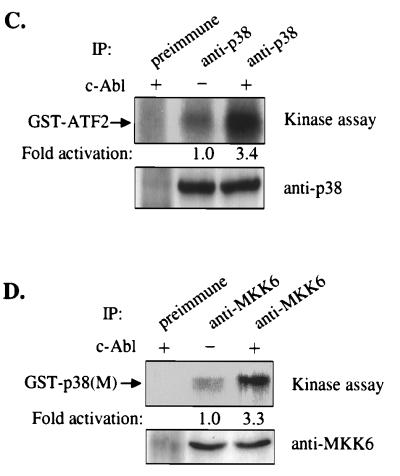
(A and B) Overexpression of c-Abl activates MKK6, but not MKK3. 293 cells were cotransfected with of MKK6HA (A) or MKK3HA (B) expression construct together with indicated c-Abl expression constructs. MKK6 or MKK3 was immunoprecipitated with anti-HA Abs. Immunoprecipitates were subjected to an in vitro kinase assay with GST-p38(M) as substrate. The reaction products were separated on a SDS/10% polyacrylamide gel, and substrate phosphorylation was detected by autoradiography and quantitated by phosphorimager analysis (Top). The expression of MKK6HA and MKK3HA was determined by blotting with anti-HA Abs (Middle). The expression of c-Abl was determined by blotting with anti-Abl (8E9) Abs (Bottom). (B) As a positive control, cells were treated with 700 mM NaCl for 30 min just before cell harvesting. (C and D) Overexpression of c-Abl activates endogenous p38 and MKK6. 293 cells were transfected with c-Abl expression construct. Endogenous p38 and MKK6 were immunoprecipitated with anti-p38 (C) and anti-MKK6 (D) Abs and subjected to an in vitro kinase assay with GST-ATF2 (C) or GST-p38(M) (D) as substrate. Substrate phosphorylation was detected by autoradiography and quantitated by phosphorimager analysis (Top). Immunoprecipitates were immunoblotted with anti-p38 (C) or anti-MKK6 Abs (D) (Bottom).
Next, we examined the effect of overexpression of c-Abl on the endogenous p38 and MKK6 in 293 cells by an in vitro kinase assay. Overexpression of c-Abl significantly activated both endogenous p38 and MKK6 (Fig. 4 C and D).
MKK6 Is Required for c-Abl-Induced Apoptosis.
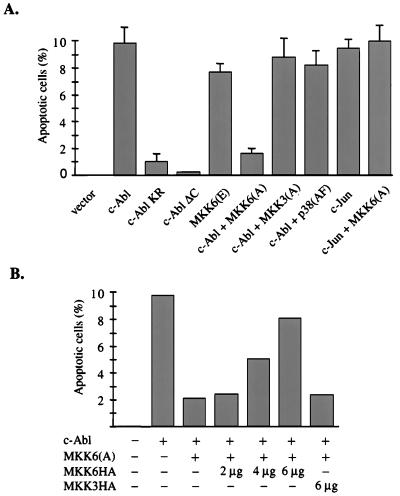
MKK6 plays a central role in Fas-mediated apoptosis, and expression of a constitutively active form of MKK6, MKK6(E), dramatically induces apoptosis in Jurkat cells (24, 25). Because c-Abl potently activated the MKK6-p38 pathway, we tested the possibility that MKK6 mediated c-Abl-induced apoptosis. NIH 3T3 cells were cotransfected with c-Abl and CD20 expression constructs. CD20-positive cells were sorted and subjected to TUNEL staining to detect cells undergoing apoptosis. No apoptotic cells were detected in cells transfected with an empty vector, while about 10% of c-Abl-transfected cells were apoptotic in this setting (Fig. 5A). The Abl kinase-deficient mutant also induced a low level of apoptosis, much lower compared with wild-type c-Abl, but clearly above the background (Fig. 5A). This finding is consistent with a recent report that high expression of the c-Abl kinase-deficient mutant is toxic to cells (39). The c-Abl C-terminal truncation mutant failed to induce apoptosis (Fig. 5A), indicating a critical role of the c-Abl C-terminal region in c-Abl-induced apoptosis.
Figure 5.
MKK6 is required for c-Abl-induced apoptosis. NIH 3T3 cells were transfected with a CD20 reporter construct, together with the indicated plasmids. CD20-positive cells were subjected to the TUNEL assay. The percentage of TUNEL-positive cells in CD20-positive cells is presented. (A) Cells were transfected with 6 μg of pMT21-Abl (or its derivatives) or c-Jun, plus 6 μg of pCDNA3, MKK6(A), MKK3(A), or p38(AF). The result represents an average of three independent experiments. (B) Cells were transfected with 4 μg of pMT21-Abl, 4 μg of MKK6(A), and various amount of MKK6HA or MKK3HA. The result represents an average of two independent experiments. All transfections included 0.6 μg of pCMV-CD20 and were balanced with the appropriate empty vector plasmids.
The abilities of different forms of c-Abl to induce apoptosis correlated well with their abilities to induce the MKK6-p38 pathway, and expression of MKK6(E) induced apoptosis in our setting. Therefore, we tested the effects of the dominant-negative forms of MKK6 and p38 on c-Abl-induced apoptosis. Cells cotransfected with c-Abl and MKK6(A) showed much less apoptosis than c-Abl alone. In contrast, MKK3(A) did not have any significant effect on c-Abl-induced apoptosis (Fig. 5A). We also found that a dominant-negative JNKK1 (MKK4) mutant did not affect c-Abl-induced apoptosis (data not shown).
Overexpression of c-Jun has been shown to induce apoptosis in NIH 3T3 cells (40). The coexpression of MKK6(A) with c-Jun had no effect on c-Jun-induced apoptosis (Fig. 5A). Further, MKK6(A) did not affect serum deprivation-induced apoptosis (data not shown). These data demonstrate that MKK6(A) does not have a general anti-apoptotic activity. To establish the specificity of MKK6(A) further, NIH 3T3 cells were transfected with a fixed amount of DNA encoding c-Abl and MKK6(A), together with an increasing amount of DNA encoding wild-type MKK6HA. Increasing levels of MKK6HA gradually relieved the inhibitory effect of MKK6(A) on c-Abl-induced apoptosis, whereas coexpression of even the highest level of MKK3HA did not affect it (Fig. 5B). Overexpression of wild-type MKK6HA or MKK3HA alone did not induce apoptosis (data not shown). These data suggest that MKK6(A) most likely inhibits c-Abl-induced apoptosis through interfering with endogenous MKK6.
We then tested whether p38 activation is required for c-Abl-induced apoptosis. To our surprise, the dominant-negative p38 mutant, p38(AF), had no obvious effect on c-Abl-induced apoptosis (Fig. 5A). Moreover, SB203580, a specific inhibitor for p38 (41), also had no obvious effect on c-Abl-induced apoptosis (data not shown). Taken together, our results suggest that c-Abl induces apoptosis through a MKK6-dependent, but p38 MAP kinase-independent pathway. This finding is similar to previous reports that the apoptosis induced by Fas receptor engagement is MKK6-dependent, but not p38-dependent (24, 25).
MKK6 and p38 Are Not Required for c-Abl-Induced Cell Cycle Arrest.
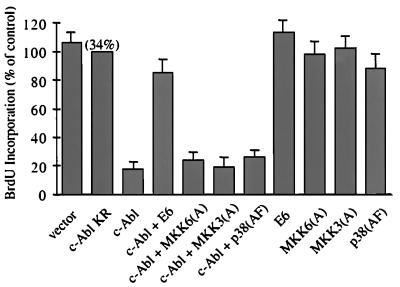
The p38 MAP kinase pathway has been implicated in cell cycle regulation. Microinjection of p38 and its upstream activators MKK3, MKK6, SEK1, or GCK inhibits cell cycle G1/S transition (42). Moreover, p38 suppresses the expression of cyclin D1 (43). Because c-Abl activates the p38 MAP kinase pathway, and both c-Abl and p38 induce cell cycle arrest, the potential role of the p38 MAP kinase pathway in c-Abl-induced cell cycle arrest was investigated. As expected, wild-type c-Abl, but not kinase-deficient c-Abl, caused a profound cell cycle arrest (Fig. 6). Consistent with the notion that c-Abl-induced cell cycle arrest is p53-dependent (4, 20), expression of HPV16 E6 protein that promotes p53 degradation (44) significantly relieved c-Abl-induced cell cycle arrest (Fig. 6). However, none of the dominant-negative MKK6, MKK3, or p38 had a significant effect on c-Abl-induced cell cycle arrest. The p38 inhibitor SB 203580 also had no obvious effect on c-Abl-induced cell cycle arrest (data not shown). These experiments suggest that the MKK6-p38 pathway is not required for c-Abl-induced cell cycle arrest.
Figure 6.
The MKK6-p38 pathway is not required for c-Abl-induced cell cycle arrest. NIH 3T3 cells were transfected with a β-galactosidase reporter construct, together with indicated plasmids. c-Abl-induced cell cycle arrest is measured by the percentage of transfected BrdUrd-positive cells relative to kinase-deficient c-Abl. The average of three experiments is shown. The absolute percentage of BrdUrd-positive cells in kinase-deficient c-Abl-transfected cells is shown in the parenthesis above the bar for this construct.
c-Abl Is Required for DNA Damage-Induced MKK6 and p38 Activation.
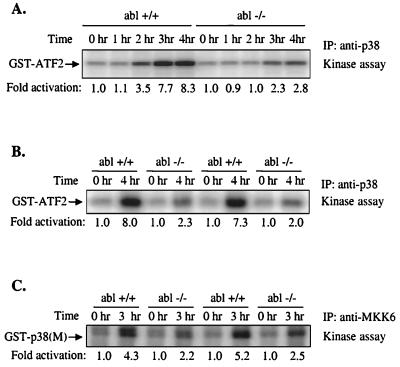
In the previous experiments, we showed that overexpression of c-Abl activates MKK6 and p38. However, it can be argued that this activation might not be physiologically relevant. To test whether the normal levels of c-Abl are required for MKK6 and p38 activation, we stimulated c-Abl kinase by treatment of wild-type and c-Abl null cells with the DNA-damaging agent cisplatin (cis-diamminedichloroplatinum or CDDP) and examined the induction of p38 and MKK6 (Fig. 7A). In wild-type MEFs, p38 activation could be detected after 2 hr and became very strong after 4 hr. In contrast, p38 activation was considerably weaker in abl−/− MEFs (Fig. 7A). Similar results were also obtained with other pairs of abl+/+ and abl−/− MEFs (Fig. 7B). On the other hand, c-Abl was not required for osmotic shock-induced p38 activation (data not shown). MKK6 was also activated by CDDP in wild-type cells, and CDDP-induced MKK6 activation was again weaker in abl−/− fibroblasts (Fig. 7C). These results are consistent with our previous finding that overexpression of c-Abl activates p38 through MKK6, and indicate that c-Abl is required for the full activation of MKK6 and p38 in response to DNA damage.
Figure 7.
c-Abl is required for full activation of p38 and MKK6 in response to DNA damage. Littermate abl+/+ and abl−/− MEFs were treated with 150 μM CDDP for the indicated times. Total cell lysates were immunoprecipitated with anti-p38 (A and B) or anti-MKK6 Abs (C) and subjected to an in vitro kinase assay with GST-ATF2 (A and B) or GST-p38(M) (C) as substrate. The amounts of immunoprecipitated p38 and MKK6 were normalized by blotting with anti-p38 or anti-MKK6 Abs (data not shown).
Discussion
The physiological function of c-Abl is still largely unknown. However, findings that c-Abl kinase activity can be activated by environmental stress and that overexpression of c-Abl induces both cell cycle arrest and apoptosis suggest that c-Abl might be causally involved in stress-regulated cell cycle arrest and apoptosis. In this study, we examined the role of stress-activated kinase cascades in c-Abl-mediated signal transduction. We find that overexpression of c-Abl selectively activates MKK6 and p38. Significantly, dominant-negative MKK6 blocks c-Abl-induced apoptosis, but not c-Abl-induced cell cycle arrest. Moreover, DNA damage-induced MKK6 and p38 activation are compromised in c-Abl-deficient cells. Our study suggests that c-Abl mediates MKK6 and p38 activation in response to DNA damage, and that the activation of MKK6 is required for c-Abl-induced apoptosis.
The stress-activated kinase cascades, which include the JNK and p38 pathways, are activated by different apoptotic stimuli. In most cases, JNK and p38 are simultaneously activated by these stimuli. In contrast, c-Abl preferentially activates p38, while it has little or no effect on JNK. The mechanism by which c-Abl preferentially targets p38 is not clear. Recently, a mammalian scaffold protein, JIP1, has been found that specifically interacts with MLK, MKK7, and JNK and ensures signaling specificity (45). Similar scaffold proteins that mediate the formation of a MKK6-p38 signaling complex might also exist. The C-terminal region of c-Abl might be responsible for targeting such a signaling complex.
The ability of different c-Abl alleles to induce apoptosis correlates with their ability to activate MKK6 and p38. Notably, the kinase-deficient c-Abl mutant, which weakly activates MKK6 and p38, also induces a very low level of cell death, consistent with a recent report that high level expression of kinase-deficient c-Abl is toxic (39). Significantly, dominant-negative MKK6, but not dominant-negative MKK3 or JNKK1, blocks c-Abl-induced apoptosis, suggesting that MKK6 is required for c-Abl-induced apoptosis. MKK6(A) exerts its effect on c-Abl-induced apoptosis most likely through negatively affecting endogenous MKK6, because this effect gradually diminished with coexpression of increasing amounts of MKK6HA. Intriguingly, the dominant-negative p38 mutant does not exert an obvious effect on c-Abl-induced cell death. The findings that the p38 inhibitors and p38(AF) fail to block Fas and MKK6-induced cell death in Jurkat cells, and that MKK3 does not induce significant cell death by itself, have led to the speculation that MKK6 induces apoptosis through a p38-independent pathway, and that p38 activation might just be an innocent bystander (24, 25). Our results on pathways required for c-Abl-induced apoptosis support this idea. We suggest that after being activated by c-Abl, MKK6 can initiate two signal transduction pathways, and through one pathway MKK6 induces apoptosis, and through the other pathway MKK6 activates p38 and modulates transcription (Fig. 8). However, we cannot exclude the possibility that some isoforms of p38, which our mutant fails to block, might play a role in the induction of cell death.
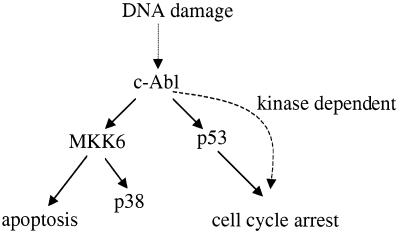
Figure 8.
The requirements of c-Abl-induced cell cycle arrest and apoptosis are different. See text.
c-Abl has been suggested as a negative regulator of the cell cycle (18): overexpression of c-Abl blocks the G1/S transition in a kinase-dependent manner, and interfering with normal c-Abl function with either the c-Abl dominant-negative mutant or antisense DNA deregulates the cell cycle (18, 19). The c-Abl-induced cell cycle arrest requires p53, and possibly Rb, but not p21Cip (4, 10, 20). Overexpression of c-Abl elevates the levels of p53 protein and its target, p21Cip. Intriguingly, the effect of c-Abl on p53 does not require the kinase activity of c-Abl (10, 20). Therefore, there must be an unidentified c-Abl kinase-dependent pathway responsible for c-Abl-induced cell cycle arrest, besides the kinase-independent p53 pathway (Fig. 8). The activation of the p38 pathway would seem to fit well as a candidate for this unidentified kinase-dependent pathway. Overexpression of p38, but not JNK, induces cell cycle arrest, possibly through inhibition of the expression of cyclin D (42, 43). However, the dominant-negative MKK6 and p38 mutants both failed to significantly relieve the c-Abl-induced G1/S block, suggesting that activation of the MKK6-p38 pathway is not the major cause of c-Abl-induced cell cycle arrest, although it might still play a minor role (Fig. 8).
The requirements of c-Abl-induced cell cycle arrest and apoptosis seem to be different (Fig. 8). Unlike c-Abl-induced cell cycle arrest, c-Abl-induced apoptosis does not require p53 and Rb, and does not require the p53 binding region on c-Abl (39). MKK6 activation seems to be required only for c-Abl-induced apoptosis. Although we show that activation of MKK6 by c-Abl plays an important role in c-Abl-induced apoptosis, the effect of c-Abl on other signal transduction pathways might also contribute to c-Abl-induced apoptosis to some extent. Indeed, c-Abl has been shown to associate with phosphatidylinositol 3-kinase (PI3-kinase) and inhibit its activity (46). Inhibition of the PI3-kinase-AKT pathway by c-Abl might also contribute to c-Abl-induced apoptosis.
c-Abl-induced MKK6 and p38 activation does not only occur after c-Abl overexpression; c-Abl is required for normal MKK6 and p38 activation in response to DNA damage. In accord with a previous report (47), we found that DNA damage-induced MKK6 and p38 activation is compromised in abl−/− MEFs. In agreement with the fact that c-Abl and MKK6 are proapoptotic kinases activated by DNA damage, cells with compromised c-Abl function are more resistant to DNA damage-induced apoptosis: MCF-7 cells expressing dominant-negative c-Abl, and c-Abl−/− fibroblasts, are less prone to apoptosis caused by DNA-damaging agents (21, 22). Our data, together with findings from other investigators, suggest that c-Abl might determine the cell fate in response to DNA damage, and one way to do so is by stimulating MKK6 and MKK6-mediated apoptosis.
Acknowledgments
We thank Jiahuai Han, Audrey Minden, Eisuke Nishida, Anning Lin, and Karl Munger for providing reagents critical to this work. We also thank Paul Rothman, Srikumar Chellappan, Ron Prwyes, Carol Prives, and Mitchell Goldfarb for critical reading of the manuscript and helpful discussion. This work was supported in part by National Institutes of Health Grant PO1 CA75399. F.C. is supported by a Hoffmann–La Roche predoctoral fellowship, and S.P.G. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- MKK
mitogen-activated protein kinase kinase
- JNK
c-Jun N-terminal kinase
- ERK
extracellular signal-regulated kinase
- GST
glutathione S-transferase
- CDDP
cisplatin
- MEF
mouse embryonic fibroblast
- HA
hemagglutinin
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling
- IP
immunoprecipitated
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Van Etten R A, Jackson P, Baltimore D. Cell. 1989;58:669–678. doi: 10.1016/0092-8674(89)90102-5. [DOI] [PubMed] [Google Scholar]
- 2.Van Etten R A, Jackson P K, Baltimore D, Sanders M C, Matsudaira P T, Janmey P A. J Cell Biol. 1994;124:325–340. doi: 10.1083/jcb.124.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J Y. Curr Opin Genet Dev. 1993;3:35–43. doi: 10.1016/s0959-437x(05)80338-7. [DOI] [PubMed] [Google Scholar]
- 4.Wen S-T, Jackson P K, Van Etten R A. EMBO J. 1996;15:1583–1595. [PMC free article] [PubMed] [Google Scholar]
- 5.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A K, Wang J Y, Hope T J. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kipreos E T, Wang J Y. Science. 1992;256:382–385. doi: 10.1126/science.256.5055.382. [DOI] [PubMed] [Google Scholar]
- 7.McWhirter J R, Wang J Y. EMBO J. 1993;12:1533–1546. doi: 10.1002/j.1460-2075.1993.tb05797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Alin K, Goff S P. Genes Dev. 1995;9:2583–2597. doi: 10.1101/gad.9.21.2583. [DOI] [PubMed] [Google Scholar]
- 9.Dai Z, Pendergast A M. Genes Dev. 1995;9:2569–2582. doi: 10.1101/gad.9.21.2569. [DOI] [PubMed] [Google Scholar]
- 10.Goga A, Liu X, Hambuch T M, Senechal K, Major E, Bert A J, Witte O N, Sawyers C L. Oncogene. 1995;11:791–799. [PubMed] [Google Scholar]
- 11.Schwartzberg P L, Stall A M, Hardin J D, Bowdish K S, Humaran T, Boast S, Harbison M L, Robertson E J, Goff S P. Cell. 1991;65:1165–1175. doi: 10.1016/0092-8674(91)90012-n. [DOI] [PubMed] [Google Scholar]
- 12.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 13.Kharbanda S, Ren R, Pandey P, Shafman T D, Feller S M, Weichselbaum R R, Kufe D W. Nature (London) 1995;376:785–788. doi: 10.1038/376785a0. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z G, Baskaran R, Lea-Chou E T, Wood L D, Chen Y, Karin M, Wang J Y. Nature (London) 1996;384:273–276. doi: 10.1038/384273a0. [DOI] [PubMed] [Google Scholar]
- 15.Lewis J M, Baskaran R, Taagepera S, Schwartz M A, Wang J Y. Proc Natl Acad Sci USA. 1996;93:15174–15179. doi: 10.1073/pnas.93.26.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baskaran R, Wood L D, Whitaker L L, Canman C E, Morgan S E, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan M B, Wang J Y. Nature (London) 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 17.Shafman T, Khanna K K, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, et al. Nature (London) 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 18.Sawyers C L, McLaughlin J, Goga A, Havlik M, Witte O. Cell. 1994;77:121–131. doi: 10.1016/0092-8674(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 19.Daniel R, Cai Y, Wong P M, Chung S W. Oncogene. 1995;10:1607–1614. [PubMed] [Google Scholar]
- 20.Yuan Z M, Huang Y, Whang Y, Sawyers C, Weichselbaum R, Kharbanda S, Kufe D. Nature (London) 1996;382:272–274. doi: 10.1038/382272a0. [DOI] [PubMed] [Google Scholar]
- 21.Yuan Z M, Huang Y, Ishiko T, Kharbanda S, Weichselbaum R, Kufe D. Proc Natl Acad Sci USA. 1997;94:1437–1440. doi: 10.1073/pnas.94.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Yuan Z M, Ishiko T, Nakada S, Utsugisawa T, Kato T, Kharbanda S, Kufe D W. Oncogene. 1997;15:1947–1952. doi: 10.1038/sj.onc.1201376. [DOI] [PubMed] [Google Scholar]
- 23.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 24.Huang S, Jiang Y, Li Z, Nishida E, Mathias P, Lin S, Ulevitch R J, Nemerow G R, Han J. Immunity. 1997;6:739–749. doi: 10.1016/s1074-7613(00)80449-5. [DOI] [PubMed] [Google Scholar]
- 25.Juo P, Kuo C J, Reynolds S E, Konz R F, Raingeaud J, Davis R J, Biemann H P, Blenis J. Mol Cell Biol. 1997;17:24–35. doi: 10.1128/mcb.17.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazona K, Gotoh Y. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 27.Takekawa M, Saito H. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 28.Kharbanda S, Pandey P, Ren R, Mayer B, Zon L, Kufe D. J Biol Chem. 1995;270:30278–30281. doi: 10.1074/jbc.270.51.30278. [DOI] [PubMed] [Google Scholar]
- 29.Moriguchi T, Toyoshima F, Gotoh Y, Iwamatsu A, Irie K, Mori E, Kuroyanagi N, Hagiwara M, Matsumoto K, Nishida E. J Biol Chem. 1996;271:26981–26988. doi: 10.1074/jbc.271.43.26981. [DOI] [PubMed] [Google Scholar]
- 30.Minden A, Lin A, Claret F X, Abo A, Karin M. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 31.Lin A, Minden A, Martinetto H, Claret F X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 32.Sonnenberg J L, Rauscher F J, III, Morgan J I, Curran T. Science. 1989;246:1622–1625. doi: 10.1126/science.2512642. [DOI] [PubMed] [Google Scholar]
- 33.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorsch M, Fan P D, Danial N N, Rothman P B, Goff S P. J Exp Med. 1997;186:1947–1955. doi: 10.1084/jem.186.12.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raingeaud J, Gupta S, Rogers J S, Dickens M, Han J, Ulevitch R J, Davis R J. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 36.Jiang Y, Chen C, Li Z, Guo W, Gegner J A, Lin S, Han J. J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 37.Han J, Lee J D, Jiang Y, Li Z, Feng L, Ulevitch R J. J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 38.Derijard B, Raingeaud J, Barret T, Wu I H, Han J, Ulevitch R J, Davis R J. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 39.Theis S, Roemer K. Oncogene. 1998;17:557–564. doi: 10.1038/sj.onc.1201973. [DOI] [PubMed] [Google Scholar]
- 40.Bossy-Wetzel E, Bakiri L, Yaniv M. EMBO J. 1997;16:1695–1709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heys J R, Landvatter S W, et al. Nature (London) 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 42.Molnar A, Theodoras A M, Zon L I, Kyriakis J M. J Biol Chem. 1997;272:13229–13235. doi: 10.1074/jbc.272.20.13229. [DOI] [PubMed] [Google Scholar]
- 43.Lavoie J N, L’Allemain G, Brunet A, Muller R, Pouyssegur J. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 44.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 45.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 46.Yuan Z M, Utsugisawa T, Huang Y, Ishiko T, Nakada S, Kharbanda S, Weichselbaum R, Kufe D. J Biol Chem. 1997;272:23485–23488. doi: 10.1074/jbc.272.38.23485. [DOI] [PubMed] [Google Scholar]
- 47.Pandey P, Kaneki M, Weichselbaum R, Davis R J, Kufe D, Kharbanda S. J Biol Chem. 1996;271:23775–23779. doi: 10.1074/jbc.271.39.23775. [DOI] [PubMed] [Google Scholar]