Abstract
Murine leukemia viruses (MuLVs) induce leukemias and lymphomas in mice. We have used fluorescence-activated cell sorter analysis to determine the hematopoietic phenotypes of tumor cells induced by a number of MuLVs. Tumor cells induced by ecotropic Moloney, amphotropic 4070A, and 10A1 MuLVs and by two chimeric MuLVs, Mo(4070A) and Mo(10A1), were examined with antibodies to 13 lineage-specific cell surface markers found on myeloid cell, T-cell, and B-cell lineages. The chimeric Mo(4070A) and Mo(10A1) MuLVs, consisting of Moloney MuLV with the carboxy half of the Pol region and nearly all of the Env region of 4070A and 10A1, respectively, were constructed to examine the possible influence of these sequences on Moloney MuLV-induced tumor cell phenotypes. In some instances, these phenotypic analyses were supplemented by Southern blot analysis for lymphoid cell-specific genomic DNA rearrangements at the immunoglobulin heavy-chain, the T-cell receptor gamma, and the T-cell receptor beta loci. The results of our analysis showed that Moloney MuLV, 4070A, Mo(4070A), and Mo(10A1) induced mostly T-cell tumors. Moloney MuLV and Mo(4070A) induced a wide variety of T-cell phenotypes, ranging from immature to mature phenotypes, while 4070A induced mostly prothymocyte and double-negative (CD4- CD8-) T-cell tumors. The tumor phenotypes obtained with 10A1 and Mo(10A1) were each less variable than those obtained with the other MuLVs tested. 10A1 uniformly induced a tumor consisting of lineage marker-negative cells that lack lymphoid cell-specific DNA rearrangements and histologically appear to be early undifferentiated erythroid cell-like precursors. The Mo(10A1) chimera consistently induced an intermediate T-cell tumor. The chimeric constructions demonstrated that while 4070A 3' pol and env sequences apparently did not influence the observed tumor cell phenotypes, the 10A1 half of pol and env had a strong effect on the phenotypes induced by Mo(10A1) that resulted in a phenotypic consistency not seen with other viruses. This result implicates 10A1 env in an active role in the tumorigenic process.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Yancopoulos G. D., Blackwell T. K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984 Jun;3(6):1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asjö B., Skoog L., Palminger I., Wiener F., Isaak D., Cerny J., Fenyö E. M. Influence of genotype and the organ of origin on the subtype of T-cell in Moloney lymphomas induced by transfer of preleukemic cells from athymic and thymus-bearing mice. Cancer Res. 1985 Mar;45(3):1040–1045. [PubMed] [Google Scholar]
- Bassin R. H., Tuttle N., Fischinger P. J. Rapid cell culture assay technic for murine leukaemia viruses. Nature. 1971 Feb 19;229(5286):564–566. doi: 10.1038/229564b0. [DOI] [PubMed] [Google Scholar]
- Beck-Engeser G., Stocking C., Just U., Albritton L., Dexter M., Spooncer E., Ostertag W. Retroviral vectors related to the myeloproliferative sarcoma virus allow efficient expression in hematopoietic stem and precursor cell lines, but retroviral infection is reduced in more primitive cells. Hum Gene Ther. 1991 Spring;2(1):61–70. doi: 10.1089/hum.1991.2.1-61. [DOI] [PubMed] [Google Scholar]
- Boyer B., Gisselbrecht S., Debre P., McKenzie I., Levy J. P. Genetic control of sensitivity to Moloney leukemia virus in mice. IV. Phenotypic heterogeneity of the leukemic mice. J Immunol. 1980 Oct;125(4):1415–1420. [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Cloyd M. W., Linemeyer D. L., Lander M. R., Rands E., Lowy D. R. Cellular origin and role of mink cell focus-forming viruses in murine thymic lymphomas. Nature. 1982 Jan 7;295(5844):25–31. doi: 10.1038/295025a0. [DOI] [PubMed] [Google Scholar]
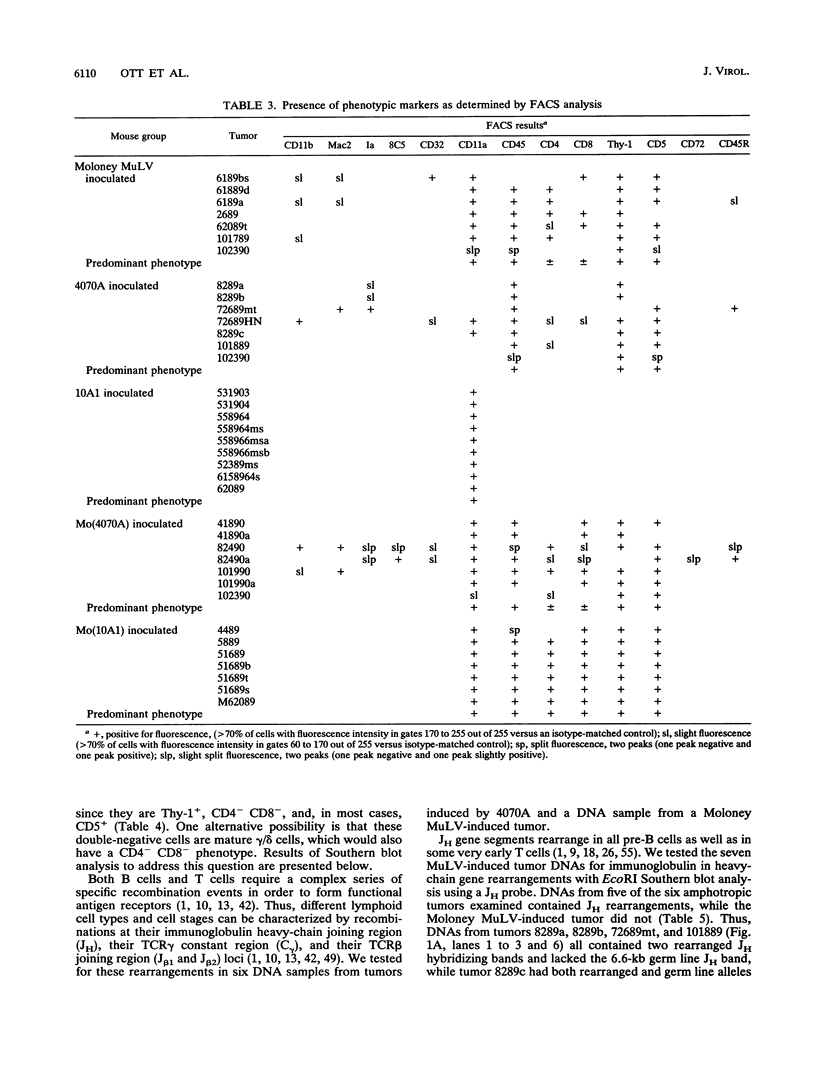
- Chattopadhyay S. K., Oliff A. I., Linemeyer D. L., Lander M. R., Lowy D. R. Genomes of murine leukemia viruses isolated from wild mice. J Virol. 1981 Sep;39(3):777–791. doi: 10.1128/jvi.39.3.777-791.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. M. T cell receptor gene diversity and selection. Annu Rev Biochem. 1990;59:475–496. doi: 10.1146/annurev.bi.59.070190.002355. [DOI] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J Virol. 1984 Nov;52(2):448–456. doi: 10.1128/jvi.52.2.448-456.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulari N. G. Murine leukemia viruses with recombinant env genes: a discussion of their role in leukemogenesis. Curr Top Microbiol Immunol. 1983;103:75–108. doi: 10.1007/978-3-642-68943-7_4. [DOI] [PubMed] [Google Scholar]
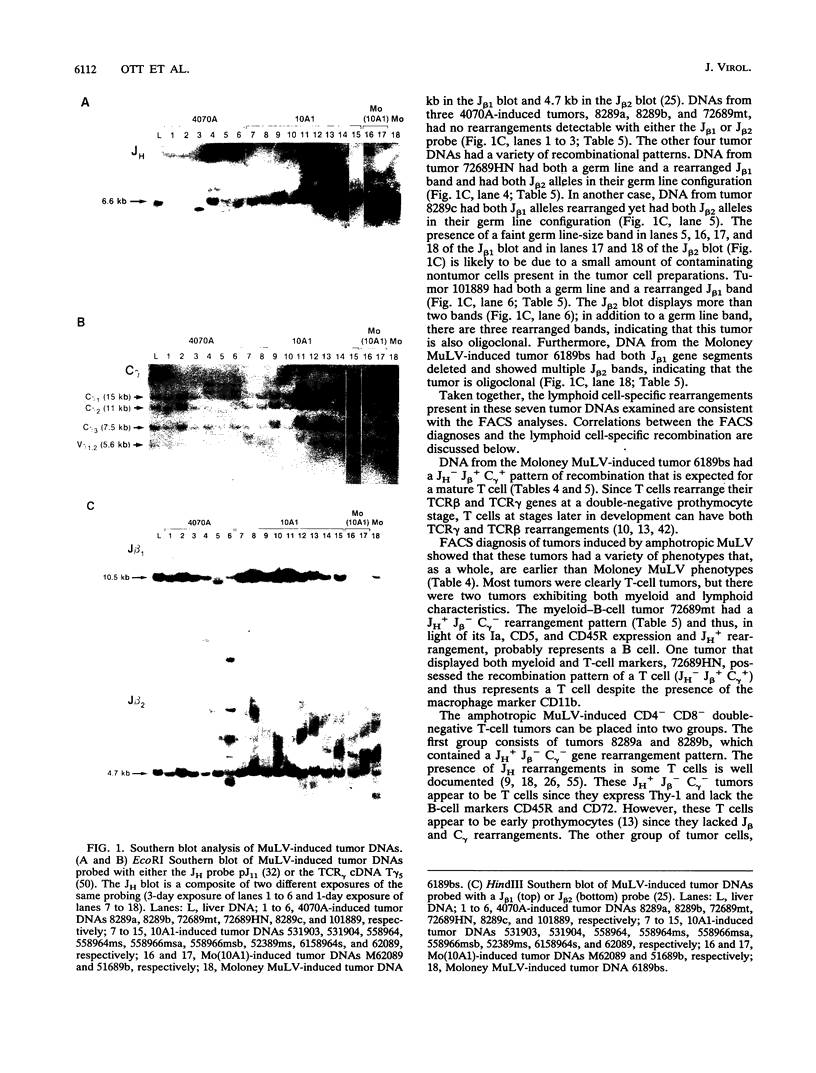
- Fowlkes B. J., Pardoll D. M. Molecular and cellular events of T cell development. Adv Immunol. 1989;44:207–264. doi: 10.1016/s0065-2776(08)60643-4. [DOI] [PubMed] [Google Scholar]
- Garman R. D., Doherty P. J., Raulet D. H. Diversity, rearrangement, and expression of murine T cell gamma genes. Cell. 1986 Jun 6;45(5):733–742. doi: 10.1016/0092-8674(86)90787-7. [DOI] [PubMed] [Google Scholar]
- Golemis E., Li Y., Fredrickson T. N., Hartley J. W., Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989 Jan;63(1):328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R. J., Henderson L. E., Hanser J. P., Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a "zinc finger-like" protein sequence. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. W., Wolford N. K., Old L. J., Rowe W. P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A. 1977 Feb;74(2):789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W., Perlmutter A. P., Gilbert W. Monoclonal AKR/J thymic leukemias contain multiple JH immunoglobulin gene rearrangements. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7433–7436. doi: 10.1073/pnas.80.24.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. At least four viral genes contribute to the leukemogenicity of murine retrovirus MCF 247 in AKR mice. J Virol. 1985 Jan;53(1):158–165. doi: 10.1128/jvi.53.1.158-165.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K. L., Langdon W. Y., Fredrickson T. N., Coffman R. L., Hoffman P. M., Hartley J. W., Morse H. C., 3rd Analysis of neoplasms induced by Cas-Br-M MuLV tumor extracts. J Immunol. 1986 Jul 15;137(2):679–688. [PubMed] [Google Scholar]
- Holmes K. L., Morse H. C., 3rd Murine hematopoietic cell surface antigen expression. Immunol Today. 1988 Nov;9(11):344–350. doi: 10.1016/0167-5699(88)91335-7. [DOI] [PubMed] [Google Scholar]
- Ishimoto A., Takimoto M., Adachi A., Kakuyama M., Kato S., Kakimi K., Fukuoka K., Ogiu T., Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987 Jun;61(6):1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg M., Goverman J., Haars R., Malissen M., Kraig E., Phillips L., Delovitch T., Suciu-Foca N., Hood L. Rearrangement and transcription of the beta-chain genes of the T-cell antigen receptor in different types of murine lymphocytes. Nature. 1985 Feb 21;313(6004):647–653. doi: 10.1038/313647a0. [DOI] [PubMed] [Google Scholar]
- Kurosawa Y., von Boehmer H., Haas W., Sakano H., Trauneker A., Tonegawa S. Identification of D segments of immunoglobulin heavy-chain genes and their rearrangement in T lymphocytes. Nature. 1981 Apr 16;290(5807):565–570. doi: 10.1038/290565a0. [DOI] [PubMed] [Google Scholar]
- Lazo P. A., Klein-Szanto A. J., Tsichlis P. N. T-cell lymphoma lines derived from rat thymomas induced by Moloney murine leukemia virus: phenotypic diversity and its implications. J Virol. 1990 Aug;64(8):3948–3959. doi: 10.1128/jvi.64.8.3948-3959.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. P., Baltimore D. Mechanism of leukemogenesis induced by mink cell focus-forming murine leukemia viruses. J Virol. 1991 May;65(5):2408–2414. doi: 10.1128/jvi.65.5.2408-2414.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. P., D'Andrea A. D., Lodish H. F., Baltimore D. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature. 1990 Feb 22;343(6260):762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- Li Q. X., Fan H. Combined infection by Moloney murine leukemia virus and a mink cell focus-forming virus recombinant induces cytopathic effects in fibroblasts or in long-term bone marrow cultures from preleukemic mice. J Virol. 1990 Aug;64(8):3701–3711. doi: 10.1128/jvi.64.8.3701-3711.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu K. B., Banerji J., Penncavage N. A., Lang R., Arnheim N. 5' flanking region of immunoglobulin heavy chain constant region genes displays length heterogeneity in germlines of inbred mouse strains. Cell. 1980 Nov;22(1 Pt 1):187–196. doi: 10.1016/0092-8674(80)90167-1. [DOI] [PubMed] [Google Scholar]
- Oliff A., Signorelli K., Collins L. The envelope gene and long terminal repeat sequences contribute to the pathogenic phenotype of helper-independent Friend viruses. J Virol. 1984 Sep;51(3):788–794. doi: 10.1128/jvi.51.3.788-794.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott D., Friedrich R., Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990 Feb;64(2):757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepersack L., Lee J. C., McEwan R., Ihle J. N. Phenotypic heterogeneity of Moloney leukemia virus-induced T cel lymphomas. J Immunol. 1980 Jan;124(1):279–285. [PubMed] [Google Scholar]
- Rasheed S., Pal B. K., Gardner M. B. Characterization of a highly oncogenic murine leukemia virus from wild mice. Int J Cancer. 1982 Mar 15;29(3):345–350. doi: 10.1002/ijc.2910290319. [DOI] [PubMed] [Google Scholar]
- Rein A. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology. 1982 Jul 15;120(1):251–257. doi: 10.1016/0042-6822(82)90024-1. [DOI] [PubMed] [Google Scholar]
- Rein A., Schultz A. Different recombinant murine leukemia viruses use different cell surface receptors. Virology. 1984 Jul 15;136(1):144–152. doi: 10.1016/0042-6822(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Ruscetti S. K., Janesch N. J., Chakraborti A., Sawyer S. T., Hankins W. D. Friend spleen focus-forming virus induces factor independence in an erythropoietin-dependent erythroleukemia cell line. J Virol. 1990 Mar;64(3):1057–1062. doi: 10.1128/jvi.64.3.1057-1062.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff R. D., Oliff A. The pathophysiology of murine retrovirus-induced leukemias. Crit Rev Oncol Hematol. 1986;5(3):257–323. doi: 10.1016/s1040-8428(86)80041-5. [DOI] [PubMed] [Google Scholar]
- Scollay R., Wilson A., D'Amico A., Kelly K., Egerton M., Pearse M., Wu L., Shortman K. Developmental status and reconstitution potential of subpopulations of murine thymocytes. Immunol Rev. 1988 Aug;104:81–120. doi: 10.1111/j.1600-065x.1988.tb00760.x. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Spira J., Asjö B., Cochran A., Shen F. W., Wiener F., Klein G. Chromosomal, histopathological and cell surface marker studies on Moloney virus induced lymphomas. Leuk Res. 1981;5(2):113–121. doi: 10.1016/0145-2126(81)90069-2. [DOI] [PubMed] [Google Scholar]
- Traunecker A., Kiefer M., Dembić Z., Steinmetz M., Karjalainen K. Rearrangements of T cell receptor loci can be found only rarely in B lymphoid cells. Eur J Immunol. 1986 Apr;16(4):430–434. doi: 10.1002/eji.1830160420. [DOI] [PubMed] [Google Scholar]
- Traunecker A., Oliveri F., Allen N., Karjalainen K. Normal T cell development is possible without 'functional' gamma chain genes. EMBO J. 1986 Jul;5(7):1589–1593. doi: 10.1002/j.1460-2075.1986.tb04400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Bear S. E. Infection by mink cell focus-forming viruses confers interleukin 2 (IL-2) independence to an IL-2-dependent rat T-cell lymphoma line. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4611–4615. doi: 10.1073/pnas.88.11.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Shepherd B. M., Bear S. E. Activation of the Mlvi-1/mis1/pvt-1 locus in Moloney murine leukemia virus-induced T-cell lymphomas. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5487–5491. doi: 10.1073/pnas.86.14.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C. C., Kan N., Dunn K. J., Papas T. S., Blair D. G. Properties of a murine retroviral recombinant of avian acute leukemia virus E26: a murine fibroblast assay for v-ets function. J Virol. 1989 Jan;63(1):205–215. doi: 10.1128/jvi.63.1.205-215.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züiga M. C., D'Eustachio P., Ruddle N. H. Immunoglobulin heavy chain gene rearrangement and transcription in murine T cell hybrids and T lymphomas. Proc Natl Acad Sci U S A. 1982 May;79(9):3015–3019. doi: 10.1073/pnas.79.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]