Abstract
Summary: ATP-binding cassette (ABC) systems are universally distributed among living organisms and function in many different aspects of bacterial physiology. ABC transporters are best known for their role in the import of essential nutrients and the export of toxic molecules, but they can also mediate the transport of many other physiological substrates. In a classical transport reaction, two highly conserved ATP-binding domains or subunits couple the binding/hydrolysis of ATP to the translocation of particular substrates across the membrane, through interactions with membrane-spanning domains of the transporter. Variations on this basic theme involve soluble ABC ATP-binding proteins that couple ATP hydrolysis to nontransport processes, such as DNA repair and gene expression regulation. Insights into the structure, function, and mechanism of action of bacterial ABC proteins are reported, based on phylogenetic comparisons as well as classic biochemical and genetic approaches. The availability of an increasing number of high-resolution structures has provided a valuable framework for interpretation of recent studies, and realistic models have been proposed to explain how these fascinating molecular machines use complex dynamic processes to fulfill their numerous biological functions. These advances are also important for elucidating the mechanism of action of eukaryotic ABC proteins, because functional defects in many of them are responsible for severe human inherited diseases.
INTRODUCTION
The ATP-binding cassette (ABC) systems constitute one of the largest superfamilies of paralogous sequences. All ABC systems share a highly conserved ATP-hydrolyzing domain or protein (the ABC; also referred to as a nucleotide-binding domain [NBD]) that is unequivocally characterized by three short sequence motifs (Fig. 1): these are the Walker A and Walker B motifs, indicative of the presence of a nucleotide-binding site, and the signature motif, unique to ABC proteins, located upstream of the Walker B motif (426). Other motifs diagnostic of ABC proteins are also indicated in Fig. 1. The biological significance of these motifs is discussed in Structure, Function, and Dynamics of the ABC. ABC systems are widespread among living organisms and have been detected in all genera of the three kingdoms of life, with remarkable conservation in the primary sequence of the cassette and in the organization of the constitutive domains or subunits (203, 420).
FIG. 1.
Conserved motifs in the ABC. Three characteristic motifs found in all ABC ATPases are represented by hatched red boxes. The Walker A motif and the Walker B motif form the nucleotide-binding fold of the P-loop ATPase family. The signature motif, also called the C loop, is unique to ABC proteins and also interacts with ATP. Other characteristic motifs, including the Q loop and the H loop (also called the switch region), contain just one highly conserved residue and are represented by hatched green boxes. These residues make contacts with the γ-phosphate of ATP. In the context of the ABC dimer, the D loop makes contacts with Walker motif A of the other monomer. Sequences between the Q loop and the signature constitute a helical domain, also referred to as a structurally diverse region (SDR), that contains residues important for the interaction of ABC proteins with their membrane partners.
ABC systems couple the energy of ATP hydrolysis to an impressively large variety of essential biological phenomena, comprising not only transmembrane (TM) transport, for which they are best known, but also several non-transport-related processes, such as translation elongation (62) and DNA repair (174). Although ABC systems deserve much attention because they are involved in severe human inherited diseases (107), they were first discovered and characterized in detail in prokaryotes, as early as the 1970s (13, 148, 238, 468). The most extensively analyzed systems were the high-affinity histidine and maltose uptake systems of Salmonella enterica serovar Typhimurium and Escherichia coli. Over 2 decades ago, after the completion of the nucleotide sequences encoding these transporters in the respective laboratories of Giovanna Ames and Maurice Hofnung, Hiroshi Nikaido and colleagues noticed that the two systems displayed a global similarity in the nature of their components and, moreover, that the primary sequences of MalK and HisP, the proteins suspected to energize these transporters, shared as much as 32% identity in amino acid residues when their sequences were aligned (171). Later, it was found that several bacterial proteins involved in uptake of nutrients, export of toxins, cell division, bacterial nodulation of plants, and DNA repair displayed the same similarity in their sequences (127, 196). This led to the notion that the conserved protein, which had been shown to bind ATP (198, 201), would probably energize the systems mentioned above by coupling the energy of ATP hydrolysis to transport. The latter was demonstrated with the maltose and histidine transporters by use of isolated membrane vesicles (105, 379) and purified transporters reconstituted into proteoliposomes (30, 98). The determination of the sequence of the first eukaryotic protein strongly similar to these bacterial transporters (the P-glycoprotein, involved in resistance of cancer cells to multiple drugs) (169, 179) demonstrated that these proteins were not restricted to prokaryotes. Two names, “traffic ATPases” (15) and the more accepted name “ABC transporters” (193, 218), were proposed for members of this new superfamily.
ABC systems can be divided into three main functional categories, as follows. Importers mediate the uptake of nutrients in prokaryotes. The nature of the substrates that are transported is very wide, including mono- and oligosaccharides, organic and inorganic ions, amino acids, peptides, iron-siderophores, metals, polyamine cations, opines, and vitamins. Exporters are involved in the secretion of various molecules, such as peptides, lipids, hydrophobic drugs, polysaccharides, and proteins, including toxins such as hemolysin. The third category of systems is apparently not involved in transport, with some members being involved in translation of mRNA and in DNA repair.
Despite the large, diverse population of substrates handled and the difference in the polarity of transport, importers and exporters share a common organization made of two hydrophobic membrane-spanning or integral membrane (IM) domains and two hydrophilic domains carrying the ABC peripherally associated with the IM domains on the cytosolic side of the membrane (26). In importers, these four domains are almost always independent polypeptide chains that come together to form a multimeric complex. In most exporters, including the E. coli hemolysin exporter HlyB, the N-terminal IM and the C-terminal ABC domains are fused as a single polypeptide chain (IM-ABC). An inverted organization in which the IM domain is C-terminal with respect to the ABC domain (ABC-IM) exists, such as in the MacB protein, involved in macrolide resistance in E. coli. No IM domain partners have been identified for ABC proteins falling into the third category, and these proteins consist of two ABCs fused together (ABC2). Since more and more proteins are being found to be related to the last category, we propose calling the whole superfamily “ABC systems” rather than ABC transporters, a term that should be restricted to systems containing typical IM domains.
The tertiary structures of many ABC monomers and dimers have been determined, and they display remarkable conservation in their global folding pattern, as expected from sequence homology (268) (see Structure, Function, and Dynamics of the ABC). IM domains are typically comprised of six TM α-helices, and several structures of intact transporters have been determined (102, 205, 285, 343, 366, 518). Input from structural biology has greatly impacted our understanding of the mechanism of action of ABC systems.
Overview of Prokaryotic Transporters
ABC transporters are one of many different types of transporters operating in bacteria and other organisms. Transporters are of critical importance for living organisms, and selective permeability to nutrients and metabolites was probably the first distinctive property of primitive cells. Functionally and structurally different transporters have been identified in living organisms (Fig. 2). It is customary to distinguish channels, primary transporters, and secondary transporters with respect to the source of energy used (411). Channels catalyze the facilitated diffusion of solutes down a concentration gradient, an energy-independent process. In the outer membranes of gram-negative bacteria, porin channels allow diffusion through a TM-spanning aqueous pore (Fig. 2a). Cytoplasmic membrane channels are gated, opening or closing in response to voltage (396) or to membrane tension, as seen for McsS, which plays a role in protecting bacteria from hypo-osmotic shock (22, 276). Primary active transporters, including the ABC transporters, couple transport against a concentration gradient to the hydrolysis of ATP (Fig. 2b, diagrams E and F, and c, diagram H). Secondary active transporters, including uniporters, antiporters, and symporters (Fig. 2a, diagrams C and D), use the energy stored in ion gradients to drive transport (411). A novel family of secondary high-affinity transporters, the TRAP (tripartite ATP-independent periplasmic) transporters (Fig. 2a, diagram B) (159, 224), which primarily catalyze the transport of C4-dicarboxylates (see reference 239 for a review), although other substrates, including sialic acid, have been described (442), was recently reported for prokaryotes. Another family of transporters unique to prokaryotes, the phosphoenolpyruvate:sugar phosphotransferase (PTS) transporters (Fig. 2a, diagram A), catalyze the uptake of sugars. Energy coupling to transport in these systems occurs via a series of phosphoryl transfer reactions (116). Transporters belong to at least 300 different protein families, and the two largest of these are the primary ABC superfamily and the secondary major facilitator superfamily (MFS) (412).
FIG. 2.
Schematic view of the organization of transport systems. In gram-negative bacteria, substrates can cross the outer membrane by facilitated diffusion through porins, which are trimeric channels. Red circles represent transported substrates, small green circles represent cotransported ions, and small blue circles represent phosphates. (a) Group translocators and secondary transporters. (A) PTSs. PTSs consist of a set of cytoplasmic energy-coupling proteins and various integral membrane permeases/sugar phosphotransferases, each specific for a different sugar. The E. coli mannitol permease consists of two cytoplasmic domains (EIIA and EIIB) involved in mannitol phosphorylation and an integral membrane domain (EIIC) which is sufficient to bind mannitol but which transports mannitol at a rate that is dependent on phosphorylation of the EIIA and EIIB domains. The two other components are common to all PTS systems. The soluble enzyme I (EI) autophosphorylates in the presence of Mg2+. The histidine protein (HPr) is the energy-coupling protein and delivers phosphoryl groups from EI to the sugar-specific transporters (EIIs). (B) TRAP transporters. A periplasmic BP, which is unrelated to an ABC BP at the sequence level but similar in secondary structure, functions in association with two membrane components, namely, a large TM subunit involved in the translocation process and a smaller membrane component of unknown function. The driving force for solute accumulation is an electrochemical ion gradient, not ATP hydrolysis. (C) Ion-driven MFS transporters. These transporters typically consist of a single cytoplasmic membrane protein with 12 TM segments that couples transport of small solutes to existing gradients of ions, such as protons or sodium ions. Symporters pump two or more types of solutes in the same direction simultaneously, using the electrochemical gradient of one of the solutes as the driving force. Antiporters (not shown) are driven in a similar way, except that the solutes are transported in opposite directions across the membrane. (D) Uniporters transport one type of solute and are driven directly by the substrate gradient. (b) ABC import systems. (E) Vitamin B12 importer. The vitamin B12 uptake system of E. coli includes a high-affinity OMR, BtuB, that translocates the substrate through the outer membrane in an energy-dependent step that requires an active TonB-ExbB-ExbD complex. Substrates are captured by the periplasmic BP BtuF in the periplasmic space and presented to a cytoplasmic complex made of two copies each of BtuC and BtuD. This complex mediates the ATP hydrolysis-dependent translocation of vitamin B12 into the cytoplasm. (F) Maltose-maltodextrin importer. The transport of maltodextrins larger than maltotriose through the outer membrane requires the trimeric maltoporin LamB. Substrates are captured by the maltose-BP MalE in the periplasmic space and presented to a cytoplasmic complex made of MalF, MalG, and two copies of MalK. (c) Comparison between secondary RND and primary ABC export systems. (G) AcrAB-TolC exporter. This hypothetical model of the RND family AcrA-AcrB-TolC drug efflux pump is based on the trimeric structures determined for TolC and AcrB. TolC is predicted to contact the apex of the AcrB trimer. Two molecules of the MFP AcrA are shown, but it is probable that this protein exists as higher-order oligomers in the complex. Hydrophobic drugs are probably pumped out of the membrane lipid bilayer coupled to the downhill movement of protons across the cytoplasmic membrane. (H) Hemolysin HlyBD-TolC exporter. This hypothetical assembled model consists of a TolC trimer, a dimer of the IM-ABC protein HlyB, and the MFP HlyD. The exact oligomeric state of HlyD is not known accurately, though it may be trimeric. The TM and ABC domains of HlyB are represented by red rectangles and green circles, respectively. Hemolysin is translocated through the envelope by an ATP hydrolysis-dependent process.
Multiple Functions of ABC Transporters
ABC transporters have a great impact on bacterial physiology, and their dysfunction can have strong deleterious effects. While some ABC transporters are clearly dedicated to the export of virulence factors, under appropriate conditions many other bacterial ABC transporters can become important for viability, virulence, and pathogenicity. An example is provided by iron ABC uptake systems, which have long been recognized as important effectors of virulence (191). Because iron exists primarily in the insoluble Fe3+ form under aerobic conditions, biologically available iron in the body is found chelated by high-affinity iron-binding proteins (BPs) (e.g., transferrins, lactoferrins, and ferritins) or as a component of erythrocytes (such as heme, hemoglobin, or hemopexin) (255). Pathogens are able to scavenge iron from these sources by secreting high-affinity iron-complexing molecules called siderophores and reabsorbing them as iron-siderophore complexes (see reference 516 for a review). Another example of involvement in virulence is the chvE-gguAB operon in Agrobacterium tumefaciens, which encodes a glucose and galactose importer (61, 241). Sugar binding to ChvE triggers a signaling response that results in virulence gene expression. Finally, a potentially lethal upshift in osmotic strength is counterbalanced by activation of osmosensing ABC transporters that mediate uptake of compatible solutes (375).
In addition to their importance in transport, ABC systems are involved in the regulation of several physiological processes. In this context, direct regulatory roles need to be distinguished clearly from indirect effects mediated through the transported substrate. Well-documented examples of cases where additional regulatory domains (RDs) in the transporter itself are involved are the C-terminal domain of MalK of the maltose-maltodextrin transporter from E. coli and S. enterica serovar Typhimurium, the tandem cystathionine-synthase domains of OpuA from Lactococcus lactis and of other members of the OTCN family, and the R domain of the human cystic fibrosis protein CFTR (reviewed in reference 26).
Inventory and Classification of ABC Systems and Evolution of the Superfamily
Computer-assisted methods have been applied to understand the complexity and the diversity of the ABC superfamily. The use of multiple sequence alignments was instrumental in the first definition of the superfamily (196). In most cases, ABC proteins of a given organism (44, 280, 385) or ABC systems with clear functional similarity (144, 211, 259) were compared. However, the presence of the highly conserved ATPase domain also allows more global comparisons (359, 420). The last publication (420), which constitutes the first global study specifically devoted to the ABC superfamily, was updated to include about 600 ABCs (87). These sequences segregate into 29 clusters or families (Table 1). A phylogenetic tree derived from the comparison of these sequences is given in Fig. 3.
TABLE 1.
Classes, families, and subfamilies of ABC systemsa
| Family and subfamilyb | No. of systems | Definition | Modelc | Other named
|
Domain(s)e | |
|---|---|---|---|---|---|---|
| HCGN | TC | |||||
| Class 1 systems (transporters with fused TM and ABC domains) | ||||||
| FAE | 99 | Very-long-chain fatty acid export | ALD_HUMAN | ABCD | P-FAT | B, E |
| DPL | 1,569 | Drugs, peptides, lipids | A, B, E | |||
| BAE | 83 | Bacteriocin and peptide export | MESD_LEUME | Pep2E | B | |
| LAE | 95 | Lantibiotic export | NIST_LACLA | Pep1E | B | |
| CHV | 13 | 1,2-beta-glucan export | CHVD_RHIME | Glucan E | B | |
| SID | 64 | Siderophore uptake | Q9R7V3_YERPE | SIUT | B | |
| HMT | 145 | Mitochondrial and bacterial transporters II | ATM1_YEAST | ABCB | HMT | B, E |
| MDL | 65 | Mitochondrial and bacterial transporters I | MDL1_YEAST | ABCB | MPE | B, E |
| LIP | 222 | Lipid A or glycerophospholipid export | MSBA_ECOLI | Lipid E | B | |
| LLP | 296 | LIP-like exporters | A, B, E | |||
| ARP | 27 | Antibiotic resistance or production | Q54203_STRGA | DrugE3 | B | |
| TAP | 38 | Peptide export | TAP1_HUMAN | ABCB | TAP | E |
| PED | 53 | Prokaryote drug export | LMRA_LACLA | DrugE2 | B | |
| HLY | 158 | RTX toxin export | HLYD_ECOLI | Prot1E | B | |
| PRT | 69 | Proteases, lipases, S-layer protein export | PRTD_ERWCH | Prot2E | B | |
| CYD | 93 | Cytochrome bd biogenesis | CYDC_ECOLI | CydDC-E | B | |
| p-gP | 135 | Eukaryote MDR and lipid export | MDR1_MOUSE | ABCB | MDR | E |
| OAD | 190 | Organic anion conjugates, anions, drugs | E | |||
| CFTR | 19 | Chloride anion channels | CFTR_HUMAN | ABCC | CFTR | E |
| MRP | 158 | Conjugate drug exporters | MRP1_HUMAN | ABCC | CT1-2 | E |
| SUR | 13 | Potassium channel regulators | SUR1_HUMAN | ABCC | E | |
| EPD | 250 | Eye pigment precursors and drugs | B, E | |||
| WHITE | 125 | Eye pigment precursors and drugs | WHIT_DROME | ABCG | EPP | B, E |
| PDR | 125 | Pleiotropic drug resistance | PDR5_YEAST | PDR | E | |
| MCM | 11 | Unknown | A | |||
| CCM | 94 | Cytochrome c biogenesis | CCMA_ECOLI | HemeE | A, B, E | |
| Class 2 systems (nontransport cellular processes and antibiotic resistance) | ||||||
| RLI | 48 | RLI | ABCE1_HUMAN | ABCE | A, E | |
| ART | 714 | Antibiotic resistance and translation regulation | A, B, E | |||
| ARE | 104 | MLS antibiotic resistance | MSRA_STAEP | DrugRA2 | B | |
| REG | 589 | Gene expression regulation | GCN20_YEAST | ABCE | B, E | |
| EF3 | 18 | Translation elongation | EF3A_YEAST | E | ||
| UVR | 245 | DNA repair and drug resistance | UVRA_ECOLI | A, B | ||
| Class 3 systems (separate TM and ATP-binding domains) | ||||||
| CBY | 269 | Cobalt, nickel, and vitamin uptake | ||||
| CBU | 62 | Cobalt and nickel uptake | CBIO_SALTY | 5 | A, B | |
| Y179 | 207 | Biotin and hydroxymethyl pyrimidine uptake | Q6GUB1_RHIET | BIOMNY | A, B | |
| MKL | 162 | Organic solvent resistance | Q9Z402_PSEPU | HCH | B, E | |
| YHBG | 129 | Translocation of LPS to the outer membrane | LPTB_ECOLI | B | ||
| o228 | 893 | Release of lipoproteins and drug resistance | LOLD_ECOLI | 6 | A, B | |
| CDI | 126 | Cell division | FTSE_ECOLI | B | ||
| ISB | 167 | Iron-sulfur center biogenesis | SUFC_ECOLI | A, B, E | ||
| DRA | 431 | Drug and antibiotic resistance | A, B, E | |||
| NOD | 31 | Nodulation | NODI_RHISM | LOSE | B | |
| DRR | 314 | Polyketide drug resistance | DRRA_STRPE | DrugE1 | A, B | |
| ABCA | 86 | Lipid transport | ABC1_HUMAN | ABCA | CPR | E |
| DRI | 854 | Drug resistance, bacteriocin and lantibiotic immunity | A, B | |||
| BAI | 44 | Bacteriocin immunity | BCRA_BACLI | B | ||
| LAI | 185 | Lantibiotic immunity | Q45404_BACSU | Pep5E | B | |
| DRB | 441 | Drug resistance (putative) | A, B | |||
| YHIH | 125 | DRB-like systems | YHIH_ECOLI | NatE | A, B | |
| NOS | 59 | Nitrous oxide reduction | NOSF_PSEST | A, B | ||
| CLS | 41 | Capsular polysaccharide, LPS biogenesis | KST1_ECOLI | 1 | A, B | |
| Class 3 systems (BPD importers) | ||||||
| MET | 293 | Metallic cations | ZNUC_ECOLI | MZT | A, B | |
| MOS | 599 | Monosaccharides and deoxyribonucleosides | RBSA_ECOLI | CUT2 | A, B | |
| DLM | 168 | d- and l-methionine | METN_ECOLI | A, B | ||
| MOI | 1,005 | Mineral and organic ions | POTD_ECOLI | 2 | A, B | |
| PAO | 708 | Polar amino acid and opines | HISP_SALTY | PAAT | A, B | |
| HAA | 399 | Hydrophobic amino acids and amides | LIVG_ECOLI | HAAT | A, B | |
| OSP | 744 | Oligosaccharides and polyols | MALK_ECOLI | CUT1 | A, B | |
| OPN | 883 | Oligopeptides and nickel | OPPD_SALTY | PepT | A, B | |
| PHN | 78 | Phosphonates and phosphites | PHNC_ECOLI | PhnT | B | |
| OTCN | 639 | Osmoprotectants taurine, cyanate, and nitrate | TAUB_ECOLI | 3 | A, B | |
| ISVH | 927 | Iron-siderophores vitamin B12 and hemin | FHUC_ECOLI | 4 | A, B | |
| NO | 334 | Unclassified systems | A, B, E | |||
The three classes of ABC systems are as follows: class 1, systems with fused ABC and IM domains; class 2, systems with two duplicated fused ABC domains and no IM domains; and class 3, systems with IM and ABC domains carried by independent polypeptide chains. There are a few exceptions to this scheme, as discussed in the text: ABC proteins of the CcmA and MCM families cluster with class 1 systems but belong to systems similar to those of class 3 and class 2, respectively.
Family names are abbreviations of the substrate or the biological process handled by the systems. For families comprised of systems of unknown function, an arbitrary name is given.
For each family or subfamily, a typical ABC protein is indicated as an example, and when available, the Swissprot identification is given.
Cross-references to the nomenclatures of ABC systems adopted by the Human Gene Nomenclature Committee (HGNC) (http://www.genenames.org/genefamily/abc.php) and by the Transport Commission (TC) (http://www.tcdb.org/tcdb/) are given. Some families described in this table are divided by the Transport Commission into subfamilies according to substrate type, as follows: 1, CPSE + LPSE; 2, PhoT + MolT + SulT + FeT + POPT + ThiT + BIT; 3, QAT + NitT + TauT; 4, B12T + FeCT; 5, CoT + NiT + NiCoT + BIOMNY; and 6, LPT + MacB + Pep4E.
A, archaea; B, bacteria; E, eukaryotes.
FIG. 3.
Simplified neighbor-joining tree of ABC domains. For clarity, only the main branches that point to ABC families are drawn. The major subdivisions of the tree correspond to the three classes of ABC systems, whose schematic structural representations are given in the right part of the figure. ABC domains are shown with green circles, and IM domains are shown with differently colored rectangles. For the sake of simplicity, accessory proteins (BPs, MFPs, and OMFs) are omitted. Class 1 (red branches) systems have fused ABC and IM domains, corresponding mainly to exporters. IM domains shown in purple represent ABC transporters with the IM domain at the N terminus, corresponding to IM-ABC and (IM-ABC)2 topologies. N- and C-terminal domains are symbolized by N and C, respectively. IM domains shown in orange represent ABC transporters with the IM domain at the C terminus, corresponding to ABC-IM and (ABC-IM)2 topologies. Class 1 contains two atypical families of systems, CCM and MCM, with a different structural organization. Class 2 (blue branches) systems have tandemly repeated ABC domains and no known TM domains (ABC2 topology), corresponding to proteins involved in nontransport processes. The UVR family was omitted during the generation of the tree because large domain insertions within the ABC domains prevent the establishment of the multiple sequence alignment. However, binary comparisons established the relationship between UVR proteins and class 2 systems. Class 3 (green branches) systems have IM (red rectangles) and ABC (green circles) domains carried by independent polypeptide chains, corresponding mainly to importers. For class 3, systems that could be exporters are shown in boxes. Family names are abbreviated according to the conventions used in Table 1 and throughout the text. See Table 1 for the abbreviations of family names and for functional descriptions. OPN-D, OPN-F, HAA-F, and HAA-G correspond to the two different ABC subunits of the OPN and HAA systems, respectively. MOS-N and MOS-C correspond to the N- and C-terminal ABC domains of MOS family ATPases. The scale at the top of the figure corresponds to 5% divergence per site between sequences.
The major finding of this study (Fig. 3 and Table 1) is that ABC cassettes diverged very early into three main subdivisions or classes that match fairly well with the three functional divisions of ABC systems, i.e., importers, exporters, and others. Class 1 is comprised of systems with fused ABC and IM domains and contains the vast majority of export systems (Fig. 2c, diagram H). Fusions can be either of the ABC-IM type or of the IM-ABC type. There are two exceptions in this scheme, since the ATPases of the CCM and MCM families cluster within class 1, though their structural organization resembles that of class 3 and class 2 proteins, respectively. The reason for this misplacement may be due to a long-branch attraction artifact of divergent sequences in the phylogenetic tree (48). Class 2 is comprised of proteins with two tandemly repeated ABC domains and no IM domains that likely do not function as transporters. Class 3 contains systems with IM and ABC domains carried by independent polypeptide chains that correspond to most binding-protein-dependent (BPD) importers (Fig. 2b) (87). ABC importers generally depend on the presence of a separate extracellular substrate-BP that recognizes substrates with high affinity. However, class 3 contains several transporters that lack BPs and that cannot conclusively be related to import. Such systems that are involved in drug and antibiotic resistance or in the biogenesis of extracellular polysaccharides have been proposed to participate in the export of such molecules. These transporters cluster with the BPD systems, suggesting either that their transport polarity has changed during evolution or that they are not directly involved in the export of these substances.
Similar studies were performed on the less conserved IM domains and BPs of ABC importers. It is notoriously difficult, if not impossible, to build multiple alignments of these proteins or domains, due to the lack of overall homology. However, clustering methods were applied to the scores of binary alignments to generate families of IM domains and BPs (421, 470). There is good agreement between families derived from the classifications of BPs, IM domains, and ABCs, suggesting that components of ABC transporters coevolve with minimal shuffling of their components (42, 259). A good correlation exists between the sequences of the ABCs and the global substrate specificity of ABC systems. This apparent relationship between sequence and function could reflect constraints imposed by the interaction of ABC proteins with their IM partners, which are thought to carry substrate recognition sites (488).
The clustering of ABCs into three classes that correspond to the topology of ABC systems is intriguing. The ABCs segregate mostly according to sequence differences in a region that lies between Walker motifs A and B and includes the helical domain (Fig. 1) (426). This region has been termed the structurally diverse region (425). Using the maltose transporter, we have demonstrated experimentally that this region is critical for the interaction between the ABC and IM domains (216, 319). Recent insight from complete structural determinations of ABC importers and exporters sheds further light on this point. In class 1 transporters, whose prototype is Sav1866, it is clear that the transmission interface between IM domains and the ABC is dual, involving both IM intracellular loops 1 and 2 (ICL1 and ICL2), which make contact with the Q loop and with a newly discovered TEVGERV motif in the helical domain, respectively (102). This new motif is conserved only in exporters. In contrast, in class 3 importers, exemplified by the Archeoglobus fulgidus ModABC transporter and the E. coli MalFGK2 transporter, a single intracellular loop, known as the EAA loop of the IM component, interacts with the Q loop of the ABC as well as with helices 3 and 5 within the helical domain (96, 205, 343). Class 2 proteins appear not to interact with a TM domain, and as exemplified by the structure of RLI (232), there is no cleft at the corresponding position to accommodate an EAA loop from an IM subunit. These differences might account, at least in part, for the differential clustering of ABC domains as importers, exporters, and others.
Proteins from all three kingdoms of life, i.e., Bacteria, Archaea, and Eukarya, are found in each class of ABC systems, and species-specific differences are observed at the very ends of the branches of the tree. These observations suggest that ABC systems began to specialize very early, probably before the separation of the three kingdoms, and that functional constraints on the ABC domain are responsible for the global conservation of sequences.
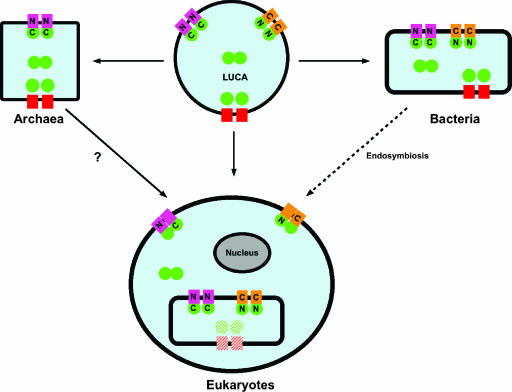
From this analysis, we propose a hypothetical scenario for the evolution of ABC systems (87, 420) (Fig. 4). The ancestor “progenote” cell or last universal common ancestor (LUCA) already had all classes of ABC systems. Prokaryotes inherited all ABC classes. However, archaea are apparently devoid of transporters of the ABC-IM type, though they have IM-ABC transporters. An alternative scenario to account for the absence of ABC-IM systems in archaea is that these systems arose in bacteria only. Eukaryotes probably acquired most IM-ABC and ABC-IM (class 1) systems and ABC2 (class 2) systems from the symbiotic bacteria that are the putative ancestors of organelles. It is noteworthy that most eukaryotic IM-ABC transporters are specifically targeted to organelle membranes, which probably descend from a prokaryotic ancestor. For instance, the mammalian TAP protein, an IM-ABC exporter involved in the presentation of antigenic peptides to the class I major histocompatibility complex, is inserted into the endoplasmic reticulum (192). The ALD protein, putatively involved in the export of very-long-chain fatty acids from the cytosol into peroxisomes, is targeted to the peroxisomal membrane (515). From genes encoding IM-ABC or ABC-IM transporters, eukaryotes developed specific systems by several independent duplication-fusion events, including those that led to the formation of the PDR (plant and fungal pleiotropic drug resistance) and P-glycoprotein-like families of proteins. Class 3 systems, particularly BPD transporters, are virtually absent from eukaryotic genomes, though there is evidence that they were acquired first and lost subsequently (see the next paragraph). The reason for the absence of class 3 transporters in eukaryotes is not clear, but it may be related to different bioenergetic requirements and environmental constraints.
FIG. 4.
Hypothetical scenario for the evolution of ABC systems. The figure uses the same topological representations and color coding as those used in Fig. 3. The hypothetical LUCA is predicted to possess all classes of ABC systems. The cell membrane is represented as a thick black line surrounding the intracellular medium (blue). The eukaryotic cell nucleus is a gray oval. The eukaryotic organelle is represented as a bacterium to symbolize its endosymbiotic origin. The arrows symbolize the evolutionary relationships between archaea, bacteria, and eukaryotes. The question mark under the arrow joining archaea and eukaryotes recalls the hypothesis that the ancestral eukaryotic cell arose by a unique endosymbiotic event involving engulfment of an archaebacterium by a gram-negative eubacterial host prior to the other endosymbiotic events leading to the appearance of organelles. Alternatively, eukaryotes may have arisen directly from the LUCA via an unidentified transient archaebacterium. A class 3 transporter, represented by a hatched pattern, recalls the hypothesis that these systems were acquired by eukaryotes and subsequently lost. See details of the scenario in the text.
Comparative Genomics of ABC Systems
The complete nucleotide sequences of more than 500 genomes are presently available, and much effort has been put forth to build complete inventories of ABC proteins in Saccharomyces cerevisiae (110), Escherichia coli (88, 280), Bacillus subtilis (385), Mycobacterium tuberculosis (44), Arabidopsis thaliana (417), Caenorhabditis elegans (449), Oryza sativa (165), and many other organisms. Global comparisons of the ABC protein contents of several genomes have also been made (161, 219, 356, 359, 485). In the course of the constitution of ABCISSE, our database of ABC systems, we have analyzed the compositions of more than 250 completely sequenced genomes.
When the total number of ABC systems is plotted against the size of prokaryotic genomes (Fig. 5), a linear relationship is seen, in agreement with the observation that the number of transporters of all categories (ion gradient-driven, PTS, ABC, and facilitator proteins) is approximately proportional to genome size (359). Bacteria with genomes in the range of 0.5 to 1.5 Mb have about 15 ABC systems. Most bacteria of this size are intracellular parasites, and the availability of intracellular metabolites or the presence of homologous host genes may have rendered some ABC genes inessential, leading to their subsequent disruption or deletion. It is therefore possible that the subset of ABC systems that are common to these species constitute the minimal requirement of ABC systems for life. The relatively small, 1.86-Mb genome of Thermotoga maritima has a very large number of ABC systems (68 systems) compared with that of species of similar genome size. This is partly due to the extensive amplification of operons encoding ABC systems putatively involved in the uptake of oligosaccharides (11 systems) and oligopeptides (12 systems). Escherichia coli (4.6 Mb) and Bacillus subtilis (4.2 Mb) have 78 and 84 ABC systems, respectively, which are typical numbers for this size of genome. In contrast, the genome of Mycobacterium tuberculosis (4.4 Mb) has only 38 systems. The lack of high-affinity import systems might be related to the intracellular lifestyle of this bacterium. This number is also significantly lower than that found in soil bacteria, such as Agrobacterium tumefaciens and Mesorhizobium loti (5.67 and 7.6 Mb, respectively), which have more than 200 ABC systems. The large number of ABC systems in this instance is probably due to highly competitive environmental conditions in the soil or within plant nodules. Eukaryotes display a smaller number of ABC proteins with respect to genome size than do prokaryotes, and this is particularly evident in the case of Saccharomyces cerevisiae, a free-living microorganism which shares with bacteria almost the same ecological niches. Indeed, ABC importers are lacking in eukaryotes.
FIG. 5.
Genomic distribution of ABC systems in living organisms. The plot shows the number of ABC ATPases versus the number of total genes in completely sequenced genomes. The number of ABC ATPases per genome (which roughly reflects the number of ABC systems) is plotted against the total number of genes (purple dots, archaea; blue dots, bacteria; green dots, eukaryotes). Selected genomes with exceptionally large or small numbers of ABC proteins are indicated with circles on the graph and are discussed in the text.
Class 1 ABC transporters (exporters with fused ABC and IM domains) are underrepresented in the genomes of bacteria and are virtually absent from the genomes of archaea. In contrast, they represent the major fraction of ABC systems in eukaryotes. Class 2 ABC proteins (ABC2 organization, with no IM domains) are found in all genomes, even the smallest ones. This observation establishes the physiological importance of this class, which contains proteins experimentally or putatively involved in regulation of gene expression and in DNA-related processes. The number of class 2 proteins per genome ranges from 1 to 8 for genomes that vary from 0.58 to 132.5 Mb. Class 3 systems (mostly importers) are found quasi-exclusively in prokaryotic genomes. Incomplete class 3 transporters are found in the genomes of plants and algae. For example, genes encoding putative thiosulfate/sulfate IM and ABC components have been identified in the DNA sequences of the chloroplast genomes of a variety of organisms, including the green algae Mesostigma viride (272), Nephroselmis olivacea (492), and Chlorella vulgaris (513) and the liverwort Marchantia polymorpha (340). The sequences of these genes share high similarity with the cysT and cysA genes of the cyanobacterium Synechococcus sp. strain PCC 7942 (265). These genes are probable remnants of BPD transporter-encoding genes present in the genome of the ancestor of organelles. Recently, the genes for a complete putative ABC-type sulfate transporter were found in the nuclear genome of the model unicellular green alga Chlamydomonas reinhardtii. Four genes code for chloroplast envelope-targeted TM proteins (SulP and SulP2), a chloroplast stroma-targeted ATP-binding protein (Sabc), and a substrate (sulfate)-binding protein (Sbp) that is localized on the cytosolic side of the chloroplast envelope (309). Antisense mutagenesis of the SulP gene demonstrated a role in chloroplast sulfate uptake (67).
Very few families of ABC systems appear to be species or kingdom specific (Table 1). For example, the MCM family of proteins with unknown function is found only in methanogenic archaea, and the PDR subfamily is found only in plants and fungi. In general, most families have been identified in more than one kingdom. Several families have been recognized from sequence analysis but have not been characterized at the functional level. All of the observations reported above establish that the ABC constitutes an ancient and universal molecular motor with a fascinating range of diverse applications.
In this review, we aim to discuss the most recent advances in the understanding of structural and functional aspects of model ABC transporters and to provide a wide overview of the diversity and versatility of bacterial ABC systems. We offer our apologies to those whose work we overlooked or were not able to include.
STRUCTURE, FUNCTION, AND DYNAMICS OF THE ABC
High-Resolution Structures of an ABC Module
Crystal structures of isolated ABC modules from many members of the ABC superfamily are now available (reviewed in reference 349). In addition, the structures of six full-length ABC transporters have been reported (102, 205, 285, 343, 366, 518). Since many of these structures have recently been reviewed (96, 349), we focus here on key characteristics and their implications in function.
All ABCs contain two domains (167), a larger domain similar to the core structure found in many RecA-like motor ATPases and a smaller, predominantly helical domain that is unique to ABC transporters (Fig. 6). The RecA-like domain typically consists of two β-sheets and six α-helices and includes the Walker A motif (GxxGxGKS/T, where x is any amino acid) and the Walker B motif (φφφφD, where φ is a hydrophobic residue). The helical domain consists of either three or four helices and a signature motif, which is also known as the LSGGQ motif, the linker peptide, or the C motif. The two domains are joined by two flexible loops, one of which contains a highly conserved glutamine residue and is known as the Q loop. In the structures of intact ABC transporters, the Q loop mediates interactions between the ABC subunits and the TM subunits (102, 205, 285, 366).
FIG. 6.
Structure of an ATP-bound ABC dimer. The structure of a MalK homodimer with two ATPs bound (PDB accession no. 1Q12) is shown. Each NBD consists of two subdomains, a RecA-like subdomain (green) and a helical subdomain (blue). A C-terminal RD (not present in all ABC proteins) is shown in yellow. Corresponding domains in the second MalK subunit follow the same color scheme but are rendered in lighter colors. Two ATPs, represented as a ball-and-stick model, are bound between the NBDs. The Walker A motif is shown in red.
Like that of other RecA-like ATPases, hydrolysis requires oligomerization. All ABC transporters have two ABCs, and ATP binding is required to obtain the dimeric state of most isolated ABCs, as first shown by Moody and coworkers (318). To obtain crystals of the ATP-bound dimers, residues in the active site were mutated (378, 457, 545) or EDTA was used (68) to circumvent the ATPase activity. In each case, two ATP molecules are bound at the interface of the dimer, interacting with residues from the Walker A motif of one subunit and the LSGGQ motif of the other (Fig. 6). This architecture for an ABC-like protein was first seen in Rad50 (206), and these structures are consistent with biochemical evidence demonstrating that ATP is in close contact with residues in both the Walker A and LSGGQ motifs during catalysis (156). Interestingly, the nature of the dimer interface was correctly predicted through an earlier analysis of the structure of the HisP monomer and patterns of degeneracy in these conserved sequence motifs (229). The adenosine ring of ATP is stabilized by a ring-stacking interaction with a conserved aromatic residue preceding the Walker A motif (12, 168). The conserved lysine residue in the Walker A motif forms hydrogen bonds with the oxygen atoms of the α- and γ-phosphates, thereby holding both phosphates in a defined orientation. A Mg2+ ion is coordinated by O atoms from the β- and γ-phosphates and residues in the Walker A motif (167, 508). A highly conserved histidine residue located in the H loop forms a hydrogen bond with the γ-phosphate and is required for hydrolysis (68, 545). The side chain of the serine and the backbone amide groups of the glycine residues in the LSGGQ motif coordinate the γ-phosphate. It was suggested that the LSGGQ motif is reminiscent of the “arginine finger” of other RecA-like ATPases, where an arginine residue provided by one subunit extends into the nucleotide-binding site of the other (540). In addition to nucleotide binding, the conserved histidine residue also contacts residues across the dimer interface in the Walker A motif and the D loop, a conserved sequence following the Walker B motif, suggestive of a tight coupling between ATP binding and formation of the dimer (68, 206, 457, 545). The sharing of nucleotide-binding sites between subunits explains why all ABC proteins have two NBDs or subunits even though one site has sometimes deviated from the consensus. Moreover, the observation of positive cooperativity in ATP hydrolysis in some systems (97, 176, 281, 463, 546) is consistent with a requirement that ATP must bind to both sites before the NBDs can assume the closed, catalytically active conformation.
Conformational Changes
One of the essential tasks for ABC transporters is to harness the energy of ATP binding and/or hydrolysis for mechanical work. This task appears to be achieved through conformational changes of the transporter. The dynamic nature of the ABCs has been revealed by comparison of structures of isolated ABC modules in apo and ATP- and ADP-bound forms (96, 349) and is supported by biochemical data sensing nucleotide-dependent changes in parameters such as protease sensitivity, fluorescence, and rates of hydrogen/deuterium exchange (427, 510). In the absence of nucleotides and TM subunits, the ABC structures show high degrees of intrinsic flexibility. When the structures of different ABCs are compared, the helical domain shows rigid-body motion relative to that of the RecA-like domain (reviewed in reference 96). In addition, two different conformations of the same protein in the apo form are reported for both Sulfolobus solfataricus GlcV (508) and E. coli MalK (68), and differences in domain orientations account for these structural differences also.
In comparison with the apo form, the ATP-bound structures in the monomeric form show that the helical domain rotates toward the RecA-like domain upon ATP binding, moving the LSGGQ loop into a position where it contacts the nucleotide across the dimer interface (68, 457, 545). It was suggested that, upon hydrolysis, the helical domain rotates away from the active site to facilitate nucleotide exchange (234). Since the ABC protein functions as a dimer, it is important to analyze the conformational changes of the dimeric structures in a transporter cycle. The nature of these changes is illustrated by the crystal structures of MalK in the resting, ATP-bound, and ADP-bound states (68, 292). While other NBDs or nucleotide-binding subunits display low affinity for each other in the absence of the TM segments, the MalK dimer is stabilized through interactions of an additional C-terminal RD, and its propensity to form a dimer in the cytoplasm was demonstrated quite early (243). A tweezer-like motion, in which the two monomers of MalK are held together at the base by the RDs and the NBDs open and close like the tips of a pair of tweezers, is revealed by crystal structures in three different dimeric configurations (Fig. 7). In nucleotide-free structures, the N-terminal NBDs are separated and the dimer is maintained solely through contacts with the C-terminal RDs. In the ATP-bound form, the NBDs make contact and two ATPs lie buried along the dimer interface. The structure of MalK in a posthydrolysis state shows that the two NBDs of the ADP-bound form are separated, similar to the case for the resting form, indicating that ADP, unlike ATP, cannot stabilize the closed form. The three different conformations are achieved principally by a rotation of the entire NBD relative to the RD about a hinge region located in the loop connecting the NBD and RD. In addition, a second rigid-body rotation within the NBDs, between the RecA-like and helical subdomains, results in inward movement of the helical subdomain toward the nucleotide-binding site on the same subunit. As we have already discussed, this second rotation is also observed by comparison of different monomer structures. In contrast to the changes at the NBD-NBD interface, the interaction between the RDs in all three homodimer structures was essentially unchanged.
FIG. 7.
Three structures of the MalK dimer. In the absence of nucleotide, the two NBDs of MalK are separated from each other, held as a dimer primarily through contacts between the C-terminal RDs. In the presence of ATP, the NBDs are closed, permitting ATP hydrolysis to occur. In the ADP-bound state, the NBDs are again separated, suggesting a possible cycle for hydrolysis-driven conformational change. Coloring is the same as that in Fig. 6. (Reprinted from reference 292 with permission of the publisher. Copyright 2005 National Academy of Sciences, U.S.A.)
A similar catalytic cycle has been presented based on structures of the isolated NBD of HlyB, which has also been crystallized in multiple liganded states (547). Comparison of nucleotide-free and ADP-bound HlyB monomers suggests an additional conformational change, not seen in the MalK dimer, involving the Walker B motif and adjoining D loop as well as helix 6 in this molecule, that may be involved in NBD-NBD communication. Furthermore, comparison of ATP-bound and Mg·ATP-bound HlyB dimers containing a mutation of the catalytic His residue revealed the presence of a putative phosphate release channel in just one of the two subunits, suggesting that the absence of Mg2+, as is the case in the majority of NBD dimer structures, may underestimate the complexity of the catalytic cycle. The significance of potential asymmetry in the nucleotide-binding sites is further addressed in “Role of Two Nucleotide-Binding Sites.”
Molecular dynamics simulations based on structures of the isolated HisP (59, 227), MalK dimer (344), BtuD (222), and MJ0796 (60, 228) proteins largely recapitulate the conformational changes suggested by the structures of nucleotide-free and ATP-bound subunits. These studies also offer insight into the function of a “complete” ATPase active site, as H2O, Mg2+, and/or a key catalytic residue lacking in the structure can be reinstated (545).
In vivo, the MalK dimer is stably assembled into a complex with TM proteins MalF and MalG, and several lines of evidence suggest that the conformational changes seen in the crystal structures of isolated MalK subunits also occur in the intact MalFGK2 transporter. When a cysteine substitution is introduced for Ala85 in the Q loop region, a cross-linked MalK dimer is observed only upon addition of ATP (216). The failure of ADP to induce cross-linking suggests that ATP, not ADP, induces closure of the NBDs in MalFGK2 (216). Photocleavage experiments using vanadate as a transition state analogue indicated that ATP hydrolysis occurs via the closed dimer (156), and the solvent accessibility of a fluorescent probe covalently attached to an amino acid in the Walker A motif is reduced in the catalytic transition state compared to that in the resting state (299), consistent with the closure of the interface between the NBDs. We discuss how nucleotide-associated conformational changes in the ABC modules may be coupled to conformational changes in the TM domains in a later section [see “Coupling of transport to hydrolysis. (iii) Conformational changes associated with transport”].
Mechanism of ATP Hydrolysis, Still an Open Question
The structures of ABCs in isolation and in the context of intact transporters show a highly conserved fold, suggesting that they hydrolyze ATP by a common mechanism. However, the precise molecular mechanism of ATP hydrolysis is still controversial. An acidic residue at the end of the Walker B motif, a glutamate in most ABC transporters, has been proposed to act as a general base polarizing the attacking water molecule (168, 318). In the crystal structures, this residue extends into the active site and makes a hydrogen bond with the putative hydrolytic water. In several ABCs, replacement of this acidic residue with the corresponding amide abolishes the ATPase activity and triggers the formation of a trapped, ATP-bound dimer (318, 347, 378). Since high-resolution structures show that the attacking water is still well positioned in the mutants (378, 457), an appealing explanation for the lack of activity is that the substituted amide cannot act as a catalytic base for hydrolysis. A very similar mechanism, by which general acids coordinate the γ-phosphate of ATP and a general base polarizes the attacking water, has been suggested for other RecA-like ATPases (540). However, some data in the literature stand in opposition to the general base mechanism. Mutation of the Walker B glutamate (E/Q) in Pgp, HlyB, and GlcV leaves some residual ATPase activity (483, 493, 508, 545), leading some to question its role as a general base. In addition, if the Walker B glutamate is the catalytic base, one would expect a change in the pH dependence of ATPase activity upon substitution of glutamine for glutamate, as glutamine is a poor base, but the pH profiles of the wild-type HlyB ABC module and the E/Q mutant both show maximal activity near pH 7.0 (545). Thus, Schmitt and colleagues proposed an alternative mechanism, namely, substrate-assisted catalysis (545), in which the H loop histidine residue plays a relatively greater role in catalysis, forming part of a catalytic dyad with the Glu following the Walker B motif. In the substrate-assisted catalysis model, the histidine acts as a “linchpin” to hold the γ-phosphate of ATP, the attacking water, Mg2+, and other catalytically important amino acids together to support hydrolysis. In contrast to its function in the general base model, the function of the Walker B glutamate proposed in this model is to restrict the flexibility of the H loop histidine so that it adopts a catalytically competent conformation (141, 349, 545). It should be noted that in a different ABC module, mutation of Asp to Glu in the catalytic dyad does alter the pH profile of ATPase activity (141), though this result does not rule out either model, as local changes in the environment at the active site can alter the pKa of a side chain (141). While substitution of the His eliminates ATP hydrolysis in the HlyB ABC module, as demonstrated clearly by the ability to obtain crystals of a Mg·ATP-bound dimer (545), 2% of the wild-type ATPase activity is retained in the intact maltose transporter with substitution of arginine for histidine (99), suggesting subtle differences in architecture of the active sites of different ABC modules. Further research is needed before we can conclude that substrate-assisted catalysis or general base catalysis is a universal mechanism underlying ATP hydrolysis of the ABC transporter family, or even if there is a uniform mechanism.
Role of Two Nucleotide-Binding Sites
A question of great interest for all ABC transporters is the role that the two nucleotide-binding sites play in the energization of the translocation process. ATP is hydrolyzed with positive cooperativity in both the maltose and histidine transporters (97, 283), indicating that the two sites interact. Since both sites lie along the dimer interface of the ABCs, the simplest explanation for the presence of cooperativity is that both sites must be occupied before the NBDs can close and hydrolyze ATP. Structural data so far are consistent with this hypothesis, since nucleotide is bound at both sites in each of the structures of ATP-bound ABC dimers (68, 457, 545). It is not yet clear, however, whether one or both ATPs are hydrolyzed during a single cycle of conformational change. In some ABC transporters, such as the ribose transporter (57), only one of the two nucleotide-binding sites retains all of the highly conserved residues thought to be essential for ATP hydrolysis, suggesting that hydrolysis at just one site is sufficient for transport. In support of this hypothesis, mutation of the catalytically important histidine residue in the nucleotide-binding site of just one of the two HisP subunits in the intact histidine transporter was relatively well tolerated (335). However, the same substitution in a single site of the maltose transporter severely impaired both transport and ATPase activity, suggesting that hydrolysis at both sites is important for function (99). Interestingly, even the glutamate substitutions that promote stable formation of the NBD dimer, when present in a single site, inactivate the intact P-glycoprotein transporter (482), though it should be mentioned that the isolated NBDs of GlcV and HlyB (509, 546), containing a similar single-site mutation, retain ATPase activity. This seeming contradiction suggests that there may be functionally important cooperative interactions between nucleotide-binding sites that are lost once the NBDs are stripped from the IM domains, although noone has compared the effects of a single-site mutation in both intact and isolated ABC subunits of the same transporter.
As first shown with P-glycoprotein, a mammalian multidrug exporter (495), trapping of Mg·ADP and vanadate in the maltose transporter occurs in just one of the two nucleotide-binding sites (447). Vanadate replaces the Pi that is formed during ATP hydrolysis and prevents the dissociation of ADP from the active site (94, 440). The Mg·ADP·Vi complex is very stable and mimics the transition state for the hydrolysis of ATP. The observation that trapping occurs at just one site suggests that hydrolysis can occur at only one site at a time. Given that both sites can hydrolyze ATP in P-glycoprotein, it is suggested that the sites alternate in catalysis and that just one ATP is hydrolyzed each time a drug is transported (440, 494). Implicit in this model is the assumption that the transporter “remembers” which site hydrolyzed last. One way to envision such a possibility is to suggest that ATP hydrolysis results in the opening and release of ADP and Pi from a single site, while the second site remains closed, with ATP bound. Alternatively, one ATP could still be hydrolyzed per catalytic cycle, but following hydrolysis at one site, ATP at the remaining site is not bound strongly enough to sustain the closed conformation during exchange of ADP for ATP in the other site. In this model, catalysis would not be ordered strictly. Finally, if both ATPs are hydrolyzed sequentially before the NBDs dissociate (the progressive clamp model [226]), then two ATPs would be expended for the transport of a single molecule. To date, no single experiment is able to convincingly rule out any one of these models. The sequential model was put forth to explain the appearance of both trapped ATP and trapped ADP in an NBD dimer stabilized by mutation of the Glu residue following the Walker B motif, which is important for catalysis (226). It is suggested that this dimer does not dissociate until ATP is hydrolyzed at both sites, suggestive of sequential hydrolysis. It is unfortunate, however, that this type of experiment can be performed only in the presence of a mutation that greatly stabilizes the ATP-bound dimer and hence may not recapitulate what occurs in the wild type.
Evidence in support of asymmetric behavior of ABC dimers whose cassettes have identical sequences has been reported. The structure of the Mg·ATP-bound HlyB ABC dimer, stabilized through mutation of His in the catalytic dyad, has a 4.4-Å tunnel in one subunit, blocked by a conserved salt bridge in the second subunit, that could allow Pi to diffuse out of the binding site prior to opening of the dimer interface (547). Sequential hydrolysis and Pi release may allow the energy of ATP hydrolysis to be used in distinct steps, possibly for separate purposes (547). Intriguingly, disruption of this salt bridge results in the loss of cooperativity in ATP hydrolysis in the isolated ABC (547).
In intact transporters, evidence for asymmetry has also been detected. Thiol-specific reagents react more readily with a Cys in one ABC in the histidine transporter than with a Cys in the other, suggesting that structural asymmetry may exist before the binding of nucleotide (257). In the maltose transporter, asymmetries are seen in the pattern of cross-linking between cysteines placed in the helical domains of the ABC MalK and the IM proteins MalF and MalG (92, 216). In both cases, the IM region is heterodimeric, which could contribute to differences. The Lol system, involved in export of lipoproteins, contains two unique IM subunits, LolC and LolE, and a dimer of the LolD ATPase. Suppressors of a dominant negative mutation in a conserved motif of LolD map to LolC and LolE, suggesting that this motif is involved in subunit interaction in the Lol system (221). Interestingly, the suppressors are in a cytoplasmic loop of LolE and a periplasmic loop of LolC, raising the possibility of asymmetries in the way that LolD interacts with the IM subunits.
In proteins that are essentially homodimers, including the exporters MsbA and BmrA and the importer BtuCD, the presence of two identical ABCs would suggest that both are able to hydrolyze ATP, even though asymmetries that restrict ATP hydrolysis to just one of the two sites at a time may arise during the catalytic cycle, as suggested by vanadate-trapping experiments (347, 447, 495). BPD importers have a second potential source of asymmetry because the BPs themselves are asymmetric (388) and could impose functional asymmetries at the nucleotide-binding sites, a possibility that has not been explored. Interestingly, the addition of the BP BtuF did induce asymmetries in the most recent structure of BtuCD (217).
Molecular dynamics simulations and normal mode analysis have also been used to investigate asymmetries in ABC transporters. A simulation beginning with a Mg·ATP/Mg·ADP-bound MJ0796 dimer, designed to mimic ATP hydrolysis at a single site, revealed movement within one helical domain that might loosen the interaction between the ADP-bound monomer and the IM domain (228). Two different groups modeled ATP into both sites in the structure of BtuCD and found that only one ATP-binding site undergoes closure (222, 345), although the idea that one site might close independently of the other runs counter to the traditional interpretation of cooperativity in nucleotide binding and hydrolysis observed in ABC transporters. The work of a third group simulating BtuCD suggests that only simultaneous opening of both ATP-binding sites triggers appropriate conformational changes in the membrane (520).
Efforts have been made to measure the stoichiometric ratio of substrate transport to ATP hydrolysis in vivo and in vitro for several ABC transporters in an effort to determine whether one or both ATPs are hydrolyzed, but no one answer is universally accepted as yet. Often, substantial levels of uncoupled ATP hydrolysis, or possibly leakage of substrate from membrane vesicles, complicate the determination. In the maltose transporter, rates of 1.4 to 17 ATPs per transported sugar are reported, varying with the concentration of ATP trapped inside (98). Stoichiometries ranging from 5 to 25 ATPs per histidine have been reported for the histidine transporter (30). In the oligopeptide transporter, where the BP OpuA is tethered to the membrane via a lipid moiety, ATP hydrolysis is tightly coupled to transport and ratios approaching 2 ATPs per peptide transporter are seen (354). In studies of maltose and glycine-betaine transport in vivo, stoichiometries approaching 1 to 2 have been reported (316). Finally, a stoichiometric ratio of 1 was reported for the maltose transporter, based on comparison of growth yields of bacteria grown on different sugars under anaerobic conditions (321). Larger substrates may offer other complications; long linear maltodextrins are transported more slowly than maltose, leading to the suggestion that maltodextrins may be fed through the maltose transporter via a ratchet-like mechanism that expends more ATP per sugar transported (118).
IMPORT INTO THE CYTOPLASM (ABC IMPORTERS)
Most ABC importers rely on the presence of a high-affinity extracytoplasmic BP for function and are also called BPD transport systems. BPs are soluble proteins located in the periplasmic space between the inner and outer membranes of gram-negative bacteria. In gram-positive organisms, which lack a periplasm, they are often lipoproteins bound to the external face of the cytoplasmic membrane by N-terminal acyl-glyceryl cysteines. BPs are also found fused to the membrane transporter itself in some gram-positive organisms (497). In archaea, BPs either are lipoproteins or display a type III signal sequence. The latter BPs are proposed to constitute an extracellular multioligomeric organelle, the bindosome (553). All BPD transporters belong to class 3 and have ABC and IM domains on independent polypeptide chains. Some BP-independent class 3 importers have been characterized, as well as a few class 1 importers with a fused IM-ABC organization. Typically, the genes encoding a given transporter are carried in an operon, but in some gram-positive organisms, a single ABC subunit is shared by multiple transporters (385).
Transport across the Outer Membrane
To be transported efficiently into the cytoplasm in gram-negative bacteria, substrates first pass through the outer membrane, using one of three different pathways (see reference 333 for a comprehensive review).
Most small substrates, with molecular masses below 650 Da, cross the outer membrane through the nonspecific (generalized) porins, such as the OmpF or OmpC porins of Enterobacteriaceae (Fig. 2a) (333). These trimeric porins vary in their preferences for solutes of different sizes and charges. Their importance in transport is highlighted by the observation that mutants lacking these proteins are pleiotropically affected in the utilization of several different substrates (23). Crystallographic analyses reveal that porins adopt a β-barrel conformation with 16 β-strands spanning the outer membrane. A large loop folds back into the barrel, forming a constriction zone about halfway through the channel that contributes to the exclusion limit and ion selectivity of the pore (251).
When the size of the substrate exceeds the size handled by generalized porins, a specific or specialized porin is used. The best example known so far is maltoporin, the lamB gene product, which is essential for the transport of maltodextrins of more than three glucose residues (Fig. 2b, diagram F) (466). In contrast to the case for general porins, the genes coding for specialized porins are often linked genetically to the regions encoding the rest of the transporter, and their expression is tightly coregulated (466). The crystal structure of maltoporin reveals an 18-stranded β-barrel with three inwardly folded loops that constrict the diameter of the channel. Aromatic residues lining the channel constitute part of a diffusion pathway for maltodextrins through the channel, which appears to be designed to facilitate translocation rather than to bind sugars tightly (133, 424). LamB also functions as a general glycoporin, whose increased expression facilitates growth in carbohydrate-limited chemostats, with glucose, lactose, arabinose, or glycerol as the carbon source (108). ScrY, the sucrose porin of Klebsiella pneumoniae, and OprB, the d-glucose and d-xylose porin of Pseudomonas aeruginosa, constitute examples of other specific porins in gram-negative bacteria (429, 490). Pseudomonas aeruginosa actually lacks general porins, and most nutrients are taken in through specific porins of the OprD family, with OprD itself mediating the uptake of basic amino acids (471, 489). The structure of OprD reveals a β-barrel with a ladder of positively charged residues that funnel into a constriction site lined by residues which are not conserved in the family (31). It is suggested that substrate specificity is mediated by variation of residues at the site of constriction in the channel.
Because the size and scarcity of certain nutrients, including vitamin B12 and the Fe3+-siderophore complexes, exceed the size limit of porins (516), these compounds are bound by high-affinity outer membrane receptors (OMRs) that also function as transporters to move the compounds into the periplasmic space (Fig. 2b, diagram E) (523). An expenditure of energy is required to release these compounds from the OMRs so they can be transported, and studies with the E. coli ferrichrome (Fhu) receptor indicate that translocation of the substrate is dependent on the cytoplasmic membrane electrochemical gradient (46). The transduction of energy from the cytoplasmic membrane to the OMR is achieved by a set of three cytoplasmic membrane proteins, ExbB, ExbD, and TonB, that form a complex in the inner membrane (377). TonB and ExbD are proteins with single TM segments and large hydrophilic periplasmic domains. It is postulated that these domains interact with a conserved region of the OMR known as the TonB box to trigger the release of the substrate and its diffusion through a channel within the OMR. The OMRs FhuE, FhuA, and IutA are highly specific for their individual siderophore substrates, i.e., coprogen, ferrichrome, and aerobactin, respectively, though all are transported into the cytoplasm by the same BPD ABC transporter, FhuBDC (143, 255). It is also remarkable that these OMRs display higher affinities for their substrates than does the periplasmic BP FhuD (256). There are currently crystal structures of the following six OMRs: FhuA (150, 286), FepA (55), FecA (149), BtuB (72), FpvA (77), and FptA (78). They show a remarkably conserved organization composed of two domains, a 22-stranded β-barrel and an N-terminal globular domain called a plug that lies within the barrel.
Some OMRs are also involved in the regulation of transcription, signaling from the cell surface to the cytoplasm via a signaling cascade (32, 186). A cascade signals the presence of the inducer in the culture medium to the cytoplasm, where gene transcription occurs. For example, when the OMR FecA binds its substrate, iron dicitrate, signaling is mediated through interaction between the N-terminal domain of FecA and the cytoplasmic membrane signaling protein FecR. Activated FecR interacts with the FecI sigma factor, leading to initiation of transcription of the fecABCDE transport genes (47).
New results are expanding the range of potential substrates that can be transported by OMRs. A very peculiar system, described for a Sphingomonas sp., was found to be expressed in alginate-induced cells. Alginate is a high-molecular-mass polysaccharide (25,000 Da) which is thought to enter the cell intact, since alginate-degrading enzymes are located exclusively in the cytoplasm (188). A specific outer membrane organelle, called the “pit,” is thought to mediate alginate uptake through the outer membrane. Alginate-induced outer membrane proteins are similar to TonB-dependent receptors, raising the possibility that some members of this family might be involved in polysaccharide uptake (187). In addition, the outer membrane protein SusC, similar to TonB-dependent receptors, is maltose inducible and essential for maltose and starch uptake in Bacteroides thetaiotaomicron (395). More recently, transport of maltose in Caulobacter crescentus was found to be mediated by MalA, a homologue of high-affinity OMRs, in an energy-dependent, TonB-independent, and ExbBD-dependent fashion (331).
Finally, mycobacteria possess an outer lipid bilayer with low permeability, rendering them intrinsically resistant to many antibiotics. Hydrophilic substances cross this membrane via porins, whose importance in phosphate uptake was highlighted recently (530).
BPD Uptake Systems
BPs bind their substrates with high affinities, in the range of 0.01 to 1 μM. This high-affinity binding is clearly responsible for the efficiency of BPD transporters at low substrate concentrations; cells can concentrate nutrients up to 106-fold when the nutrients are present at submicromolar concentrations in the external milieu (118). However, BPs are still essential for transport even at high substrate concentrations, as demonstrated by deletion of the gene encoding the maltose-BP in E. coli (451). BPs located in the periplasmic space of gram-negative bacteria can be released by a cold osmotic shock, and this procedure also leads to inactivation of transport due to the loss of the protein. In either circumstance, transport can be restored if BPs are introduced back into the periplasmic space (45).
The range of substrates that are transported by BPD transporters is extremely diverse, including mono- and oligosaccharides, organic and inorganic ions, amino acids and short peptides (124), iron-siderophores, metals, polyamine cations, opines, and vitamins. Most transporters are specific for a single substrate or for a family of structurally related substrates, such as maltose and maltodextrins. However, some BPD transporters are more versatile, handling structurally unrelated substrates. Versatility can be achieved in two ways. First, a single BP can have a wide substrate specificity, as illustrated by the multiple-sugar transporter Msm of Streptococcus mutans, which recognizes melibiose, sucrose, raffinose, isomaltotriose, and isomaltotetraose (409, 474). Second, multiple BPs with different binding specificities can interact with a single transporter, as illustrated by the histidine, lysine, and arginine transport system in Enterobacteriaceae (194) and the oligopeptide/muramyl peptide transport system of E. coli (353).
BPs are monomeric; structural and kinetic analyses indicate that there is only one substrate-binding site per BP (387), and the structures of the ModABC, BtuFCD, and MalEFGK2 transporters reveal just one BP interacting with the membrane protein complex (205). However, unusual BPD transporters, in which either one (OpuA) (27) or two (GlnPQ) (434) extracytoplasmic substrate-binding domains, analogous to BPs (497), are fused to the termini of the inner membrane transport proteins, have been described for Lactococcus lactis. These transporters, which function as dimers, therefore have either two or four substrate-binding domains tethered to the transporter. This unique architecture offers the potential to enhance rates of substrate delivery to the transporter via improvements in proximity and orientation. While deletion experiments revealed that just one substrate-binding domain is absolutely required for transport in the OpuA system, if both substrate-binding domains are present, they interact in a cooperative manner to enhance transport activity (27). The transporter displays positive cooperativity with respect to the transported substrate concentration when both substrate-binding domains are present and functional, and interestingly, transport rates remain high when reversible substrate binding to one substrate-binding domain is blocked through modification by a covalent substrate mimic. It is possible that the physical presence of one domain facilitates either the docking of the second substrate-binding domain or the transfer of the substrate to the TM domain.
Research into BPs has been driven by the desire to understand (i) how a BP recognizes its substrate, (ii) the thermodynamics of the binding interaction, and (iii) the nature of the conformational changes that take place upon substrate binding. Several excellent reviews discuss the structure and ligand-binding interactions in detail (146, 388, 389, 524). More recent work addresses these questions in greater detail and also begins to address the question of how BPs interact with transporters, as they are clearly integral to the mechanism of translocation.
Conformational changes in periplasmic BPs.
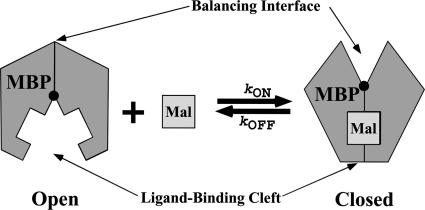
Most of the knowledge on substrate-BPs comes from the study of proteins from gram-negative bacteria, as only a few substrate-binding lipoproteins from gram-positive organisms and archaea have been characterized, either biochemically or structurally (242, 275). High-resolution structures reveal that all periplasmic BPs characterized to date adopt a similar folding pattern made of two globular domains or lobes, called the N and C lobes since they contain the N and C termini of the protein. These lobes are connected by one or more polypeptide chains, and the substrate binds between them (388, 524). Each lobe is composed of an alpha-beta fold consisting of pleated β-sheets surrounded by α-helices and connected by loops. Polypeptide is distributed between the two domains and in the number and order of strands in each domain (162). Type I BPs include simple sugar (arabinose and ribose)- and branched-chain-amino-acid-BPs, and type II BPs include the maltose-maltodextrin-, phosphate-, and sulfate-BPs (524). In the ligand-free conformation, the domains are well separated, with an open, solvent-accessible cleft between them. The ligand occupies this cleft and induces a substantial domain rotation, resulting in closure of the two domains (Fig. 8). More recently, a novel BP type, type III, was recognized, encompassing both the divalent cation (Mn2+ and Zn2+)-BPs and iron/siderophore/vitamin B12-BPs. In this class, the two lobes consist of a central five-stranded β-sheet surrounded by α-helices and the domains are connected by a single α-helix spanning the length of the protein (37, 271). While ligand binding in type III BPs also occurs in the cleft, high-resolution structures suggest that binding is not accompanied by a large domain movement (235, 435). Molecular dynamics simulations, however, suggest that these BPs undergo a breathing motion, similar to that seen in other families, that may allow binding and release of ligand (231). Intriguingly, a recent structure of BtuF, the vitamin B12-BP, determined in complex with the cognate transporter, reveals a more open conformation (217).
FIG. 8.
Structures of periplasmic maltose-BP. Structures of maltose-BP in the open, unliganded (blue; PDB accession no. 1ANF) and closed, maltose-bound (pink; PDB accession no. 1OMP) conformations are aligned based on the positions of α-carbons in the C lobe. The offset of the N lobes illustrates the domain rotation induced by ligand binding to class I and class II BPs.
A substantial body of evidence demonstrates that type I and II BPs are open in the absence of substrate and closed in the presence of substrate, undergoing a ligand-induced conformational change that is central to the mechanism of translocation. In addition to crystal structures of proteins in open and closed conformations, a variety of other techniques have been used to monitor ligand binding and conformational change, including the use of intrinsic (tryptophan) fluorescence (468, 519), extrinsic fluorescent probes (113), and fluorescence energy transfer (115). Recent developments in assaying conformational changes by using fluorescence have been driven by the desire to develop nanoscale biosensors (135). For example, the phosphate-BP, with an extrinsic fluorophore attached to Cys197, is routinely used as a sensor for Pi in ATPase assays (51, 200). Changes in distance between domains have been measured using site-directed spin labeling and electron paramagnetic resonance (EPR) (184). Small-angle X-ray scattering detects changes in protein size associated with closure (341, 478), and nuclear magnetic resonance (NMR) analysis can measure the average angle between domains via dipolar couplings (456).
(i) Bending at the hinge.
Several BPs have been crystallized in multiple open conformations, making it possible to visualize the trajectory of domain closure. For the leucine, isoleucine, valine (LIV)-BP, comparison of open and closed forms reveals that the conformational change is essentially a domain rotation mediated almost exclusively by a change in the backbone torsion angles of just two residues, Asp123 in the first interdomain strand and Phe329 in the third interdomain strand (486). Both the ribose-BP and the allose-BP have been crystallized in three different open conformations (33, 298). In both examples, the three structures lie along a single trajectory, indicating that the multistranded hinge restricts the degrees of freedom of motion. In contrast to the LIV-BP, embedded H2O also participates in the torsional changes in the hinge associated with movement in these proteins (33, 298). Despite 35% primary sequence identity between ribose-BP and allose-BP, domain opening occurs in different directions and involves torsional changes in different residues in the hinges (298). This difference in direction may reflect constraints imposed by differences in the mode of ligand binding and/or the ligand structure. Because the hinge-bending motion consists of both a rotation and a twist, the ligand is expected to bind initially to just one lobe of the open BP, and the pathway of closure must not dislodge the ligand from its binding site. The trajectory is also likely to be important in the interaction of the BP with a transporter, as the BP and transporter interact and may move in unison during the transport process (69).
(ii) Energetics of domain closure.
An interesting question is whether BPs remain open until substrate binds or have open and closed forms that are always in equilibrium. The crystal structure of an unliganded BP in the closed conformation suggests that at least some BPs can close in the absence of ligand (158). The presence of multiple open conformations in crystal structures of LivJ, allose-BP, and ribose-BP also supports the notion of conformational flexibility in the absence of ligand (33, 158, 298, 486). For maltose-BP, time-resolved tryptophan fluorescence anisotropy analysis found substantial orientational fluctuations of the two domains in the absence of ligand that disappeared upon maltose binding (128). Using paramagnetic NMR, Clore and colleagues were able to detect a small fraction (5%) of a partially closed species of maltose-BP in rapid equilibrium with open BP in the absence of maltose (472). Compared to the closed liganded maltose-BP, the N- and C-terminal lobes in this partially closed species are offset by a 6° translation, avoiding highly unfavorable interactions that would occur in the closed sugar-binding cleft in the absence of maltose.
A thermodynamic cycle of BP similar to that shown in Fig. 9 is often presented for the ligand-induced closure of a periplasmic BP (94, 305). A key point to make is that the observed Kd (dissociation constant) for binding is the product of a binding event and a conformational change. As a consequence of the fact that the equilibrium between closed and open forms is strongly influenced by ligand binding, the ligand will be held less tightly by the open than by the closed form. In fact, because substrate is completely buried in the closed conforma-tion, substrate binding is likely to precede closure in the binding pathway.
FIG. 9.
Thermodynamic cycle for ligand binding to a periplasmic-BP. Munliganded and Mliganded are the equilibrium allosteric constants for the interconversion of the open and closed forms of the BP in the absence and presence of ligand (filled circles). Kopen and Kclosed are the theoretical equilibrium association constants for the binding of ligand to open and closed forms of the BP. (Reprinted from reference 94.)
New insights into the energetics of domain closure have been gained in recent years from studies directed at perturbing the equilibrium between the open and closed forms of BPs. In maltose-BP and ribose-BP, though not all BPs, a set of residues located in both domains on the opposite side of the hinge from the substrate-binding cleft have been identified that are closer together in the open conformation and move apart in the closed conformation to create a surface crevice (305), later designated a “balancing interface” (478) (Fig. 10). Importantly, mutations in this region are distant from the site of substrate binding and are not likely to perturb binding directly. Placement of a fluorophore at position 95 in this interface results in a strong fluorescence increase upon maltose binding and closure (304), which was later traced to the loss of specific hydrogen-bonding interactions between the fluorophore and two tyrosine residues upon opening of the balancing interface (90). Insertion of residues with side chains of increasing size into the balancing interface created a set of maltose-BP mutants that bound maltose with increasing affinities (94, 305), leading to the suggestion that the bulkier residues destabilized the open form and shifted the equilibrium from open to closed. Since the ligand-binding affinity is a function of both ligand binding and domain closure, a shift in the equilibrium toward the closed state would be expected to increase the affinity for maltose in this system (305). In an elegant NMR-based experiment, Kay and colleagues clearly demonstrated that this series of mutations dramatically affects the average degree of opening of the cleft of the apo-maltose-BP and that there is a linear relationship between the decrease in angle opening and an increase in maltose-binding affinity (315). The orientation of the two domains in solution relative to the applied magnetic field was determined by examining residual dipolar couplings and applying the data to the known X-ray crystal structures of maltose-BP in the open or closed conformation (320, 456). Intriguingly, there was also a decrease in stability of the apoproteins in this series of mutants, as judged by the ΔG of unfolding, that correlated with the decrease in angle opening, suggesting an energy cost associated with closure that is normally balanced by the gain in energy realized through increased binding of maltose to both lobes of maltose-BP (315). In the absence of the necessity to overcome this energy cost to close, the mutant proteins bound maltose with a higher affinity, a change that is clearly associated with a decrease in the rate of dissociation of maltose from maltose-BP (305). Mechanistically, these observations may be quite important, as in the absence of maltose, this energy cost would keep maltose-BP predominantly in the open conformation, an important regulatory feature to avoid inappropriate activation of the transporter in the absence of substrate. The stabilizing influence of the balancing interface in maltose-BP was also compromised by either deleting residues lining the cleft or substituting smaller side chains at these positions (478). These mutations also increased the affinity for maltose to the same degree as the bulky substitutions that likely prevented full opening did, although, somewhat surprisingly, they did not alter the average conformation of the apo-maltose-BP, as judged by small-angle X-ray scattering. As discussed above, it may be disruption of the conformational equilibrium of the liganded maltose-BP rather than the apo-maltose-BP that is most important, since ligand cannot bind to closed maltose-BP (Fig. 9).
FIG. 10.
Balancing interface in a BP. Cartoons of maltose-BP in open (unliganded) and closed (liganded) conformations depict the ligand-binding cleft and balancing interface. (Reprinted from reference 478 with permission of the publisher.)
Specificity of substrate-BPs.
Substrate-binding specificity is mediated primarily by H bonding in most BPs (388, 524). Some BPs display high specificity for one ligand over another, as illustrated by the oxyanion-BPs, while others bind a narrow range of similar substrates, as illustrated by the amino acid-BPs.
Despite similarities between the oxyanions phosphate, sulfate, and molybdate, the respective BPs exhibit exquisite specificity for their substrates (209, 225, 297, 387, 511). In each of the BPs, these molecules are accommodated through ion-dipole interactions consisting of hydrogen bonds with backbone NH and side chain OH groups of the protein. These interactions efficiently overcome the hydration energy of the ions, as the ligands are desolvated when buried in the cleft (362). The basis for discrimination between phosphate and sulfate relies on the protonation of phosphate at physiologic pH. Phosphate-BPs contain one or more negatively charged amino acids that will accommodate only protonated oxygens, thereby excluding sulfate, which is fully ionized at all pHs (387, 511). The basis for discrimination between sulfate and molybdate relies on the difference in size of these oxyanions (209, 266). An extended array of hydrogen bonds serves to exactly position the dipoles to bind just one substrate. An example of amino acid binding is illustrated by the LIV-BP LivJ (486). Three separate structures have been determined, each with one of these three branched-chain amino acids bound. In each case, the cleft has closed, completely dehydrating and engulfing the ligands. Superposition of the three closed structures reveals that the amino acids are bound in very similar configurations, with the amino and carboxyl groups of the amino acids held by hydrogen bonds, with six donated from domain I and two donated from domain II. In contrast, the aliphatic side chains lie in a hydrophobic pocket, and 80% of the contacts are mediated by nonpolar van der Waals interactions, mediated mainly through contacts with domain I. The binding pockets are designed to hold only l-amino acids, as shown experimentally (10).
The periplasmic maltose-BP binds and transports both maltose and linear maltooligosaccharides. In structures of closed liganded maltose-BP, the sugars are buried in the groove between the two domains, and binding specificity is mediated through hydrogen bonding interactions with residues located predominantly in one domain and through nonpolar interactions with aromatic residues located in the other domain (446). Structures of open ligand-bound maltose-BP reveal that the sugars bind exclusively to the domain with the aromatic residues (130, 131). Initial binding of ligand to just one of two lobes prior to closure is seen in a second example, where crystals of an open BP were soaked in leucine (410), and may be a general feature important for BP function. Simulation of domain movement by molecular dynamics and normal-mode analysis also predicts that the ligand remains bound to one domain upon opening of a BP (486).
Oligopeptide-BPs are designed to accommodate peptides of different compositions and lengths. In gram-negative organisms, where peptides must first traverse the outer membrane through porin proteins, dipeptide- and oligopeptide-BPs (Dpp and Opp, respectively) optimally bind either dipeptides or peptides of up to five amino acids in length (124). In contrast, a nonapeptide was found in the crystal structure of the peptide-BP AppA from a gram-positive organism (275). Peptides are bound within the cleft via hydrogen bonds connecting the main chain of the protein to the main chain of the peptide. In AppA, the N-terminal α-amino group of the peptide is bound by an Asp and the C-terminal α-carboxyl group interacts with an Arg, positioned where it can also interact with the C termini of shorter peptides. The side chains are hydrated and lie in distinct “voluminous” pockets that make fewer direct contacts with the peptide and do not restrict the type of side chain. Residues that do contact substrate side chains can function as both hydrogen bond acceptors and donors, increasing flexibility, and they can mediate contact directly or via H2O. These binding sites function to combine high-affinity binding with broad substrate specificity to transport almost any peptide. The nonspecific nature of substrate binding was demonstrated through binding of peptides generated as combinatorial libraries to OppA of the gram-positive organism Lactococcus lactis (114). In these experiments, almost all peptides were bound, although some preferences for particular side chains were observed, in particular at positions 4, 5, and 6 in a BP that favors nonapeptides as substrates. In contrast to most BPs, the family of oligopeptide-BPs contains a third domain and a portion of the nonapeptide binding site in AppA extends into this region, providing the first suggestion of a possible function for this domain (275). OppA from L. lactis has only six pockets to accommodate side chains (264) but can bind and transport peptides of as large as 35 amino acids. Therefore, peptide substrates are not always completely enclosed by the BPs (123).
Substrate recognition by membrane transporters.
Does BP specificity dictate transport specificity, or do the membrane transporters also contribute to specificity? In in vitro assays with the oligopeptide transporter OppA, every peptide that is bound by OppA, even peptides of up to 35 amino acids in length, is also transported, suggesting that the membrane transporter imposes few or no further restrictions on specificity (123). In vivo studies contradicting this conclusion for the Opp transporter (65) have been challenged (123) on the basis that the metabolic breakdown and release of some transported peptides may have obscured their uptake. In several systems, including the histidine system, two BPs with different substrate specificities interact with the same transporter, demonstrating that the membrane transporters accommodate greater substrate diversity than the BPs do (194). In the maltose system, a variety of maltooligosaccharides can bind to maltose-BP, and transport specificity may be dictated by which ligands trigger closure of maltose-BP (183, 184). There may also be a limit on the length of maltodextrins that can be transported; while maltose-BP can bind longer dextrins, transport has been demonstrated only for sugars of up to seven glucose residues (147).
Evidence that the membrane components have a binding site for substrates comes from the isolation and characterization of mutant transporters that have gained the ability to transport substrate in the absence of their BPs (451, 488). These mutants were first isolated from the maltose system, and the membrane transporter still displayed specificity for maltose in the absence of BP. The apparent Km for maltose in the transport reaction was, however, greatly increased, confirming the importance of BPs for high-affinity uptake. BP-independent mutants of the histidine transporter supported growth on both histidine and arginine, and although rates of transport were too low to measure, the affinity for histidine decreased from 10 μM to 0.5 mM, suggesting the presence of a low-affinity substrate-binding site in the membrane (459). Random and site-directed mutagenesis of the membrane-spanning domains of the maltose transporter has generated mutants with altered specificities (137, 464, 475, 517), some of which transport maltose but not maltodextrins and vice versa. In one of the most remarkable alterations in substrate specificity, an amber mutation that led to the premature termination of the TM protein MalF permitted growth of genotypically Lac-negative cells on lactose (313). Transport required maltose-BP, though lactose is not bound by maltose-BP, suggesting that the truncation allowed access of lactose to the TM translocation pathway. Intriguingly, residues responsible for maltose binding in the TM subunits are found exclusively in MalF, not in the second TM subunit, MalG (343).
Coupling of transport to hydrolysis.
Since ABC transporters functioning in efflux do not use BPs, BPs are sometimes viewed as accessory proteins that impart upon bacteria the ability to transport nutrients with a high affinity. BPs are, in fact, integral parts of the translocation machinery for importers and are essential for function because they play an essential role in coupling of transport to ATP hydrolysis.
(i) Interaction of BP with transporter.
The first step in transport can be viewed as substrate recognition by the BP. Since binding triggers a conformational change that generally engulfs the substrate, features of closed BPs that are distinct from those of open BPs are likely recognized by the transporters. The surface of maltose-BP that interacts with the transporter has been defined by mapping mutations that affect transport (134, 303, 467, 487) onto the structure of maltose-BP. These residues lie on either side of the substrate-binding cleft, defining a potential binding site for the transporter that would differ in the open and closed forms. A genetic suppressor analysis further suggested that the C lobe of maltose-BP interacts with MalF and the N lobe of maltose-BP interacts with MalG (80, 207, 487), which would position the substrate-binding cleft close to the interface between MalF and MalG. Indeed, both the structures of ModA in complex with ModB2C2 and of maltose-BP in complex with MalFGK2 reveal just such an interaction (205, 343). The chemotactic response to maltose is based on a similar recognition strategy.
Curiously, both His-BP and maltose-BP bind to their transporters with similar affinities in the presence or absence of substrate (14, 18, 312, 380), and excess free BP competes with liganded BP, inhibiting transport (379). In early studies probing the interaction between His-BP and the transporter, the extents of cross-linking with one reagent were similar for both ligand-bound and ligand-free His-BP, while another reagent cross-linked much more strongly with ligand-bound than with ligand-free His-BP, suggesting a difference in the mode of interaction (380). The pattern of immobilization of spin labels attached to the surface of maltose-BP suggests that just the N lobe mediates the initial interaction of maltose-BP with the transporter, and this observation may account for the similar affinities of open and closed BPs (18). Consistent with this idea, the N lobe of maltose-BP could be cross-linked to MalG in both the presence and absence of maltose (91).
(ii) BPs stimulate ATP hydrolysis.
Insight into the interaction between BPs and transporters comes from the observation that the addition of BP stimulates the in vitro ATPase activity of the transporters (100, 281). In the maltose and histidine transport systems, rates of ATP hydrolysis are 6- to 10-fold higher in the presence of the transported substrates than in their absence, although the ligand-free BP can also stimulate hydrolysis (100, 361). Maltose analogues that fail to induce closure of maltose-BP do not stimulate ATPase activity and fail to be transported, suggesting that maltose-BP needs to be in the closed conformation to initiate a cycle of transport and hydrolysis (183). The ability of ligand-free BP to stimulate activity could reflect its ability to close in the absence of substrate and indicates a potential for futile cycling of ATP hydrolysis in vivo if the BPs are expressed but not saturated with substrate.
A transition state analogue for ATP hydrolysis, consisting of Mg, ADP, and vanadate, traps a high-affinity complex of maltose-BP and MalFGK2, suggesting that maltose-BP stimulates ATP hydrolysis by stabilizing the catalytic transition state of the nucleotide (69). In the absence of the analogue, the affinity between maltose-BP and MalFGK2 is estimated to be in the range of 25 to 100 μM, based on the Km for maltose-BP in the transport reaction (106, 300). Hence, the transporter must pass only transiently through this high-affinity conformational state to stimulate hydrolysis. Maltose is most logically transferred from maltose-BP to MalFGK2 while these proteins are tightly bound, thereby coupling transport to ATP hydrolysis.
(iii) Conformational changes associated with transport.
A considerable amount of data, both structural and biochemical, supports a model in which nucleotide binding induces closure of the NBDs. Distance measurements between lobes of maltose-BP, obtained from spin-spin interactions in EPR experiments (184), indicate that maltose-BP is open when it is tightly bound to the transporter (18). [14C]maltose is not tightly bound in this conformation, consistent with the fact that maltose-BP is open (18). From the accumulated data, a model for transport by BPD ABC transporters evolves (Fig. 11), in which, in the absence of substrate, maltose-BP is open and the transporter rests in a periplasmic closed (P-closed) conformation, even in the presence of ATP. We suggest that binding of maltose triggers the closure of maltose-BP and that interaction of the closed, liganded BP stimulates ATP hydrolysis by allowing the NBDs to close. In our model, we believe that these changes occur in one concerted action in which maltose-BP becomes progressively more tightly bound to the transporter as it opens and as the NBDs close. Though little is known about the nature of conformational change in the TM domains, this second conformation has been designated P-open, to indicate that conformational changes known to occur in the peripherally associated proteins (maltose-BP and the MalK dimer) may promote movement of adjacent helices in similar directions. In this way, a cytoplasmic exit from the translocation pathway closes while the periplasmic entrance opens to receive the substrate from the BP (Fig. 11). Tight binding between the BP and transporter would ensure efficient transfer of maltose to the transporter when maltose-BP is open. Schneider and colleagues have detected, by protease susceptibility analysis, conformational changes in the extracellular loops of MalF and MalG associated with this conformational transition (93).
FIG. 11.
Concerted model for maltose transport. In the absence of maltose, the MalFGK2 transporter rests in a conformation in which the NBD dimer interface is open and the translocation pathway is exposed only to the cytoplasm (P-closed conformation). On binding of maltose, the periplasmic maltose-BP undergoes a conformational change from open to closed, and interaction with the closed maltose-BP in the presence of ATP triggers a global conformational change in which the NBDs close to promote ATP hydrolysis, maltose-BP becomes tightly bound to MalFGK2, and both maltose-BP and MalFGK2 open at the periplasmic surface of the membrane to facilitate the transfer of substrate from maltose-BP to a binding site in the membrane (P-open conformation). Following ATP hydrolysis, which destabilizes the NBD dimer, the transporter returns to the resting state and maltose completes its translocation across the membrane. (Adapted from reference 69 with permission of the publisher. Copyright 2001 National Academy of Sciences, U.S.A.)
A site of interaction between the IM domains and the NBDs in the maltose transporter was first identified by genetic and biochemical approaches as a short, loosely conserved pattern, the EAA motif, located in a cytoplasmic loop of MalF and of MalG (319). Secondary structure predictions suggested that the EAA region is structured, forming two short α-helices separated by a loop containing a highly conserved glycine residue (421). Mutations in the EAA motif of either MalF or MalG slightly affect or do not affect transport, but a synthetic defective transport phenotype arises when both mutations are present in the same transporter. The observation that MalK is found in the cytosol in some of these maltose-negative mutants, as seen in strains in which the malF and malG genes are deleted, suggests a role in subunit interaction. Suppressor mutations in malK that restore transport were selected from these maltose-negative mutants, and they mapped mainly in the helical domain of MalK, consistent with the idea that the EAA regions constitute a recognition site for the helical domains of ABC ATPases. The proximity of residues in the EAA motifs of MalF and MalG to MalK was further confirmed through cross-linking experiments (215). The hypothesis that the EAA motif constitutes a site of interaction between the IM proteins and the conserved ABC modules (89, 244) is clearly proven correct by structural studies (205, 285, 343, 366).
The simplest model for coupling of ATP hydrolysis to conformational changes in the IM domains would involve the movement of the EAA loops, in concert with the helical domains of MalK, further apart and closer together as the MalK dimer opens and closes. This type of loop motion may be sufficient to mediate the gating of the IM region, although other mechanisms, which may involve nucleotide-dependent changes in the nature of the interaction of each individual NBD with its corresponding IM domain, may also come into play.
Intriguingly, incubation of ATP with the intact transporter in the absence of Mg2+ to prevent hydrolysis also triggers formation of a complex of transporter with maltose-BP bound in the open conformation (18), though a stable complex is not formed, as the addition of Mg2+ will allow turnover and return the system to the resting state. The nonhydrolyzable ATP analogue Mg·AMP-PNP also triggers the formation of this conformational state (18). The conformational change from the P-closed to P-open state is therefore not solely a property of the catalytic transition state; the binding energy obtained from both ATP binding under noncatalytic conditions and maltose-BP binding is sufficient to stabilize the P-open conformation.
If BPs and transporters are in contact when they open at the periplasmic surface of the membrane, then binding of different ligands could lead to subtle variations in the trajectory of rotation of the BP or in the initial degree of closure, which could affect the rate of transport. In the histidine transporter, where both the histidine-BP (HisJ) and the lysine-arginine-ornithine-BP bind multiple ligands, the Kd for binding of substrate to the BP does not correlate with the Km for substrate transport (528), suggesting that other factors do influence translocation (527). Although crystallization experiments do not reveal differences in the conformational states of BPs bound to different substrates (339, 554), conformational differences in HisJ bound to different ligands are measurable by several other criteria (258, 527). While no ligand-dependent differences are found in the equilibrium between open and closed forms of BP, one factor that correlates with the ability of different ligands to stimulate ATPase activity is the rate of decay of intrinsic (tryptophan) fluorescence anisotropy, a measure of protein dynamics (258). It was suggested that there may be only one orientation of the two domains that induces ATPase activity and that different ligands alter the degree of interdomain motion in the closed conformation, thereby affecting the frequency with which the proper orientation is achieved.
For type III BPs, those that do not obviously undergo ligand-induced domain rotations, including BtuF (235) and FhuD2 (435), the mechanism of coupling of transport to hydrolysis described herein may differ. In proteoliposome-based assays, the first reported for a type III BP (38), the addition of the BtuF protein does stimulate the ATPase activity of the transporter, though only ∼2-fold, as the transporter has a high basal activity. BtuF displays a high affinity for the transporter, suggesting that it may not cycle on and off the transporter as depicted for maltose-BP. Also in contrast to the case for maltose-BP, the presence or absence of vitamin B12 has no effect on the ability of BtuF to stimulate ATPase activity, a result that is consistent with the idea that BtuF does not undergo major conformational changes in response to substrate binding.
BP-independent mutants may mimic the P-open conformation.
Several lines of evidence suggest that mutations in the maltose and histidine transporters that allow transport in the absence of BP have altered the conformational state of the transporter. In contrast to the case for the wild type, where BP stimulates ATPase activity, these mutant transporters have gained the ability to hydrolyze ATP in the absence of BP (100, 361). The addition of substrate without BP does not affect the rate of ATP hydrolysis, consistent with the hypothesis that the conformational state of the BP, rather than the interaction of the transporter with the transported substrate, influences the rate of ATP hydrolysis (100). Fluorescence and protease sensitivity studies of the MalF500 mutant revealed that the BP rests in a conformational state that is more similar to that of the vanadate-trapped transporter than to the nucleotide-free conformation (93, 299). These mutations may allow the BP-independent transporters to undergo conformational changes necessary for transport (closure of NBDs and opening of the periplasmic surface) without the added energy obtained from BP interaction that is essential for the wild type. Maltose-BP binds with higher affinity to the MalF500 transporter than to the wild type in the absence of nucleotide (299). In keeping with the model in Fig. 11, we suggest that the P-closed conformational state presents a low-affinity binding site for BP, while the P-open conformation presents a high-affinity binding site for BP. A functional consequence of the untimely exposure of this high-affinity binding site in MalF500GK2 is that maltose-BP inhibits maltose transport by MalF500 (488) without drastically altering the ATPase activity (100). The ability of open maltose-BP to dock to the BP-independent transporter may disrupt the design of the coupling mechanism. Interestingly, reduced maltodextrins, which bind in a mode that favors the open over the closed conformation of maltose-BP in solution, are transported by the BP-independent MalG511 transporter but not by the wild-type transporter (130, 183). In this example, partially or fully open maltose-BP may be accommodated by the transporter either partially or fully in the P-open conformation, thereby restoring a productive interaction.
BP-independent mutants of the histidine transport system also gain the ability to hydrolyze ATP in the absence of BP. However, the addition of BP to BP-independent mutants consistently restores high-affinity transport (459) and further increases the rate of turnover of ATP (361). Since the rates of transport by these mutants were too low to be measured readily, they may represent a population of transporters in which the energy barrier associated with conformational change is just marginally overcome by mutation. Interestingly, the point mutations leading to BP-independent transport in this system are located exclusively in the ATPase subunit, HisP, while those in the maltose system are located exclusively in the TM subunits, MalF and MalG (80, 459). Another difference is that only single point mutations were observed in HisP, while almost all BP-independent mutants in MalF/MalG contained two point mutations. Several of the mutations in HisP lie in a hinge between the helical domain and the RecA-like domain that could affect the energetics of ATP-induced closure of NBDs to allow transport in the absence of BP (227). One distinction between the maltose and histidine systems may lie in the presence of the RD of MalK, which could impact the energetics of domain closure, requiring a different mechanism to overcome the need for BP. Alternatively, the stringency of the screen may have differed substantially, as the mutants for maltose had mutations in the chromosome and displayed robust transport, while the mutants in the histidine transporter had mutations in an overexpressing plasmid, and marginal increases in ATPase activity might have been sufficient to allow growth in the absence of BP.
Structures of BPD importers.
The desire to obtain high-resolution structural data has fueled an interest in archaeal ABC transporters, as thermostable proteins may stand up better to the rigors of crystallization than their bacterial counterparts do (177, 423, 508). Several recent reviews discuss the unique properties of the archaeal transporters (4, 74, 270). The molybdenum transporter from Archaeoglobus fulgidus is the first such success for an intact ABC transporter (205). Atomic-resolution structures of three other bacterial importers have also been determined (285, 343, 366). All of these structures provide detailed pictures of how the IM domains interact with the ABC domains and with each other. Furthermore, they reveal two different conformations of the transporter, with putative substrate pathways opening in two opposite directions (Fig. 12). Structural comparison of these conformations provides invaluable information on how substrates are alternately exposed to opposite sides of the membrane, a key feature proposed for all transporters.
FIG. 12.
Structures of four importers. All transporters consist of four subunits, including a homodimer or heterodimer of IM subunits and a homodimer of NBD subunits. The ModB2C2 transporter is cocrystallized with ModA, a periplasmic BP in a closed, liganded conformation (cyan). The ligand, molybdate, is represented with a space-filling model. Maltose-BP (purple) is trapped in complex with MalFGK2 by using an E159Q substitution in MalK to prevent ATP hydrolysis (343). Nucleotide (ATP) is present only in MalFGK2E. A space-filling rendition of MalFGK2 is cut away to reveal an occluded pocket containing bound maltose. (Lower panel modified from reference 343.)
The IM subunit of BtuCD (285), the vitamin B12 importer, contains 10 TM helices rather than the 6 TM helices seen in most ABC families. Interestingly, the TM helices vary in length, ranging from 12 to 35 residues in BtuCD. Three helices are too short to span the membrane and are compensated for by the presence of several hydrophobic loops folded into the center of the predicted lipid bilayer. The main contacts between the IM and ABC subunits are through a cytoplasmic loop between two TM helices and the Q loop in the ABC, as first demonstrated by Dassa and colleagues (319). The two ABCs are folded as expected from the structures of isolated ABCs, and the nucleotide-binding interface is open, as would be anticipated in the absence of ATP (Fig. 7).
The crystal structure of a BtuCD homolog, the putative metal-chelate transporter of Haemophilus influenzae, HI1470/1 (366), was recently reported. The structures of BtuCD and HI1470/1 appear to represent two different conformational states of an ABC transporter (Fig. 12). The predicted translocation pathway in BtuCD is open to the periplasm and closed at the cytoplasmic side of the lipid bilayer, whereas the translocation pathway in HI1470/1 faces the opposite direction and is open only to the cytoplasm. Intriguingly, this seemingly dramatic change in the orientation of the translocation pathway actually involves rather small changes in the IM and ABC subunits. Comparison of one IM subunit between the two structures reveals that seven TM helices can be superimposed and only three helices shift substantially in their orientation (366). Comparison of both IM subunits in both structures reveals an additional 9° twist of one IM subunit relative to the other in one of the two structures. The two ABC subunits of both structures are different from each other, though both display open conformations compared to the closed, ATP-bound form of MalK (68). The HI1470 dimer closely resembles the semiopen form of MalK, and the BtuD dimer exhibits a more closed conformation.
The complete structure of an archael importer, the putative molybdate transporter ModB2C2 from Archaeoglobus fulgidus, has been determined in complex with its BP, ModA (205). This structure lacks nucleotide, and in keeping with other structures (68, 285, 366) and proposed models (457), its nucleotide-binding interface is open. Compared to the BtuCD and HI1470/1 structures, the IM ModB subunits are smaller, with just six TM helices per subunit, and the architecture diverges from the side-by-side architecture of the former in that TM helix 1 of one ModB subunit crosses over to form part of the IM domain that includes TM helices 2 to 6 of the second ModB subunit. The presumed translocation pathway through the center of the molecule is closed at the external surface and open to the cytoplasm and is shielded from the membrane bilayer. Intriguingly, the BP is bound to the transporter in an orientation that places the substrate-binding cleft over the presumed gate to the translocation channel, and both lobes of the BP contact the transporter, consistent with earlier predictions for maltose-BP (80, 282, 303). Residues involved in gating access of the translocation channel to the outside of the cell reside in TM helix 3 and TM helix 5 and consist of two highly conserved motifs in the MOI family of molybdate, sulfate, and phosphate importers (205).
The structure of the maltose transporter MalFGK2 from E. coli has been determined at 2.8-Å resolution in an intermediate conformation that likely resembles the catalytic transition state for ATP hydrolysis (343). Substitution of Gln for the catalytically important Glu residue (E159Q) trapped a conformation with ATP bound in a closed MalK dimer and maltose-BP tightly bound on the opposite side of the transporter in an open state, forming part of the boundary of an occluded, water-filled cavity that reached halfway into the TM regions of MalF and MalG (Fig. 12). Maltose was bound in a binding site at the base of this cavity. Interestingly, a periplasmic loop of MalG had inserted into the open substrate-binding cleft of maltose-BP, obscuring the binding site for maltose in maltose-BP, thereby ensuring that maltose was released into the cavity as the nucleotide-binding subunits closed to hydrolyze ATP. This structure is largely consistent with the experimentally derived model shown in Fig. 11 and reveals a conformation of the TM helices which closes the translocation pathway facing the cytoplasm and opens a translocation pathway facing the periplasm. The structure offers a framework for understanding the unidirectional nature of transport anticipated for ABC transporters if the BPs block exit to the periplasm during ATP hydrolysis. Likewise, it raises the question of whether a BP-independent mutant functions in just one direction or if substrate could exit during a cycle of ATP hydrolysis. Interestingly, whole-cell assays monitoring uptake and efflux of radiolabeled amino acids suggest that several BPD transporters are bidirectional and may have a role in the secretion of amino acids accumulating in the cell as a result of metabolism (208). It remains to be seen how these results will be reconciled with the emerging model for BPD uptake.
One feature that is conserved between transporters is the placement of one helix in the EAA loop (319) in a cleft on the surface of the NBDs. This portion of the EAA loop, also referred to as the coupling helix (205), forms the major contact between IM domains and NBDs in ABC transporters and therefore must mediate the coupling of transport to hydrolysis. A similar organization was found for the EAA regions of BtuCD (285), ModB2C2 (205), MalFGK2 (343), and HI1470/1 (366). Interestingly, the conserved Glu residue of the EAA motif forms a salt bridge with an Arg located just after the Walker A motif of MalK (343).
Further comparison of the structures of MalEFGK2 and ModAB2C2 revealed very similar folds for the TM domains (343), consistent with the close positioning of the OSP and MOI families in the phylogenetic tree shown in Fig. 3. Given this similarity, it is reasonable to assume that these two structures are representative of the two conformational states predicted for ABC transporters, namely, an inward-facing resting state (Mod) and an outward-facing transition state (Mal).
Biased molecular dynamics simulations of BtuCD are consistent with the idea that closure of the BtuD dimer, as seen in the ATP-bound MalK dimer, would lead to an opening trend of the IM BtuC dimer toward the periplasm, while further opening of the BtuD dimer would lead to an opening trend toward the cytoplasm (458). Interestingly, in separate simulations of BtuCD with ATP bound (222), NBD closure did not occur unless the BP BtuF was docked at the periplasmic surface, and as alluded to earlier (see “Role of Two Nucleotide-Binding Sites”), only one of the two ATP-binding sites actually closed, though outward movement of the lobes of this type III BP was coupled to this closure, as depicted in the model for maltose transport (Fig. 11).
BP-Independent ABC Importers
While the vast majority of importers are class 3 BPD importers, several exceptions have been discovered. In an earlier bioinformatic analysis, we identified a family of ABC transporters sharing similarity with the putative cobalt transporter of Salmonella (87). These transporters form two subfamilies in our phylogenetic classification, namely, CBU for cobalt transporters and Y179 for transporters of unknown substrate specificity. In addition to sequence conservation, these transporters share the overall organization of their subunits, justifying their assignation to the same family (CBY). Their distinguishing characteristic is that they apparently lack a typical extracytoplasmic BP. The union of functional genomics and experimental investigation has revealed an unexpected complexity in these two subfamilies of systems, as discussed below.
Cobalt and nickel transporters (CBU subfamily).
The presence of an ABC transporter for cobalt was recognized when the complete sequences of the loci encoding cobalamin (vitamin B12) biosynthetic enzymes in Salmonella were determined (407). The putative core transporter is comprised of an ATP-binding protein, CbiO, and a cytoplasmic membrane protein, CbiQ, with seven TM helices. In addition, a set of two membrane proteins, CbiM and CbiN, are predicted to form a complex with CbiO and CbiQ. While heterologous expression of CbiMNOQ in E. coli catalyzes cobalt uptake, expression of the CbiMN subunits by themselves also mediates significant cobalt transport, suggesting that the CbiQO ABC transporter may be required for additional energization of a core CbiMN transporter (402).
In a systematic in silico analysis of transition metal transporters, Rodionov and coworkers identified other genes similar to those encoding the CbiMNOQ transporters (402). Analysis of upstream regulatory elements revealed two groups of transporters, those that were likely to be controlled by the availability of vitamin B12 (cobalt transporters) and those that were likely to be controlled by nickel. The putative nickel transporters were renamed NikMNOQ. Heterologous expression of the nikMNOQ operon of Rhodobacter capsulatus in E. coli confirmed the nickel transport activity (402). These transport systems are different from the previously described NikABCDF transporter, which is a “classical” BPD ABC importer similar to the oligopeptide ABC importers (328).
Biotin and hydroxymethyl pyrimidine uptake systems (Y179 subfamily).
In Sinorhizobium meliloti, mutations in the bioM and bioN genes lead to reduced biotin uptake. These two genes share strong similarity with the cbiO and cbiQ genes described above (140). Disruption of the Rhizobium etli bioM gene results in a mutant that takes up biotin at a lower rate than that of the wild type. The R. etli bioMN operon comprises a third gene, bioY, that encodes a putative membrane protein whose inactivation also leads to reduced biotin uptake (181). Heterologous expression of the bioMN-bioY operon and of the single bioY gene of R. capsulatus confers biotin transport activity on recombinant E. coli cells (190). Kinetic analyses led to the notion that although BioY alone displays significant transport activity, the additional expression of BioMN leads to a transporter with a higher affinity for biotin. The Rodionov group also detected Y179 subfamily ABC transporters under the control of the THI element, involved in thiamine regulation, predicted to be involved in the transport of hydroxymethyl pyrimidine, a precursor in the biotin biosynthetic pathway (403).
Class I IM-ABC transporters that may be importers.
Yersinia pestis, the causative agent of plague, makes a siderophore termed yersiniabactin that is used to obtain iron during growth at 37°C. The genes required for the synthesis and utilization of yersiniabactin are located within a chromosomal locus called pgm that contains ybtP and ybtQ, two genes that encode proteins similar in sequence and topology to MsbA and other drug exporters with an IM-ABC organization. Strains containing mutations in these genes are unable to grow on iron-deficient media at 37°C and cannot be cross-fed by culture supernatants from a yersiniabactin-producing strain of Y. pestis. They still produce yersiniabactin, suggesting that YbtP and YbtQ are needed for siderophore uptake (155). This would be the first example of a bacterial importer with fused IM and ABC domains. A very similar system has been found in Mycobacterium tuberculosis, where IrtA and IrtB do not participate in siderophore synthesis or secretion but are required for efficient utilization of iron from Fe-carboxymycobactin (405). However, the substrates and polarity of transport of such systems are still open questions, since a recent report on IroC, which displays the same structural organization as YbtPQ and IrtAB, strongly suggests a role for this ABC transporter in the export of the siderophore salmochelin (81).
EXPORT OUT OF THE CYTOPLASM BY CLASS 1 TRANSPORTERS
Prokaryotic ABC exporters of class 1 (Fig. 2c, diagram H) are abundant and have close homologues in eukaryotes. Two types of systems have been studied extensively, one involved in protein export and the other involved in drug efflux. In both prokaryotes and eukaryotes, ABC proteins have gained notoriety because they can contribute to the resistance of cells to anticancer agents (66) and antibiotics by pumping them out of cells. Proteins secreted from bacteria include toxins, hydrolytic enzymes, S-layer proteins (28, 542), lantibiotics, bacteriocins, and competence factors (350). They also play important roles in biosynthetic pathways, including extracellular polysaccharide biosynthesis (552) and cytochrome biogenesis (374).
Cytochrome biogenesis provides an interesting example of how ABC proteins can be involved in bacterial metabolism in unique ways. Two ABC systems impact the biogenesis of cytochromes c and/or bd in E. coli. While apo-cytochrome c is secreted via the traditional Sec-dependent pathway, the heme group required for function is transported separately and assembly of the holoenzyme occurs in the periplasm (Fig. 13). While early work implicated both CydCD and CcmAB ABC transporters in heme export (374), more recent work has shown that neither is essential for heme transport (79). CydCD may mediate the export of reducing equivalents (cysteine and reduced glutathione) into the periplasm, influencing biogenesis by maintaining a redox environment favorable to the assembly of heme into the heme-binding motif (Cys-X-X-Cys-His) in cytochrome c (172, 173, 367, 368). In the Ccm system, CcmE appears to act as a heme chaperone, and the most recent hypothesis for function is that ATP hydrolysis by CcmAB may mediate the release of holo-CcmE from CcmC, allowing transfer of heme to cytochrome c (145).
FIG. 13.
Cytochrome c-type biogenesis through the CcmA to CcmH and CydDC pathways. CcmA and CcmB (CcmA2B2) may form an ABC transporter for the transport of an as yet undiscovered molecule, and CcmC was proposed to export heme (represented as a cross with a dot) into the periplasm. Alternatively, a CcmA2BC complex may be involved in the export of heme. After transport, heme is transferred to the heme chaperone CcmE. CcmE is proposed to shuttle between CcmC and CcmF, and the protein CcmD has been shown to facilitate the interaction between CcmC and CcmE. Heme is transferred from CcmE to CcmF, which, in association with CcmG and CcmH, forms a heme-lyase complex. CcmG and CcmH maintain the apocytochrome (apo-cytc; green oval) in a reduced state. CcmG interacts with a periplasmic domain of the DsbD protein, which transfers electrons from the cytoplasm to the periplasm. Export of reductants such as cysteine or glutathione by the CydDC ABC transporter would help to maintain periplasmic redox homeostasis. This figure is based on data summarized in references 75 and 480.
Protein Trafficking between and across Membranes
Components of a type I protein secretion system.
Type I secretion systems in gram-negative organisms consist of three components, an IM-ABC-type ABC transporter, a membrane fusion protein (MFP) (117), and an outer membrane factor (OMF) (357), that function together to secrete virulence proteins, including bacteriocins (protein toxins targeting other bacteria), across both the inner and outer membranes, bypassing the periplasm (Fig. 2c, diagram H). OMFs also interact with other types of IM transporters, including resistance-nodulation-cell division (RND) and MFS proteins, to mediate the extrusion of low-molecular-weight hydrophobic substances, including antimicrobials, from the cell (Fig. 2c, diagram G) (see reference 138 for a review). Following substrate recognition, the ABC transporter presumably transports the proteins across the inner membrane and directly into a tunnel formed by the OMF. Some of the most important new insights into function have been obtained through high-resolution structures of these accessory proteins, shown in Fig. 14 modeled in complex with AcrB, an IM-RND-type transporter (199). Based on images obtained by electron microscopy, the periplasm is thought to be 13 to 25 nm wide (132, 175), making the model in Fig. 14 feasible. Several excellent reviews on type I systems are available (111, 203, 204, 542).
FIG. 14.
Model depicting export across two membranes. Both RND and ABC transporters interact with OMFs and MFPs to form a continuous translocation pathway across both the inner and outer membranes of gram-negative bacteria. This model offers one possible juxtaposition, using the structures of the OMF TolC trimer, the MFP MexA, and the RND AcrB trimer. (Reprinted from reference 199 with permission of the publisher. Copyright 2004 National Academy of Sciences, U.S.A.)
Similar protein secretion systems are operative in gram-positive bacteria. Although they lack an OMF, they often have a protein similar to MFP that is essential for export (see reference 314 for a review). Competence-stimulating peptides are one class of proteins secreted from gram-positive bacteria via these systems (213). Lantibiotics, antimicrobial peptides containing posttranslationally modified amino acids such as dehydrated amino acids and lanthionine residues, are secreted only by gram-positive bacteria, and no MFP homologue has been detected in operons carrying lantibiotic biosynthetic genes (350).
In gram-negative organisms, three protomers of the OMF protein join to form a single β-barrel that spans the outer membrane and a channel of 30 Å in diameter that is bounded by long helical coiled coils and extends into the periplasm (1, 254) (Fig. 14, green section). In TolC, the β-barrel domain appears wide open, whereas in the OprM structure, internal loops constrain the size of the opening, suggestive of a gating property. In both structures, the channels taper closed at their periplasmic entrances. Opening of this entrance appears to be achieved by an iris-like motion of the helices (1, 254); mutations have been isolated that increase the current through TolC, perhaps by loosening interactions between helices (16), and disulfide cross-linking between helices designed to restrict such a motion inhibits activity in vivo (142).
The MFP is thought to promote the attachment of the IM-ABC protein to the OMF, though it may also form part of the translocation channel. Crystal structures of the MFP MexA reveal an elongated monomer with an extended α-helical coiled-coil domain and two globular domains consisting primarily of β-strands (2, 199). Substantial biochemical evidence suggests that MFPs exist as oligomers (329, 334, 549), and in crystals, MexA protomers oligomerize in spirals, forming the basis for the model in Fig. 14 (blue section). In contrast to earlier predictions that suggested that MFPs might span the periplasm with their N and C termini bound to the inner and outer membranes, respectively (548), the structures suggest that both the N and C termini are likely to lie in close proximity to the inner membrane. Mutagenesis and suppressor analyses suggest that the C termini of the MFPs interact with the inner membrane transporters (at least in the case of the RND transporters Mex and Acr) (139, 329, 330), while the N terminus may be involved in oligomerization of the MFPs (329).
The IM-RND-type transporter AcrB, which has 12 TM domains and a large periplasmic domain, is a trimer in crystallographic form (323, 376, 544) (Fig. 14, white section). In contrast, IM-ABC components do not appear to have large periplasmic domains, so substantial differences in the mode of attachment with MFPs are anticipated. It may be that the modular structure of MFPs aids in their ability to facilitate interaction between IM proteins of different families and structurally similar OMFs.
Mechanism of action of type I protein secretion.
The pathway by which proteins traverse the inner membrane is not well studied, but two interesting possibilities can be suggested. First, the translocation pathway may lay within the presumed TM helix bundles, as proposed for BtuCD based on the presence of a cavity along the dimer interface large enough to accommodate the substrate (285). Alternatively, multiple ABC proteins could come together in the membrane to form a central translocation pathway, as seen for AcrB. In AcrB, a translocation pathway through the periplasmic domain of the monomer that releases substrates into a centrally located funnel-shaped cavity formed by the trimer has been identified (322, 436). This cavity is closed to the periplasm in the AcrB structure and presumably opens in concert with TolC to pass substrates into the exit channel. The appeal of such a model is based on a symmetry argument, as it would be easier to dock the TolC trimer to an inner membrane trimer of HlyB dimers than to a single HlyB dimer.
(i) Signal for protein secretion.
Most proteins and peptides that are secreted from the cell contain a special signal sequence that allows for recognition by cell secretory systems. For example, proteins destined for secretion by the general, or Sec-dependent, secretory pathway in E. coli and other organisms have a distinctive hydrophobic N-terminal signal sequence that is cleaved off during the translocation process by a leader peptidase (86). In type I secretion systems, secreted protein toxins, including hemolysin (HlyA), have signal sequences at the C terminus that are not cleaved during transport. HlyA belongs to a repeat-in-toxin (RTX) family that is defined by a conserved glycine-rich repeat. This repeat is located just before the signal sequence in the linear sequence of the protein (111). The C terminus of the HasA protein from Serratia marcescens, which is involved in heme acquisition, is cleaved, but by secreted proteases after the transport step (223). While the structure and function of the C-terminal signal have been studied extensively and some conserved primary and secondary structural elements have been identified in this region, a clear consensus on what is required for substrate recognition has not been reached (202, 212, 261); NMR analyses suggest that this region of the protein is largely unstructured (223).
Smaller antibacterial proteins and peptides, including colicin V, bacteriocins, and lantibiotics, carry a signal at the N terminus that is cleaved either by cysteine protease activity associated with a cytosolic N-terminal extension of the cognate ABC transporter or by a dedicated extracellular serine protease (9, 189, 314, 499). These sequences contain a double-glycine motif surrounding the cleavage site.
(ii) Signal recognition by type I machinery.
In contrast to the case for periplasmic BPD transporters, where substrate recognition is understood at the molecular level, the region(s) of the type I secretion proteins that interacts with the signal sequence is unknown. Genetic suppressors of a deletion within the signal sequence of HlyA mapped to regions in the ABC protein HlyB that appear to correspond to the intracellular domain (ICD) of importers, suggesting that this region may be involved in signal recognition (551). More recently, direct evidence of binding between the isolated NBD of HlyB and a truncated HlyA protein containing the C-terminal signal was detected by surface plasmon resonance (24). Interestingly, binding of the cytoplasmic N terminus of the MFP HlyD to HlyA has also been hypothesized as an initial or early step in recognition, since these proteins can be cross-linked in vivo, even in the absence of HlyB (19, 479).
(iii) Protein substrates are unfolded during secretion.
Given the small size of a typical ABC transporter, it is unlikely that folded proteins pass through ABC proteins in the cytoplasmic membrane. SecB, a cytoplasmic chaperone that functions to slow the rate of folding of proteins destined for secretion by the general secretory pathway, slows the folding of HasA and is required for secretion of HasA, suggesting that HasA must be unfolded to be transported (112, 418, 529). If HasA is allowed to fold in the cytoplasm, it inhibits secretion of newly synthesized molecules, suggesting that the folded protein retains affinity for the transporter but is not competent for transport (109). Most proteins that are secreted by type I systems do not require SecB; instead, it has been proposed (204) that the conserved glycine-rich repeat (RTX), present in most type I substrates but absent from HasA, may serve the same function by delaying the folding process. The glycine-rich motifs bind Ca2+, and the scarcity of Ca2+ in the interior of the bacterial cell may prevent the folding of the repeat, facilitating the initiation of the translocation process. Premature folding, as judged by cytoplasmic disulfide bond formation, prevents translocation (152). Furthermore, folding of the same disulfide-containing protein was not affected by the addition of competing sulfhydryl alkylating agents outside the cell, suggesting that folding may commence as the protein traverses the periplasm inside TolC (152). The dimensions of the TolC tunnel (140 Å by 30 Å) are similar to those of other protein folding chambers, such as GroEL and GroES (202, 254). It has also been suggested that Ca2+ needed for folding might be obtained from the lipopolysaccharide (LPS) present in the outer membrane as the proteins leave the cell (204).
(iv) Interactions between components of the type I machinery.
The type I secretion apparatus is not a stable resident of the cell envelope, but rather, it may assemble to allow transport only following binding of the substrate to be transported. A three-step model for translocation emerged from the HasA system. Different combinations of secretory components were coexpressed in E. coli and purified by affinity chromatography, taking advantage of the high affinity of the transport substrate HasA, a heme-binding protein, for heme. Expressed individually, only the ABC protein copurified with HasA, but in combination, the MFP and ABC components copurified with HasA on the heme column, and OMF was found in assembly with HasA only if both the MFP and ABC proteins were also present (274). A similar model was proposed for hemolysin export. Binding of the substrate HlyA to the HlyB-HlyD assembly recruits TolC to the secretion assembly (479). Deletion of portions of the N-terminal cytoplasmic domain of HlyD eliminated secretion concomitant with loss of both cross-linking to HlyA and the resultant interaction with TolC, without disrupting the HlyB-HlyD interaction (19, 365), suggesting that the transport substrate-induced recruitment of TolC is mediated directly through HlyD.
ATP hydrolysis by the ABC transporter HlyB is required for transport but not for the HlyA-dependent recruitment of TolC, since assembly still occurs in a mutant unable to hydrolyze ATP (479). Documented strategies to lock TolC into a closed conformation also did not interfere with transport substrate-induced assembly (16, 142), leaving open the possibility that ATP hydrolysis by HlyB might be harnessed to open the transperiplasmic tunnel to the outside of the cell. Mechanistically, ATP binding and/or hydrolysis has been documented to disrupt the binding of the isolated NBD to the C-terminal signal in HlyA (24), and if this is true, it may serve to promote translocation of HlyA across the membrane after the initial binding step that recruits TolC. Translocation into the periplasm is prevented in the absence of TolC (253), indicating that a tight coupling of transport across the inner and outer membranes is also a part of the regulatory mechanism of this secretion machine.
Drug Efflux Pumps in Bacteria
Drug resistance in bacteria.
Since the discovery of antibiotics in the early 20th century, many pathogenic bacteria have developed resistance mechanisms that abolish the efficiency of therapies, and other species are intrinsically resistant to some antibiotics (5). Bacterial drug resistance has become an increasingly major health problem, reaching a crisis in the last 10 years (277, 514). Several sophisticated mechanisms are involved in these resistance phenotypes, including (i) enzymatic inactivation of antibiotics (101, 531), as seen for β-lactams (43); (ii) modification of the antibiotic target (for example, enterococci synthesize a cell wall precursor with a modified terminal dipeptide, leading to vancomycin resistance) (399); (iii) a decrease in membrane permeability to the antibiotic, resulting from mutation or modification of porin expression (104); and (iv) an increase of antibiotic efflux from the bacterial cell.
Active efflux.
In 1980, Levy and colleagues presented the first evidence that antibiotic resistance was caused by active efflux of a drug (308), although drug efflux mediated by the P-glycoprotein in mammalian cancer cells was described even earlier (230). Because P-glycoprotein is perhaps the best-studied efflux pump and has offered important insights into the mechanism of bacterial pumps (see references 8, 290, 419, 448, and 539 for a review), we will periodically refer briefly to work on this eukaryotic system. Efflux systems are now thought to play an important role in bacterial drug resistance (278, 288, 372). Whereas some efflux pumps are selective for a given substrate, many transporters are able to extrude a plethora of structurally unrelated drugs, conferring a multidrug resistance (MDR) phenotype (263, 278). In addition to a role in antibiotic resistance, some drug efflux pumps also allow the bacterium to survive in the presence of natural substances produced by its host, including bile in the intestinal tract and molecules mediating self-defense (363). Bacterial MDR transporters thus participate in the persistence of bacteria in their ecological environment. Furthermore, some transporters are involved in pathogenicity and are suspected to export virulence factors important for colonization and infection of mammalian cells (363). Interestingly, some bacteria, such as the Actinomycetes, produce antibiotics that target other bacteria. These antibiotics are secreted through drug transporters, which also contribute to self-resistance (or immunity) against the antibiotic (25, 310).
The overwhelming majority of MDR efflux pumps identified and characterized to date are energized by proton motive force, and depending on their size, similarity in primary structure, and topology, they fall into one of four distinct transporter families (383), namely, the MFS (301), the SMR (small MDR) family (358), the RND family (Fig. 2c, diagram G) (413), and the MATE (multidrug and toxic compound extrusion) family (50). However, several ABC transporters mediating MDR efflux in bacteria have also been identified. An excellent review on the distribution and physiology of these bacterial transporters was recently published (294).
Drug transport and specificity: an overview of classical methodologies.
Export of drugs by ABC transporters is demonstrated with several types of assays. A cellular resistance assay is important to establish the physiologic relevance of a drug transporter. The growth of cells expressing or overexpressing the putative MDR gene is compared to the growth of “control” cells in the presence of increasing concentrations of drug. The most appropriate control cells are generally the ones that express a nonfunctional mutant of the MDR protein, but cells that do not express the protein have also been used.
Whole bacterial cells have also been used to monitor in real time the transport of ethidium, which is often a substrate of both ABC (463, 504) and proton-dependent (35, 178, 332, 381, 416, 541) multidrug efflux pumps. Ethidium diffuses into the bacterial cells, and its fluorescence is greatly enhanced when it binds to intracellular nucleic acids. Therefore, in the presence of an active efflux pump and under energized conditions (usually glucose is added to the cell suspension), the accumulation of ethidium in the cells is reduced. As a supplementary control, an inhibitor of the efflux pump, such as reserpine, can be used in the experiment (11, 332, 463, 504).
Gene expression can be quantified by reverse transcription-PCR for bacterial strains cultivated in the presence or absence of select drugs. If a drug or several drugs are able to increase the expression of a given ABC transporter, it is reasonable to hypothesize that the drug(s) is a substrate for efflux. Also, comparison of gene expression between drug-resistant strains and wild-type strains can be very useful for identifying proteins involved in drug resistance (293).
Many in vitro assays have been developed to monitor drug efflux. Inverted membrane vesicles can be prepared from cells overexpressing a drug pump, and transport of either radioactive or fluorescent drugs can be assayed. In these vesicles, the NBDs are facing out, and the efflux pump will transport radioactive substrate into the vesicles in the presence of Mg·ATP. By the use of rapid filtration techniques, the vesicles can be retained and washed on a filter, and the radioactivity associated with the vesicles can be quantified. The alternative use of fluorescent drugs offers the advantage of visualizing transport in real time. One of the most commonly used compounds is the fluorescent dye Hoechst 33342. Due to its hydrophobicity, Hoechst 33342 concentrates in the membrane bilayer, where it is highly fluorescent. In contrast, it is only weakly fluorescent in aqueous medium (444, 445). When Mg·ATP is added to the system, Hoechst 33342 is transported out of the membrane and into the vesicle via the ABC protein, resulting in a decrease of its fluorescence. As illustrated by the experiments with LmrA discussed below (496), the choice of “control” membranes can be critical to understanding the mode of transport. Anthracyclins, including daunomycin and doxorubicin, constitute another class of fluorescent drugs. Their transport into DNA-loaded inverted vesicles can be monitored, since their fluorescence is quenched upon interaction with DNA (182). Rhodamine 123 transport into inverted membrane vesicles is monitored as a decrease in fluorescence due to a concentration-dependent self-quenching of fluorescence (445).
Inverted membrane vesicles have also been used to demonstrate that, in P-glycoprotein, low concentrations of Hoechst 33342 stimulate the transport of rhodamine 123, and low concentrations of rhodamine reciprocally stimulate the transport of Hoechst 33342 (445). This effect demonstrates that the transporter can accommodate the simultaneous binding of several drugs. Based on an analysis of drug-drug interactions, Shapiro and Ling proposed a functional model of P-glycoprotein containing at least two positively cooperative sites, the H site and the R site, for Hoechst 33342 and rhodamine 123, respectively. The binding sites of some P-glycoprotein substrates, including actinomycin D, appear to overlap the H and R sites (Fig. 15). Reciprocal stimulation of transport by low concentrations of drugs is also seen with a bacterial MDR protein, LmrA, suggestive of at least two binding sites (503), whereas at higher concentrations these drugs compete, suggesting that they can bind to the other site, though with lower affinity, as seen also for P-glycoprotein (445). This property may be unique to MDR transporters and to their transcriptional activators, and the nature of a multiple-drug-binding site is revealed in the high-resolution structures of these activators with different drugs bound (430-433, 543, 544).
FIG. 15.
Schematic representation of the drug-binding sites in P-glycoprotein, as proposed by Shapiro and Ling. Some drugs bind exclusively in the H site (blue) or in the R site (yellow), while others may be accommodated in both sites (red). This figure is based on data from reference 445.
The ATPase activity of an ABC transporter is usually, though not always, stimulated by its drug substrates, so this property is useful for identifying drug transporters as well as putative substrates for a known drug transporter. The drug stimulation of ATPase activity is relatively low for bacterial MDR transporters, however, usually only up to twofold (500). Furthermore, the ATPase activity of ABC transporters is well known to be inhibited by orthovanadate. Orthovanadate is often used to inhibit ATPase activity, to trap ADP in a catalytic site, or to block a transporter in its transition-state conformation.
LmrA, the first bacterial ABC MDR transporter discovered.
In 1996, Konings's group identified the first bacterial ABC MDR transporter, LmrA (Lactococcus MDR ATP), in Lactococcus lactis (504). LmrA (64 kDa) has a predicted topology of six α-helical TM segments followed by a single ABC. The transporter functions as a homodimer, with a topology similar to that of the human MDR pump P-glycoprotein (503). The primary sequence of LmrA shares 34% identity with each half of the P-glycoprotein (25% in the IM domain and 45% in the NBD). The homology between the two proteins was functionally confirmed, since transient transfection of LmrA in human lung fibroblasts conferred an MDR phenotype (500). Cells transfected with wild-type LmrA are resistant to many substrates of P-glycoprotein, including daunomycin, doxorubicin, vinblastine, vincristine, ethidium, colchicines, and rhodamine 123. Interestingly, resistance is reversed by several P-glycoprotein modulators, including verapamil and cyclosporine. Overexpression of LmrA in a drug-hypersensitive (TolC-negative) E. coli strain (465) increases resistance to a large variety of antibiotics (382). In addition, the rate of accumulation of ethidium in whole bacterial cells is reduced by the expression of LmrA (302, 504). Inverted membrane vesicles with and without LmrA have been used to monitor the transport of radioactive daunomycin (504) or fluorescent Hoechst 33342 (302). Daunomycin transport in vesicles was also inhibited by compounds which are well known as inhibitors of P-glycoprotein (verapamil) and of many MDR pumps (reserpine) (11). In addition to drugs, the purified, reconstituted LmrA protein has been demonstrated to transport fluorescently labeled phosphatidylethanolamine but not fluorescently labeled phosphatidylcholine (302). Complementation of a temperature-sensitive mutation in another bacterial lipid transporter, MsbA, also suggests that LmrA has the capability to transport at least some lipids (397).
Using radioactive vinblastine in conjunction with equilibrium binding and rapid filtration techniques, the drug-binding sites of LmrA have been investigated in detail in a membrane vesicle system (503). Based on these results, an “alternating two sites (two-cylinder engine)” mechanism for transport was proposed (for reviews, see references 501 and 502). In this model, LmrA contains two drug-binding sites, each of which alternates in accessibility, displaying high affinity in the inner leaflet of the membrane and low affinity in the outer leaflet of the membrane, from which the drug is released. The interconversion of these two sites is driven by ATP hydrolysis, and in the transition state, the high-affinity site is occluded.
Recently, several studies on LmrA revealed the possibility of a surprising alternative mode of transport. First, according to van Veen's group, LmrA can mediate the reverse transport (or uptake) of ethidium in ATP-depleted cells in the presence of an inwardly directed drug concentration gradient (20). This reverse transport correlated with synthesis of ATP (20). The authors proposed that the LmrA-mediated transport reaction is not too far from equilibrium under ATP-depleted conditions and can be driven in the forward or reverse direction depending on the direction of the substrate gradient.
Second, a truncated version of LmrA, lacking the NBDs (LmrA-MD), was engineered to study protein-drug interactions (506). Surprisingly, expression of LmrA-MD increased the sensitivity of L. lactis to several drug substrates, including ethidium, whose uptake was inhibited by both nigericin and valinomycin, chemicals that dissipate the TM proton gradient (ΔpH) and the TM potential (Δψ), respectively. The authors proposed that truncated LmrA-MD can function as a secondary active MDR system, using either ΔpH or Δψ of the proton motive force (Δp). Mutation of residue E314 in the truncated LmrA-MD protein eliminated ΔpH-driven ethidium transport in a proteoliposome-based assay (450, 506), although it still allowed Δψ-dependent transport of ethidium (450), supporting the notion that ethidium uptake was mediated by LmrA-MD. The authors suggested that LmrA may have evolved from a secondary active transporter precursor by acquiring an NBD and that ATP hydrolysis by full-length LmrA is able to drive the efflux of ethidium and protons from the cell against a proton concentration gradient. Interestingly, a region of conservation, containing a Glu residue, between MDR efflux pumps of the ABC and RND superfamilies has been identified (246), although E314 is not located within this conserved region.
van den Berg van Saparoea et al. (496) questioned whether LmrA actually functions as a multidrug efflux pump in L. lactis after they demonstrated that expression of LmrA in cells lacking chromosomally encoded efflux pumps increased rather than decreased the cytotoxicity of the commonly used efflux pump substrate Hoechst 33342, whereas expression of a second ABC transporter from L. lactis, LmrCD, did mediate resistance. In addition, while transport of Hoechst 33342 in inverted membrane vesicles depended on both ATP and the overexpression of LmrA, a mutation in LmrA eliminating ATP hydrolysis had little impact on the rate of drug extrusion (496). The observation that inhibition of ATP hydrolysis by vanadate or by mutation eliminated transport in vesicles containing LmrCD but not in those containing LmrA further suggested that Hoechst 33342 transport was not dependent on ATP hydrolysis by LmrA. These researchers speculated that the ATP dependence of Hoechst 33342 transport in vesicles containing LmrA relates not to ATP hydrolysis by LmrA but rather to the generation of a pH gradient via ATP hydrolysis by the FoF1 ATPase. The pH change, in turn, expels the drug from the membrane via the proton-drug symport activity inherent in LmrA. They also noted that changes in pH may alter the protonation state of Hoeschst 33342 and hence its fluorescence and solubility in the membrane. The group of van Veen later suggested, using purified LmrA reconstituted into proteoliposomes, that either ΔpH or ATP hydrolysis supports Hoechst 33342 transport (507). These studies leave us with many unanswered questions, including the physiological function of LmrA and the preferential mode of action of this interesting transport protein.
LmrCD from Lactococcus lactis.
The ydaG and ydbA genes of Lactococcus lactis are adjacent open reading frames (ORFs) that encode two half-ABC transporters. Using a nisin-inducible system, these proteins were either expressed separately or coexpressed in L. lactis (295). Only when both proteins were coexpressed were ethidium, daunomycin, and BCECF-AM [2′-7′-bis-(2-carboxyethyl)-5,6-carboxylfluorescein-acetoxymethyl ester] extruded from cells. Moreover, the two proteins could be purified as a stable heterodimer, and only the heterodimer had significant ATPase activity in detergent. Importantly, a Lactococcus lactis strain lacking these genes is hypersensitive to several toxic compounds (293, 294), and the genes are up-regulated in MDR strains of L. lactis selected for growth in the presence of several drugs. YdaG and YdbA were therefore renamed lactococcal MDR proteins C (LmrC) and D (LmrD), respectively. LmrCD is the first heterodimeric ABC MDR transporter to be reported for bacteria. Interestingly, the two NBDs differ with respect to the conservation of several important residues, including the conserved glutamate following the Walker B motif, which is known to be involved in ATP hydrolysis (347). Mutation of this residue suggested a structural and/or functional asymmetry in the NBDs of LmrCD (296). Residue Glu587 in LmrD proved to be essential for both drug transport and ATPase activity of LmrCD, whereas mutation of the equivalent residue, Asp495, in LmrC has a less severe effect on the activity of the heterodimer. Furthermore, photolabeling/cross-linking with 8-azido-nucleotides shows a preferential cross-linking of 8-azido-ATP to LmrC, while AlFx-induced trapping of the hydrolyzed nucleotide induces cross-linking mainly to LmrD. Given that AlFx, like vanadate, functions as a transition state analogue for ATP hydrolysis, these results strongly suggest that LmrD is more catalytically active than LmrC. This observation is interesting because a similar catalytic asymmetry occurs in eukaryotic ABC transporters, such as MRP1, CFTR, SUR1, PDR5, and TAP (296, 347).
BmrA from Bacillus subtilis.
In 1997, the complete sequence of the Bacillus subtilis genome revealed a preponderance of ABC transporters, and the functions of most of them are still unknown (262). This bacterium has 78 transporters, 38 of which are importers and 40 of which are exporters (385). A primary sequence comparison performed with the BLAST program (385) identified a previously unknown protein, YvcC, as being most similar to LmrA (462). The 65-kDa YvcC protein has 42% identity and 66% similarity with LmrA. Biochemical characterization of YvcC demonstrated its capacity to transport several structurally unrelated drugs, and it was accordingly renamed BmrA (Bacillus MDR ATP) (463). BmrA is expressed constitutively in Bacillus subtilis, and a knockout mutant had a lower rate of ethidium efflux than did the wild-type strain. Overexpression of BmrA in E. coli allowed the preparation of highly enriched inverted membrane vesicles that actively and specifically transport classical P-glycoprotein substrates, including Hoechst 33342, doxorubicin, and 7-amino-actinomycin D. Interestingly, each of these substrates binds a different site in the P-glycoprotein multisite model of Shapiro and Ling (445), and Hoechst 33342 reciprocally stimulates the transport of doxorubicin, demonstrating that these two drugs do not bind to the same site.
The transport of drugs by BmrA is strictly dependent on its ATPase activity; either inhibition of ATPase activity by vanadate or mutation of key catalytic residues in BmrA prevents transport (463). BmrA has been purified to homogeneity as a functional dimer in detergent (393) and can bind many P-glycoprotein substrates and modulators (463). Upon reconstitution into liposomes, BmrA displays positive cooperativity in ATP hydrolysis (Hill number of 1.34), a property that was previously described only for some ABC importers (463). Consistent with this, time-resolved fluorescence energy transfer experiments demonstrated that BmrA functions as a homodimer in liposomes (84). The high basal ATPase activity of reconstituted BmrA (6.5 μmol/min/mg) is vanadate sensitive and slightly stimulatable by drugs, usually in the range of 25%. This rather modest activation might be an intrinsic property of the transporter, since similar extents of activation are observed in proteoliposomes of different lipid compositions and in inverted membrane vesicles of E. coli overexpressing BmrA (463).
Structural similarities noted between the ABC and helicase superfamilies (168) triggered an examination of the role of the conserved glutamate adjacent to the Walker B motif in BmrA (347). This Glu was mutated to Asp, Ala, Gln, Ser, and Cys, and all mutants were devoid of ATPase activity. Use of radioactive 8-N3-ATP showed that the mutants were unable to perform hydrolytic turnover, consistent with a role of Glu as a catalytic base in ATP hydrolysis. Furthermore, upon addition of the nucleotide that drives the closure of NBDs (348), the mutant transporters reach a conformation in which one molecule of 8-azido-ATP per transporter is tightly trapped, a stoichiometry in agreement with results obtained later with P-glycoprotein (484). The importance of this residue in hydrolysis is clear, although its exact role is still under debate. As discussed above (see “Mechanism of ATP Hydrolysis, Still an Open Question”), it may function either as a catalytic base (347) or as a residue that stabilizes the orientation of a conserved His also shown to be essential for catalysis (545).
A low-resolution (25 Å) three-dimensional structure of BmrA reconstituted into a lipid bilayer has been obtained (63). Interestingly, the NBDs of BmrA lie far from the membrane, separated by a stalk region that likely corresponds to the ICD seen in the high-resolution structure of the Sav1866 drug exporter (102). Fluorescence transfer experiments also suggested a fairly long distance of 37 Å between the Hoechst 33342 binding site in the inner leaflet of the membrane and the ATP sites of P-glycoprotein (384). Sequence alignments suggest that the ICD may be a general feature of all exporters, while NBDs lie closer to the membrane in importers.
Spontaneous formation of an intramolecular disulfide bond between cysteines introduced into the Q loop of the NBD, at position 428, and the first intracellular loop of the IM domain, at position 123, residues selected based on the modeling of BmrA on other structures available at the time, indicates that these loops are close together in the tertiary structure of BmrA (85). Interestingly, formation of the disulfide bond prevents both drug transport and ATPase activity, and the addition of dithiothreitol restores function. The addition of Mg·ATP and vanadate prevents formation of the disulfide bond in the wild type, and Mg·ATP alone prevents disulfide bond formation if the E504Q mutation is introduced to prevent hydrolysis. These results are indicative of a substantial motion affecting the first intracellular loop and the Q loop during a cycle of catalysis and transport in BmrA.
Other class 1 ABC bacterial drug resistance proteins.
VcaM, a half-ABC transporter from Vibrio cholerae, was cloned into the drug-hypersensitive E. coli strain KAM32, which is deficient in the proton-dependent MDR pumps AcrAB and YdhE (210). The transformed strain had elevated levels of resistance to a number of dissimilar drugs, including tetracycline, norfloxacin, and ciprofloxacin, which are commonly used for cholera treatments, doxorubicin and daunomycin antibiotics, and the drug 4′,6′-diamidino-2-phenylindole. Fluorescence experiments demonstrate efflux of Hoechst 33342 and doxorubicin from whole cells, which is inhibited when reserpine or sodium orthovanadate is added to the cell suspension.
Using the same strategy, the same group identified EfrAB as an MDR transporter in Enterococcus faecalis (269). Resistance to several compounds, including norfloxacin, ciprofloxacin, doxycycline, acriflavine, and tetraphenylphosphonium, increased four- to eightfold following transformation of EfrAB into a drug-sensitive strain. In addition, energy-dependent acriflavine efflux was detected as a decrease in fluorescence, and this transport was inhibited by reserpine, verapamil, and orthovanadate.
Serratia marcescens is a gram-negative bacterium that causes nosocomial infections and has a high level of intrinsic resistance to many antimicrobial agents. Recently, a new heterodimeric ABC transporter from this microorganism, SmdAB, was cloned, and the plasmid harboring the genes was able to confer resistance to several agents in a hypersensitive strain of E. coli (307).
Lactobacillus brevis is a major contaminant in spoiled beer, despite the presence of bacteriostatic compounds derived from the hop plant that are added to beer to give a bitter taste. These iso-α-acids act on most gram-positive bacteria and are able to dissipate the proton motive force (414). A hop-tolerant strain has been isolated and shown to carry a plasmid harboring the horA gene (415), which encodes an ABC transporter whose sequence is 53% identical to that of LmrA. Unfortunately, this transporter remains poorly characterized, although a biochemical study is available (414).
Structure of the bacterial exporter Sav1866.
The structure of the bacterial exporter Sav1866 from Staphylococcus aureus was recently determined to 3.0-Å resolution (102). While stimulation of ATPase activity by several drugs has been demonstrated, the ability of Sav1866 to mediate MDR still needs to be confirmed. The transporter is a homodimer of two subunits, each consisting of an N-terminal IM domain and a C-terminal NBD. Although the protein was crystallized in the presence of ADP, the two NBDs exhibit the ATP-sandwiched dimer conformation seen in the closed form of MalK, the ATP-bound dimers of MJ0796(E171Q) and HlyB(H662A), and the intact MalEFGK2 complex (68, 343, 457, 545). It was suggested that the purification and crystallization conditions may have shifted the protein to the ATP-bound conformation. In fact, Sav1866 with bound AMP-PNP, a nonhydrolyzable ATP analogue, has the same conformational state (103). The TM helices of Sav1866 are 70 to 80 Å long and extend beyond the lipid bilayer. As a result, the NBDs are 25 Å away from the membrane (Fig. 16). The TM helices split into two “wings” in the middle of the membrane, forming a putative outward-facing translocation pathway between them. The cavity in the interior of the protein, where drugs presumably bind, is largest in the inner leaflet of the inner membrane, though it appears inaccessible to substrate from either the cytoplasm or the inner leaflet of the inner membrane in this conformational state. It was noted that the lining of this cavity is not very hydrophobic, a feature that may help to expel drugs into the periplasm or the outer leaflet of the membrane. Intriguingly, each wing consists of TM helices from both subunits. One of the characteristic features of Sav1866 is the way in which the two subunits twist around each other. Not only do the TM helices twist with each other, but the NBDs interact with both their own IM domain and the IM domain of the second subunit. Despite these additional contacts, the overall structure of the nucleotide-bound dimer is much the same as that seen for importers (343). This architecture is quite different from the “side-by-side” conformation shown for the BPD importers (Fig. 12). As discussed earlier (see “Inventory and Classification of ABC Systems and Evolution of the Superfamily”), the transmission interface between the TM domains and the NBDs occurs mainly through the intracellular loops ICL1 and ICL2, both of which contain a small helix. The NBDs contact the IM domains through the Q loop and a novel TEVGERV motif, seen only in exporters, that precedes the ABC signature motif in the NBD.
FIG. 16.
Structure of an exporter. Sav1866 is a homodimer of two IM-ABC subunits. IM domain 1 (green) is fused to NBD1 (blue), and IM domain 2 (red) is fused to NBD2 (yellow). Bound ADP is represented with a ball-and-stick model. The front view (left) reveals a cleft in the IM region that would be exposed to the outer leaflet of the lipid bilayer. TM helices are predicted to extend well into the cytoplasm, creating an ICD. The side view (right) reveals the intertwisting of TM helices that places the coupling helix of one subunit into the cleft of the NBD of the second subunit in an example of domain swapping.
The structure of Sav1866 has had a major impact in the ABC field, because superimposition of the TM helices of Sav1866 on another exporter structure, that of MsbA, revealed an incompatibility of arrangement, leading Chang and coworkers to retract earlier publications, notably three X-ray structures of MsbA (64). New structures have now been released (518), but a significant amount of work published since 2001 that was based on the retracted structures will require reevaluation.
Drug-binding site: localization and substrate recognition.
A striking characteristic of MDR ABC transporters is their ability to recognize a large variety of structurally unrelated substrates. The observation that P-glycoprotein lacking NBDs retains its capacity to bind substrates indicates that the drug-binding sites are located in the IM domains (291). Loo and Clarke undertook an extensive analysis of the drug-binding sites of P-glycoprotein based on the cross-linking of thiol-reactive substrates, such as dibromobimane, methanethiosulfonate-verapamil, and methanethiosulfonate-rhodamine, to a series of single cysteine mutants of the transporter (290). The results from these thiol modification studies, performed in conjunction with drug substrate protection studies, show that TM segments 4, 5, and 6 in the N-terminal IM domain and TM segments 9, 10, 11, and 12 in the C-terminal IM domain contribute residues to the drug-binding pocket. These results further suggest that the drug-binding pocket may lie at the interface between the two IM domains. Using a series of propafenone-based photoaffinity substrates (propafenone is an antiarrhythmic drug) and mass spectroscopy, Pleban and coworkers showed that the most labeling was observed in TM segments 3, 5, 8, and 11 (369). Applying the same methodology to LmrA, Ecker et al. observed preferential labeling at TM segments 3, 5, and 6 (136). In the structure of Sav1866 (Fig. 16), the large cavity that is present at the interface of the two IM domains is actually lined by residues from each TM helix, and it was proposed that it constitutes the substrate translocation pathway (102).
Despite the fact that MDR transporters recognize a striking variety of structurally unrelated substrates, Seelig and coworkers suggested that a general pattern of P-glycoprotein substrates consists of two or three electron donors (hydrogen bonding acceptors) with a fixed spatial separation (437, 439). In addition, the interaction of substrates with MDR transporters strongly depends on their ability to interact with the lipidic bilayer. Therefore, physical-chemical properties of the interacting molecule and the membrane are critical for interaction between the putative ligand and the transporter (154, 166). Seelig and colleagues investigated the role of the membrane in the interactions of P-glycoprotein with 15 structurally unrelated substrates. They determined the free energy of drug binding from the lipid membrane to the activating binding site of the transporter as the difference between the free energy of drug binding from water to the binding site and the free energy of drug partitioning into the lipid membrane. Thus, by taking into consideration the lipid-water partition coefficients of these compounds, they determined the membrane concentrations of drugs at half-maximum P-glycoprotein activation, which were 180- to 16,000-fold higher than the corresponding aqueous concentrations (166). These high drug concentrations in the membrane suggest relatively weak interactions between the substrates and the transporter. Analysis of the data suggested a modular binding concept rather than a “key-lock” model, where the binding module in drugs consists of two hydrogen bond acceptor groups.
Recently, Omote and Al-Shawi used molecular dynamics to study how substrates are recognized by P-glycoprotein in the lipid membrane (346), giving new insight into membrane-mediated substrate-transporter interactions (438). They proposed that substrates diffuse laterally in the cytosolic leaflet of the bilayer, exchanging their H bonds with water molecules for H bonds with the transporter as they bind. Importantly, molecular dynamics simulations were used to predict H bond interaction energies that are not directly measurable, and an inverse relation was found between the rate of transport and the H bonding potential of the substrates. Based on this study, a solvation exchange mechanism was proposed in which both polar and hydrophobic residues inside the drug-binding chamber are involved in sequential drug dehydration, solvation (with any solvent present in the drug-binding chamber of the transporter), and rehydration to promote the translocation of the drug across the membrane (346).
Translocation (Flipping) of Lipids and Lipid-Linked Oligosaccharides
In addition to hydrophobic drugs, ABC transporters have also been implicated in the transport (or flipping) of lipids and lipid-linked oligosaccharides. At least two different ABC transporters are involved in the complex process of LPS biosynthesis. LPS is the major lipid component of the outer leaflet of the outer membrane in gram-negative bacteria, serving essential functions in the structure, protection, and virulence of the cell. LPS consists of the following three parts: lipid A (a glucosamine-based saccharolipid), core oligosaccharide (3-deoxy-d-manno-octulosonic acid [KDO]), and O antigen (an extended oligosaccharide chain). Separate ABC transporters “flip” the undecaprenyl-linked O antigen precursor and the lipid A-linked KDO core from the cytosol to the periplasm (Fig. 17). Several excellent reviews describe the biosynthesis pathway in detail (39, 119, 390, 391).
FIG. 17.
Undecaprenyl-linked O antigen polysaccharide precursors (O-PS), initiated by a WecA homologue and elongated at the nonreducing end (black dots), are polymerized in the cytosol by the Wbd glycosyltransferases, shown in purple. The ABC transporter formed by Wzm and Wzt is required for transfer of these undecaprenyl-linked polymers to the periplasmic face of the membrane (left side of the figure). The core-KDO-lipid A complex is synthesized by the Lpx proteins, and the addition of sugar residues (blue dots) that constitute the core polysaccharide is catalyzed by the Waa proteins. KDO-lipid A is flipped from the inner face to the outer face of the membrane by the MsbA ABC transporter (right side of the figure). The WaaL ligase, shown in green, ligates O-PS to KDO-lipid A to form LPS. Export of LPS to the outer membrane is dependent on the LptABC proteins (formerly called YhbN, YhbG, and YrbK), forming an ABC transporter with LptFG (formerly called YjgP and YjgQ) (middle part of the figure). Additional proteins (?) might participate in this step. The LptDE proteins (formerly called Imp and Rlp) are required to translocate LPS to the outer face of the outer membrane. This figure is based on data from references 41, 391, 408a, and 522.
A gene (pglK) encoding an IM-ABC transporter has also been discovered in a locus involved in the N-linked glycosylation pathway of Campylobacter jejuni (160, 512). PglK is required for glycan assembly (3, 240). The pglK gene is able to complement a mutation in the wzx gene that encodes one of the E. coli LPS O antigen flippases (391), and PglK likely plays a similar role, flipping the lipid-linked oligosaccharide precursors of N-linked glycosylated proteins in C. jejuni (3).
Lipid A flippase MsbA.
A role for MsbA in LPS biogenesis was first realized when the msbA gene was isolated as a multicopy suppressor of a mutation that interfered with a late step in lipid A biosynthesis (76, 233). The E. coli msbA gene is essential for bacterial viability (371, 552), and a role in translocation of LPS was implied from an experiment in which radioactivity from N-acetyl-[3H]glucosamine, a precursor of LPS, accumulated in the inner membrane in an msbA conditional lethal strain at the nonpermissive temperature. Translocation of the 3H-labeled LPS precursor to the outer membrane was restored when a wild-type copy of the msbA gene was restored in trans (371). Likewise, LPS synthesized de novo in E. coli spheroplasts cofractionates with the outer membranes, and this proper localization is dependent on a functional MsbA protein (477).
The observation that both lipid A and glycerophospholipids accumulate in the inner membrane in E. coli strains in which MsbA synthesis is selectively shut off at 42°C suggests that both lipid A and glycerophospholipid transport to the outer membrane may depend on MsbA (122, 552). The idea that MsbA actually flips lipid A from the inner to the outer leaflet of the inner membrane is supported by the observation that modifications to the lipid A head group known to occur in the cytoplasm still occur at the nonpermissive temperature, while modifications known to occur in the periplasm are blocked (120).
Although the role of MsbA in lipid A trafficking is well accepted, its role as a general lipid flippase has been challenged by both in vitro studies suggesting that ATP is not necessary for the TM movement of phospholipids across the inner membrane of E. coli (214, 252) and in vivo experiments on msbA mutants of Neisseria meningitidis (476). In contrast to the case for E. coli, an N. meningitidis msbA mutant is viable, although it contains very small amounts of LPS. Electron microscopy reveals the presence of an outer membrane in the mutant that actually has higher levels of phospholipid than the wild-type membrane does, showing that phospholipid transport must occur (476). It is also notable that MsbA is found exclusively in gram-negative bacteria that synthesize lipid A. Therefore, the accumulation of phospholipids in the E. coli msbA conditional mutant may be a secondary effect of a toxic accumulation of lipid A precursor in the cytoplasm (119).
E. coli MsbA has been purified and reconstituted in E. coli phosholipid vesicles (121). Its basal ATPase activity (∼37 nmol/min/mg) was stimulated fourfold by hexa-acetylated lipid A, supporting a role in transport of LPS precursors (121). Surprisingly, MsbA also exhibits drug binding and transport abilities in vitro (397, 526), and the lmrA gene is able to complement the temperature-sensitive allele of msbA (397), indicating that LmrA and MsbA have overlapping specificities.
The corrected crystal structures of MsbA isolated from several different bacterial species (518) reveal the following three different conformations: an open-apo (nucleotide-free) conformation, solved at 5.3-Å resolution; a closed-apo form, solved at 5.5- Å resolution; and a closed, nucleotide-bound form similar to the Sav 1866 structure, with either one Mg·ADP·Vi (at 4.2 Å) or two AMP-PNP (at 3.7 Å) species bound. In the open-apo form, the NBDs are well separated and IM domains are in contact only near the periplasmic surface of the membrane, creating a V-shaped molecule. The large variations between the three structures suggest that substantial conformational changes may take place during the catalytic cycle of an ABC exporter (518). There is strong interest in determining whether the conformational states appearing in the crystals are representative of those occurring in an efflux pump in vivo.
Site-directed spin-labeling and EPR spectroscopy experiments (36, 52-54, 125), designed to examine the conformation of MsbA, show that apo-MsbA exists in an equilibrium that includes several conformers (125) and that ATP binding/hydrolysis promotes conformational changes during the transport cycle (36, 53, 54, 125). These experiments are consistent with the presence of an inward-facing chamber accessible to H2O in apo-MsbA that closes upon ATP binding (125). ATP binding also increases the accessibility of the periplasmic regions to H2O, and these differences are consistent with the idea of ATP triggering an alternation in the exposure of a chamber from the interior to the exterior side or leaflet of the membrane (125). Aqueous solvation in the predicted IM domain was also deduced in previous studies on LmrA (370) and P-glycoprotein (289). Furthermore, the distance between NBDs was estimated to change as much as 33 Å in both detergent-soluble and reconstituted MsbA, consistent with the formation of an NBD dimer from well-separated domains in the vanadate-trapped species (36).
CLASS 3 ABC SYSTEMS THAT ARE PROBABLY NOT IMPORTERS
To complete our discussion of efflux pumps, we now focus on several subfamilies of class 3 proteins that appear to function in export rather than import. Similar to the class 1 exporters we have been discussing, these proteins mediate the export of peptides, proteins, drugs, and polysaccharides, and many play an important role in drug resistance and immunity to naturally occurring bacterium-killing agents. In keeping with their classification, and in contrast to the bulk of exporters, the NBDs and IM proteins are encoded by separate genes. In addition, another well-studied class 3 system, the Lol system, functioning in the trafficking of lipoproteins from the inner to the outer membrane, is discussed.
Systems Involved in Resistance and Immunity to Antibiotics, Drugs, Lantibiotics, and Bacteriocins
DrrAB proteins (DRA family) from streptomycetes and their homologues.
Several bacterial drug transporters have been described to display a fairly restricted substrate specificity, in contrast to the MDR ABC transporters already discussed. Doxorubicin and daunorubicin are two antibiotics produced by Streptomyces peucetius that are also commonly used in chemotherapy. S. peucetius itself is resistant to these drugs because of an operon which encodes the DrrA and DrrB proteins (180). DrrA consists of an ABC-type NBD, while DrrB is an IM protein. Using gene fusions, a topology model has been proposed for DrrB, consisting of eight membrane-spanning segments with both N and C termini in the cytoplasm (164). Cysteine cross-linking revealed a motif within the N-terminal tail of DrrB that may be a modified version of the EAA motif present in ABC importers (236, 392). These two components are biochemically coupled and are thought to form an efflux pump (236, 237). Several homologues of DrrAB have been described for different species of drug-producing Streptomyces, conferring resistance to oleandomycin, tetronasin, mithramycin, and kasugamycin (151, 220, 279, 404).
Despite the general assumption that proteins in the Dra family confer resistance to specific drugs, there is some suggestion that they are also able to function as MDR determinants. When expressed in the drug-sensitive species Streptomyces lividans, the Streptomyces rochei msrAB genes confer resistance to multiple unrelated drugs, such as oleandomycin, erythromycin, spiramycin, doxorubicin, and tetracycline (153).
Homologues of the DrrAB systems are present in mycobacteria, and the operons also contain a third gene, drrC, encoding a protein with sequence similarity to DrrB. The drr genes are located within a large, 50-kilobase fragment of the chromosome containing 13 genes involved in biosynthesis of the phthiocerol dimycocerosate (PDIM). PDIM is a complex lipid residing in the mycobacterial capsule and is an important virulence determinant. The drrC gene is important for the proper localization of PDIM. In a drrC mutant strain, PDIM is localized in the cytoplasmic membrane rather than in the cell wall. The mutant also exhibits greater cell wall permeability and is more sensitive to sodium dodecyl sulfate than the wild-type strain is (58).
Expression of Mycobacterium tuberculosis DrrAB in E. coli confers increased resistance to a number of structurally unrelated compounds, including ethidium bromide, doxorubicin, daunorubicin, chloramphenicol, and puromycin. Expressed in Mycobacterium smegmatis, DrrAB conferred resistance to tetracycline, erythromycin, ethambutol, norfloxacin, streptomycin, and chloramphenicol. The MDR phenotype could be reversed by verapamil and reserpine, two inhibitors of ABC transporters (73).
The DRI family, providing immunity to bacteriocins and lantibiotics.
Bacteriocins are protein or peptide toxins produced by bacteria that are designed to kill similar bacteria. Lantibiotics are a special class of peptide antibiotics produced by gram-positive organisms that target other gram-positive organisms and are posttranslationally modified (see reference 129 for a review). In regions determining the synthesis of bacteriocins or lantibiotics, several transporters similar to those found in antibiotic resistance clusters are present and mediate immunity to the bacteriocin (400, 537) or lantibiotic (248, 452). The lantibiotic epidermin kills bacteria by forming membrane potential-dependent pores in cytoplasmic membranes. Expression of the epiEFG genes, encoding an epidermin ABC transporter, confers immunity by extruding epidermin from the membrane to the external medium (351).
ABC Transporters Mediating Antibiotic Resistance and Lipoprotein Release from the Cytoplasmic Membrane
The o228 family of proteins share strong primary sequence conservation, including a highly conserved domain of unknown function, DUF274, in the membrane protein (245). The most well-studied members of this family are the MacAB and LolCDE systems of E. coli. In addition, the family includes MbrAB of Streptococcus mutans, a bacitracin resistance determinant (491), and DevABC of Anabaena, a putative glycolipid exporter (157).
The MacA-MacB-TolC system in Enterobacteriaceae.
The ybjYZ genes of E. coli conferred erythromycin resistance when they were overexpressed in a hypersensitive strain lacking the MDR pump AcrAB (250). Because the uptake of macrolides was reduced in this strain, these genes were renamed macAB to reflect the likelihood that they encode a macrolide-specific ABC efflux pump. MacB is an integral membrane protein with an N-terminal NBD fused to a C-terminal IM domain composed of four TM segments (249). MacA is a peripheral membrane protein that belongs to the MFP family. Remarkably, in proteoliposomes, the ATPase activity of MacB is strictly dependent on MacA, displaying a 45-fold increase in catalytic efficiency. A similar MFP-mediated stimulation of ATPase activity has never been detected in type I protein secretion systems, such as the hemolysin secretion system. In contrast, the addition of macrolide antibiotics has no effect on the ATPase activity (481). MacAB requires the outer membrane channel protein TolC for its transport function in vivo, as do many MFP-dependent efflux systems in E. coli (324, 373, 550), suggesting that drug stimulation of ATPase, if it exists, may be dependent on the presence of the entire transporter complex.
The importance of MacAB in drug resistance has been questioned in studies of other gram-negative bacteria. Deletion of the macAB genes in S. enterica serovar Typhimurium has no effect on erythromycin resistance, and overexpression of these genes leads to only a modest increase in resistance (336). Although overexpression of the gonococcal MacAB efflux pump in an E. coli background leads to a twofold increase in bacterial resistance to macrolides, its loss in a gonococcal clinical isolate only slightly decreased bacterial resistance to azithromycin and erythromycin (408). Thus, the relevance of the MacAB system to clinical resistance appears to be marginal, bringing into question the role of an erythromycin efflux pump in bacteria intrinsically resistant to this antibiotic.
Lol, an ABC transporter involved in lipoprotein trafficking.
An ABC system designated Lol (lipoprotein localization) is essential for growth in E. coli and is responsible for mediating the trafficking of lipoproteins from the inner to the outer membrane (327, 535). The TM ABC complex LolCDE recognizes lipoproteins attached to the periplasmic surface of the inner membrane and transfers them to the LolA protein, a periplasmic carrier protein. LolB, a lipoprotein tethered to the inner surface of the outer membrane, acts as a receptor for lipoproteins and releases them into the membrane. Both LolA and LolB have a hydrophobic cavity consisting of a β-barrel and an α-helical lid that sequesters the lipid moiety of the lipoprotein when it is outside the membrane (469). Since the lid of LolA is closed in the crystal structure, it has been suggested that the energy from ATP hydrolysis is used for both the opening of the lid to receive the lipid moiety and the detachment of lipoproteins from the membrane (469). Intriguingly, a mutation preventing lid opening results in high-affinity binding between LolA and the LolCDE complex (317). A progressive increase in affinity for the lipoprotein substrate ensures energy-independent transfer from LolA to LolB and from LolB to the outer membrane (473).
The Lol system also has the ability to sort lipoproteins destined for the outer membrane from those that belong in the inner membrane, and in E. coli, only lipoproteins lacking an Asp at residue 2 in the mature sequence (residue 1 is always a lipid-modified cysteine) are recognized as substrates and transferred to the outer membrane (443, 536). The Asp at position 2 is best visualized as an avoidance signal, since proteins containing all other amino acids at position 2 are recognized as substrates for transfer and compete for recognition, whereas those with Asp do not compete (306). All lipoproteins tested, with the exception of one containing the Asp residue at position 2, stimulated the ATPase activity of LolCDE in the absence of LolA (306), suggesting that LolCDE carries the recognition site for the modified N terminus of the substrate. Consistent with this hypothesis, a mutant LolC protein with a C40P substitution has been isolated and shown to allow translocation of Asp-containing proteins (326).
Further characterization of the avoidance signal indicates that positively charged lipids in the cytoplasmic membrane are required if the Asp-containing lipoprotein is to avoid transfer to the outer membrane by Lol and that a negative charge is needed at a fixed distance from the α-carbon of residue 2, since Glu will not substitute for Asp (185). The tight association of Asp with positively charged phosphatidylethanolamine may present an unrecognizable lipoprotein molecule, which appears to have five rather than the usual three acyl chains, to LolCDE. Intriguingly, sorting signals in other bacteria appear to differ from that in E. coli, suggesting that more than one mechanism for recognition has evolved (327).
Biogenesis of Extracellular Polysaccharides
Polysaccharide transporters of the CLS family.
Extracellular polysaccharides are important virulence determinants in most bacteria. The biosynthetic regions for group 2 and 3 capsular polysaccharides (454), some O antigens (49), and teichoic acids (267) have been shown to comprise genes encoding ABC transporters and to cluster within the CLS family. Capsular polysaccharides are targeted to the outer membranes of gram-negative bacteria. The minimal composition of these transporters is one IM protein and one ABC protein that likely dimerize to form the ABC transporter (454). Mutations in either protein lead to reduced production of the polysaccharide and to accumulation in the cytoplasm (34, 364).
Capsular polysaccharide-specific ABC transporters are comprised of KpsM (IM protein) and KpsT (ABC protein) and require the following two additional proteins to function properly in gram-negative bacteria: KpsE, a cytoplasmic membrane protein of the MPA2 family that is presumed to play the same role as MFPs in exporters; and KpsD, an outer membrane protein of the OMA family (355, 521). Mutations in the genes encoding KpsE and KpsD lead to an accumulation of the polysaccharide in the periplasm, suggesting that these proteins are involved in polysaccharide export through the outer membrane (360, 453).
The rfb (wb*) locus, which encodes the O antigen biogenesis pathway in gram-negative bacteria, includes components required for transfer across the plasma membrane. O antigen oligosaccharides that are polymerized at the nonreducing terminus (for example, those of E. coli serotypes O8 and O9a) are completely synthesized in the cytosol, and the fully elongated chain is probably flipped across the membrane via an ABC transporter comprised of Wzm (IM protein) and Wzt (ABC protein) (Fig. 17) (522). In contrast, the first repeat unit of O antigen oligosaccharides that are polymerized at the reducing end (like those from Shigella dysenteriae type 1) is translocated by the Wzx flippase, and subsequent polymerization is catalyzed in the periplasm by Wzy (391). The actual role of these ABC transporters is not known in detail, but mutagenesis and heterologous expression experiments suggest that they are essential for the constitution of fully assembled, smooth-type LPS (49, 401). The O8- and O9a-specific Wzm/Wzt transporters are specific for their cognate substrates, in contrast to capsular polysaccharide KpsM/KpsT transporters, which can export different types of polysaccharides. The IM Wzm components of the O8 and O9a transporters are functionally interchangeable, albeit with reduced efficiencies, but the cytosolic Wzt components are not, indicating a specific role for Wzt in substrate specificity (83). The Wzt ABC proteins contain a C-terminal domain not present in KpsT, and the region involved in substrate specificity resides in this C-terminal domain (82).
In gram-positive bacteria, genes highly similar to those involved in the biogenesis of capsular polysaccharide and LPS are found (267, 534). In B. subtilis, limited expression of the tagGH genes leads to anomalous morphology and a decrease in cell wall galactosamine and phosphate, two components of teichoic acids. It has been suggested that the products of these genes might be involved in the translocation of teichoic acid through the cytoplasmic membrane (267).
In Bacillaceae, S-layer glycoproteins constitute the major portion of the cell surface. S-layer glycans usually possess a tripartite structure reminiscent of that known for the LPS of gram-negative bacteria. This comprises an O-antigen-like polysaccharide chain, a core region, and a glycosylation site replacing lipid A. The S-layer glycan biosynthesis region contains an ABC transporter highly similar to the Wzm/Wzt transporters of gram-negative bacteria (338).
Translocation of LPS to the outer membrane.
In the course of our phylogenetic analyses, we identified a putative class 3 ABC system belonging to the YHBG family that is highly conserved in gram-negative bacteria. This system is comprised of three genes, yrbK (encoding a putative TM protein with just one TM segment), yhbN (encoding a putative periplasmic protein), and yhbG (encoding an ABC protein). These genes are located just downstream of the genes encoding two enzymes involved in the biogenesis of the KDO component of LPS, i.e., kdsD (yrbH) and kdsC (yrbI), encoding d-arabinose 5-phosphate isomerase and 3-deoxy-d-manno-octulosonate 8-phosphate phosphatase, respectively (311, 532). The yrbK, yhbN, and yhbG genes are essential for E. coli viability (461). Mutants depleted of YhbN (LptA) and/or YhbG (LptB) produce anomalous LPS, are defective in LPS transport to the outer membrane, and accumulate de novo-synthesized LPS in a novel membrane fraction of intermediate density between the inner membrane and the outer membrane. LptA is located in the periplasm, and expression of the lptAB operon is controlled by the extracytoplasmic sigma factor RpoE. When viewed in the context of the global pathway for LPS biogenesis (Fig. 17), these results are consistent with a model in which the LptA/LptB complex is responsible for conveying newly synthesized LPS through the periplasmic space to the outer membrane. TM components LptF (YjgP), LptG (YjgQ), and LptC (YrbK) are necessary to form a complete ABC transporter (408a, 460). Alternatively, the LptABCFG system may guide the LPS molecule along putative membrane adhesion sites between the cytoplasmic and outer membranes. Final insertion of LPS into the outer membrane and its flipping to the outward face are mediated by the Imp/RlpB complex (40, 533).
CLASS 2 ABC SYSTEMS INVOLVED IN NONTRANSPORT CELLULAR PROCESSES AND IN ANTIBIOTIC RESISTANCE
An increasing number of cytosolic proteins carrying the ABC signature are being discovered, and significant progress has been made in understanding their functions. These proteins share significant sequence similarity and consist of two tandemly repeated ABC domains, but they lack detectable IM domains. They belong to several families, whose members have been shown to participate in several non-transport-related processes.
The UVR Family, Involved in Nucleotide Excision Repair and Drug Resistance
The UvrA protein is involved in the excision of damaged DNA. In this system, a UvrA dimer binds to UvrB, and the resulting complex interacts with the damaged site on the DNA (see reference 174 for a review). Bacterial genomes carry several paralogues of the UvrA protein. DrrC has been shown to participate in the resistance of Streptomyces peucetius to daunorubicin and doxorubicin (287). The drrC gene encodes a DNA-binding protein whose expression is induced by daunorubicin. ATP and daunorubicin, a DNA-intercalating agent, stimulate DrrC DNA-binding activity. It is proposed that DrrC releases the drug from DNA by using the energy of ATP hydrolysis (163).
The crystal structure of Bacillus stearothermophilus UvrA (352) reveals the presence of composite ATP-binding sites containing residues from both NBDs, just like the case for ABC transporters. UvrA also harbors two unique domains, one of which is required for interaction with UvrB. Analysis of the surface properties of UvrA in combination with biochemical studies on mutant proteins led to the identification of a DNA-binding surface that lies on the ventral side of the UvrA dimer.
The RLI Family, Involved in Ribosome Biogenesis
The RNase L inhibitor (RLI) family is conserved among eukaryotes and archaea. No data are available on the archaeal proteins. The mammalian RLI (ABCE1) binds to RNase L, modulating interferon production and the stability of MyoD and mitochondrial mRNAs (29, 273). Depletion of RLI1 in vivo leads to a cessation of growth, a lower polysome content, and a decrease in the average size of a polysome (126). In independent studies, RLI1 was found to be associated with both pre-40S particles and mature 40S subunits and with eukaryotic translation initiation factor 3 (eIF3), eIF5, and eIF2 (70, 538). Recent biophysical, biochemical, and yeast genetic analyses suggested that RLI1 harbors two essential [4Fe-4S] clusters. Only seven of the eight conserved cysteines coordinating the [Fe-S] clusters are essential for cell viability (21). RLI1 is associated with ribosomes and with Hcr1p, a protein involved in rRNA processing and translation initiation. Depletion of RLI1 causes a nuclear export defect of the small and large ribosomal subunits and, subsequently, a translational arrest (247). The crystal structure of the Pyrococcus furiosus Mg·ADP-bound RLI at 1.9-Å resolution, devoid of its [Fe-S] binding sites, shows the canonical ABC dimer with two composite active sites. The linker between the two ABC domains and the C terminus of the protein constitutes a hinge at the interface of the ABCs opposite the active site cleft, and mutations in the linker eliminate function. The first ABC domain contains a helix-loop-helix insertion in its helical domain. Because of its conformation in the crystal, the helix-loop-helix is proposed to interact with the missing [Fe-S]-binding domain. The residues constituting the signature sequence are 11 Å away from the computed position of the γ-phosphate of ATP, suggesting that RLI undergoes a large conformational change upon ATP hydrolysis. RLI is likely an ABC mechanoenzyme, whose ATP-driven conformational states mediate the assembly, disassembly, and maturation of RNA-protein complexes in the cell (232).
ART Family of Proteins with Diverse Functions
Phylogenetic analyses distinguish a large class 2 family of proteins that can be subdivided into three subfamilies. The EF-3 subfamily is involved in translation elongation, REG subfamily members participate in several nontransport processes, and the ARE subfamily is involved in antibiotic resistance.
The EF-3 subfamily, involved in translation elongation.
The yeast elongation factor EF-3 is required for in vitro translation and for in vivo growth. It interacts with ribosomes to stimulate EF-1 alpha-dependent binding of aminoacyl-tRNA to the ribosomal A site when the E site is occupied by deacylated tRNA (see reference 62 for a review). The crystal structure of EF-3 was recently solved, providing new information on how this elongation factor interacts with the ribosome (17).
The REG subfamily.
The yeast ABC protein GCN20 is an essential protein that associates with the GCN1 protein and stimulates the activity of GCN2, a kinase that phosphorylates the translation initiation factor eIF2, leading to increased translation of the transcriptional activator GCN4 in amino acid-starved cells (505).
Several ORFs detected in complete bacterial genomes are homologous to GCN20, implying a role in regulatory processes. Indeed, the Agrobacterium tumefaciens ChvD protein was inactivated in mutants selected for reduced transcription of the virA and virG genes (284, 525).
Inactivation of the gene encoding Uup in E. coli leads to a dramatic increase in precise excision of transposons and in deletions of chromosomal sequences between short direct repeats. Uup putatively functions via a DNA polymerase slippage mechanism independent of RecA and transposase (394). Purified Uup protein binds to DNA in a non-sequence-specific fashion. This finding suggests that Uup protein might be involved in helping the DNA replication machinery to correctly copy DNA sequences located between direct repeats (325).
ARE subfamily proteins that confer resistance to MLS antibiotics.
Interestingly, class 2 ABC proteins homologous to those just described are involved in antibiotic resistance (42, 87, 310). They confer resistance to macrolide, lincosamide, and streptogramin (MLS) antibiotics (398). MLS antibiotics are a large group of antibacterial agents used clinically against a wide range of infectious bacteria, and they function by binding to the 50S ribosomal subunit and inhibiting protein synthesis. It is still not clear whether these ABC proteins mediate drug resistance by interacting with as yet unknown membrane partners to achieve efflux or whether they prevent the MLS antibiotics from binding their target site, the 50S ribosome (398).
Class 2 ABC proteins involved in MLS antibiotic resistance differ in their specificities. The Msr(A) protein from Staphylococcus epidermidis (406) is responsible for resistance to erythromycin and type B streptogramins but not for that to lincosamides and 16-membered-ring macrolides (398). The VgaA proteins from staphylococci (6, 7, 71) confer resistance to lincosamides and streptogramin A compounds (71). Disruption of the lsa(A) gene from Enterococcus faecalis increases susceptibility to quinupristin (streptogramin B), dalfopristin (streptogramin A), and clindamycin (lincosamide) >40-fold (455).
Interestingly, a significant number of class 2 ABC proteins have been found in antibiotic-producing organisms, especially actinomycetes (310). These organisms need efficient resistance mechanisms to protect them against the antibiotics they produce. Several macrolide resistance genes have been isolated, including car(A) from Streptomyces thermotolerans, srm(B) from Streptomyces ambofaciens, and tlr(C) from Streptomyces fradiae, which confer resistance to the 16-membered-ring macrolides carbomycin, spiramycin, and tylosin, respectively (428). In Streptomyces antibioticus, which produces the 14-membered-ring macrolide oleandomycin, the ole(B) gene confers resistance to oleandomycin in the heterologous hosts Streptomyces albus and S. lividans (342). In addition to class 2 ABC proteins, organisms producing antibiotics often express ribosomal methyltransferases that confer resistance by modifying the macrolide target site on the ribosome (310, 398).
Very few studies have attempted to address the molecular mechanism of drug resistance in this family. Binding of oleandomycin to a maltose-BP-OleB fusion protein was detected by intrinsic fluorescence (56), but the apparent affinity for the antibiotic was low, in the millimolar range, and no saturation of the signal was reached due to competing absorbance of the compound itself. Given that no interacting TM domains have been identified in this family, substrate specificity may be determined by the ABC proteins. Indeed, a Vga(A) variant with a four-amino-acid substitution in the linker between the two ABC domains shifted the drug resistance profile from streptogramin A to lincosamides (337).
To our knowledge, just one study addresses the possibility that these proteins function as efflux pumps. Cells expressing Msr(A) exclude [14C]erythromycin, suggestive of active efflux (398, 406). In these experiments, S. aureus cells transformed with the S. epidermidis resistance gene msr(A) were incubated with [14C]erythromycin, and time-dependent drug uptake was measured. After an initial period of uptake, the radioactivity associated with these cells decreased with time, while radioactivity remained constant in control cells (406). It was suggested that these proteins may be able to “hijack” the TM domains of other ABC transporters to mediate efflux (398). Consistent with this hypothesis, the Vga(A) protein colocalizes mainly in the membrane fraction of its host cell (71). However, no membrane partners for Msr(A) or related proteins have yet been identified, nor has there been any analysis comparable to the in vitro studies on class 1 efflux pumps to support a role for ARE proteins in active efflux of drugs. Furthermore, reserpine, a well-known inhibitor of multidrug pumps, has no effect on Msr(A) (170). Since nonprotonated erythromycin is able to diffuse through the cellular membrane down a concentration gradient, displacement of erythromycin from its ribosomal binding site might allow the antibiotic to simply diffuse out of the cell. In this case, Msr(A) would undergo an ATP-dependent conformational change that leads to dissociation of erythromycin from the ribosome.
CONCLUSION
Bacterial ABC transporters are clearly involved in very diverse functions, as best illustrated by the class 2 proteins. We are far from completing an overview of bacterial ABC systems, since <30% of ABC systems identified so far have been characterized functionally in even a cursory way. Among these, only about 50 are actively investigated at the molecular level. Investigation of the remaining uncharacterized ABC systems constitutes a wide wild “terra incognita” for the discovery of unprecedented functions in living organisms.
Common Themes Emerging from Studies of ABC Transporters with Diverse Functions
Genomic analysis clearly indicates an early divergence of exporters (class 1) from importers (class 3), which would permit divergence in the mechanism of translocation between these classes. However, there is also evidence for inversion in the direction of translocation within classes, with at least one example of a class 1 protein mediating import and several examples of subfamilies within class 3 mediating export, suggesting that the mechanisms of action will not be too different. The homology of ABCs in all classes and their similarity in structure strongly suggest that these components of ABC transporters all work in the same way. The major global conformational change taking place in the ABC is a domain rotation accompanied by closure of the nucleotide-binding interface upon binding and hydrolysis of ATP. The diversity in sequence and size of the IM regions, however, may allow for variation in the way that the motions of the ABC are coupled to the actual translocation process.
For importers, it appears relatively straightforward to fit models for translocation based on biochemical data into the emerging structures. For example, structures of the BPD importers show evidence of a central translocation channel that alternates in exposure to the inner or outer face of the membrane and that is positioned directly below a well-characterized high-affinity binding site for substrate in the BP, a protein whose substrate-dependent conformational changes are well defined.
For exporters, this task may prove to be a far greater challenge. While alternating access is again likely to be a key feature, the exporter structure is quite distinct from the importer structures, with the addition of an extramembranous domain that distances the ABCs from the membrane, an intertwisting of TM helices, and a swapping of NBDs, features that may permit distinct differences in the nature of conformational changes in the membrane region in response to closure of the NBDs. The Sav1866 structure has a large internal cavity, but it is not yet clear how hydrophobic substrates enter or exit the cavity. In contrast to the case for importers, unambiguous identification of the substrate-binding site may be challenging due to the hydrophobic and polyspecific nature of the substrates, which may also bind nonspecifically to the hydrophobic ABC protein.
The Challenge of Membrane Protein Crystallography
While it is preferable to compare structures of the same transporter captured in different conformational states, membrane proteins are notoriously difficult to crystallize, and the degree of structural conservation between BtuCD and HI1470/1 or ModB2C2 and MalFGK2 suggests that it should be possible to draw conclusions about the nature of global conformational changes taking place during the transport cycle. However, several cautions are in order. First, as suggested by Schmitt and colleagues (547), the finer details of mechanism, including the physical nature of mechanical switches that operate to mediate these larger global domain movements, may well be specific to a given transporter, requiring structures of the same protein in different conformational states for elucidation. Second, as indicated by Rees and colleagues (366), ABC transporters are crystallized following solubilization from the native membrane environment with detergents and subsequent purification, and under these conditions, they may not behave in the same way. Discrepancies between expectations based on biochemical results and structures include the fact that the translocation pathways in BtuCD and HI1470/1 face opposite sides of the membrane despite the lack of bound nucleotide in either structure, while it is the binding and hydrolysis of ATP that are thought to promote such a conformational change. In addition, two structures of Sav1866, with ADP and AMP-PNP bound (102, 103), reveal essentially the same closed ABC dimer, inconsistent with biochemical results suggesting that ATP but not ADP promotes closure of the ABC dimer interface. Finally, several of the reported structures of intact transporters are of limited resolution, highlighting the difficulties associated with the crystallization of membrane proteins. For these reasons, it will be important to find independent means to verify details revealed by new crystal structures and to test hypotheses based on the structures. Biochemical approaches carried out on transporters in their native environment, combined with structural analysis, should provide validation of future structures, resulting in the elucidation of the mechanism of ABC transporters.
Understanding the Mechanism: Areas for Future Investigation
While great advances have been achieved in our understanding of the mechanism of translocation by ABC transporters over the last decade, there is still much work to be done before we have a complete understanding of how ABC transporters work. The fact that the ABC module is highly conserved raises the possibility that the basic mechanism for hydrolysis is conserved throughout the family. However, the existence of degeneracy in the conserved motifs indicates that not all ABC transporters require two functional hydrolytic sites. Several mechanisms have been proposed, involving either sequential or alternating hydrolysis at the two sites, and the events occurring before, during, and after catalysis have not been correlated with movements within the NBDs. For example, does the ABC dimer open after a single hydrolytic event or after both ATPs are hydrolyzed? Does the timing of the opening coincide with phosphate release or ADP release? Do both nucleotide-binding sites open simultaneously, or can they open independently? What is the significance of the cooperativity between ATP-binding sites?
There are still many open questions about the nature of the translocation pathways and the identification of substrate-binding sites in the IM domains. Evidence suggests that in exporters, conformational changes lead to decreased affinities for substrates, but no explanatory details are available.
As shown in Fig. 11, we propose a very simple two-state model for coupling of transport to hydrolysis in which the closure of the ABC modules to bind and hydrolyze ATP coincides with the reorientation of the translocation pathway. Closure may be sufficient to promote ATP hydrolysis, and following ATP hydrolysis, the closed ABC dimer may no longer be stable, triggering a return to the initial conformational state. Viewed in this way, the transport process may be no more complicated than a G protein process, in which GTP hydrolysis promotes the switch between two different conformations. Schmitt and colleagues offer the possibility of a far more intricate catalytic cycle, in which different steps are coupled to different aspects of the translocation process (547). They emphasize the fact that the ATPase can function as a dual-mode engine wherein ATP binding provides mechanical motion (perhaps reorientation of TM helices) and the sequential hydrolysis of ATP and stepwise release of Pi provide additional chemical energy that can be used for separate (as yet undefined) purposes. Is a simple two-state model adequate to explain transport, or are more complex mechanisms involved to ensure unidirectional transport and efficient coupling of transport to hydrolysis? These questions and more ensure many more years of stimulating research and discussion in the field of ABC transporters.
Acknowledgments
Work in our labs was supported by the National Institutes of Health (J.C. and A.L.D.), the Welch Foundation (A.L.D.), and the Pew Scholar Program (J.C.). C. Orelle is a recipient of a postdoctoral fellowship from Fondation pour la Recherche Médicale.
E.D. thanks Olivier Chesneau for reviewing parts of the manuscript. J.C. and A.L.D. thank Michael Oldman for preparing figures.
REFERENCES
- 1.Akama, H., M. Kanemaki, M. Yoshimura, T. Tsukihara, T. Kashiwagi, H. Yoneyama, S. Narita, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 27952816-52819. [DOI] [PubMed] [Google Scholar]
- 2.Akama, H., T. Matsuura, S. Kashiwagi, H. Yoneyama, S. Narita, T. Tsukihara, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 27925939-25942. [DOI] [PubMed] [Google Scholar]
- 3.Alaimo, C., I. Catrein, L. Morf, C. L. Marolda, N. Callewaert, M. A. Valvano, M. F. Feldman, and M. Aebi. 2006. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 25967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albers, S. V., S. M. Koning, W. N. Konings, and A. J. Driessen. 2004. Insights into ABC transport in archaea. J. Bioenerg. Biomembr. 365-15. [DOI] [PubMed] [Google Scholar]
- 5.Alekshun, M. N., and S. B. Levy. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 1281037-1050. [DOI] [PubMed] [Google Scholar]
- 6.Allignet, J., and N. El Solh. 1997. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202133-138. [DOI] [PubMed] [Google Scholar]
- 7.Allignet, J., V. Loncle, and N. el Sohl. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 11745-51. [DOI] [PubMed] [Google Scholar]
- 8.Al-Shawi, M. K., and H. Omote. 2005. The remarkable transport mechanism of P-glycoprotein: a multidrug transporter. J. Bioenerg. Biomembr. 37489-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altena, K., A. Guder, C. Cramer, and G. Bierbaum. 2000. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol. 662565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amanuma, H., and Y. Anraku. 1974. Transport of sugars and amino acids in bacteria. XII. Substrate specificities of the branched chain amino acid-binding proteins of Escherichia coli. J. Biochem. (Tokyo) 761165-1173. [DOI] [PubMed] [Google Scholar]
- 11.Ambudkar, S. V., S. Dey, C. A. Hrycyna, M. Ramachandra, I. Pastan, and M. M. Gottesman. 1999. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 39361-398. [DOI] [PubMed] [Google Scholar]
- 12.Ambudkar, S. V., I. W. Kim, D. Xia, and Z. E. Sauna. 2006. The A-loop, a novel conserved aromatic acid subdomain upstream of the Walker A motif in ABC transporters, is critical for ATP binding. FEBS Lett. 5801049-1055. [DOI] [PubMed] [Google Scholar]
- 13.Ames, G. F., and J. Lever. 1970. Components of histidine transport: histidine-binding proteins and hisP protein. Proc. Natl. Acad. Sci. USA 661096-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ames, G. F.-L., C. E. Liu, A. K. Joshi, and K. Nikaido. 1996. Liganded and unliganded receptors interact with equal affinity with the membrane complex of periplasmic permeases, a subfamily of traffic ATPases. J. Biol. Chem. 27114264-14270. [DOI] [PubMed] [Google Scholar]
- 15.Ames, G. F.-L., C. Mimura, and V. Shyamala. 1990. Bacterial periplasmic permeases belong to a family of transport proteins operating from Escherichia coli to human traffic ATPases. FEMS Microbiol. Rev. 75429-446. [DOI] [PubMed] [Google Scholar]
- 16.Andersen, C., E. Koronakis, E. Bokma, J. Eswaran, D. Humphreys, C. Hughes, and V. Koronakis. 2002. Transition to the open state of the TolC periplasmic tunnel entrance. Proc. Natl. Acad. Sci. USA 9911103-11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen, C. B., T. Becker, M. Blau, M. Anand, M. Halic, B. Balar, T. Mielke, T. Boesen, J. S. Pedersen, C. M. Spahn, T. G. Kinzy, G. R. Andersen, and R. Beckmann. 2006. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 443663-668. [DOI] [PubMed] [Google Scholar]
- 18.Austermuhle, M. I., J. A. Hall, C. S. Klug, and A. L. Davidson. 2004. Maltose binding protein is open in the catalytic transition state for ATP hydrolysis during maltose transport. J. Biol. Chem. 27928243-28250. [DOI] [PubMed] [Google Scholar]
- 19.Balakrishnan, L., C. Hughes, and V. Koronakis. 2001. Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J. Mol. Biol. 313501-510. [DOI] [PubMed] [Google Scholar]
- 20.Balakrishnan, L., H. Venter, R. A. Shilling, and H. W. van Veen. 2004. Reversible transport by the ATP-binding cassette multidrug export pump LmrA: ATP synthesis at the expense of downhill ethidium uptake. J. Biol. Chem. 27911273-11280. [DOI] [PubMed] [Google Scholar]
- 21.Barthelme, D., U. Scheele, S. Dinkelaker, A. Janoschka, F. MacMillan, S. V. Albers, A. J. M. Driessen, M. S. Stagni, E. Bill, W. Meyer-Klaucke, V. Schunemann, and R. Tampe. 2007. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J. Biol. Chem. 28214598-14607. [DOI] [PubMed] [Google Scholar]
- 22.Bass, R. B., P. Strop, M. Barclay, and D. C. Rees. 2002. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science 2981582-1587. [DOI] [PubMed] [Google Scholar]
- 23.Bavoil, P., H. Nikaido, and K. von Meyenburg. 1977. Pleiotropic transport mutants of Escherichia coli lack porin, a major outer membrane protein. Mol. Gen. Genet. 15823-33. [DOI] [PubMed] [Google Scholar]
- 24.Benabdelhak, H., S. Kiontke, C. Horn, R. Ernst, M. A. Blight, I. B. Holland, and L. Schmitt. 2003. A specific interaction between the NBD of the ABC-transporter HlyB and a C-terminal fragment of its transport substrate haemolysin A. J. Mol. Biol. 3271169-1179. [DOI] [PubMed] [Google Scholar]
- 25.Bernard, R., P. Joseph, A. Guiseppi, M. Chippaux, and F. Denizot. 2003. YtsCD and YwoA, two independent systems that confer bacitracin resistance to Bacillus subtilis. FEMS Microbiol. Lett. 22893-97. [DOI] [PubMed] [Google Scholar]
- 26.Biemans-Oldehinkel, E., M. K. Doeven, and B. Poolman. 2006. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 5801023-1035. [DOI] [PubMed] [Google Scholar]
- 27.Biemans-Oldehinkel, E., and B. Poolman. 2003. On the role of the two extracytoplasmic substrate-binding domains in the ABC transporter OpuA. EMBO J. 225983-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binet, R., and C. Wandersman. 1995. Protein secretion by hybrid bacterial ABC-transporters: specific functions of the membrane ATPase and the membrane fusion protein. EMBO J. 142298-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bisbal, C., M. Silhol, H. Laubenthal, T. Kaluza, G. Carnac, L. Milligan, F. Le Roy, and T. Salehzada. 2000. The 2′-5′ oligoadenylate/RNase L/RNase L inhibitor pathway regulates both MyoD mRNA stability and muscle cell differentiation. Mol. Cell. Biol. 204959-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bishop, L., R. Agbayani, Jr., S. V. Ambudkar, P. C. Maloney, and G. Ames. 1989. Reconstitution of a bacterial periplasmic permease in proteoliposomes and demonstration of ATP hydrolysis concomitant with transport. Proc. Natl. Acad. Sci. USA 866953-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biswas, S., M. M. Mohammad, D. R. Patel, L. Movileanu, and B. van den Berg. 2007. Structural insight into OprD substrate specificity. Nat. Struct. Mol. Biol. 141108-1109. [DOI] [PubMed] [Google Scholar]
- 32.Biville, F., H. Cwerman, S. Letoffe, M. S. Rossi, V. Drouet, J. M. Ghigo, and C. Wandersman. 2004. Haemophore-mediated signalling in Serratia marcescens: a new mode of regulation for an extra cytoplasmic function (ECF) sigma factor involved in haem acquisition. Mol. Microbiol. 531267-1277. [DOI] [PubMed] [Google Scholar]
- 33.Bjorkman, A. J., and S. L. Mowbray. 1998. Multiple open forms of ribose-binding protein trace the path of its conformational change. J. Mol. Biol. 279651-664. [DOI] [PubMed] [Google Scholar]
- 34.Bliss, J. M., C. F. Garon, and R. P. Silver. 1996. Polysialic acid export in Escherichia coli K1: the role of KpsT, the ATP-binding component of an ABC transporter, in chain translocation. Glycobiology 6445-452. [DOI] [PubMed] [Google Scholar]
- 35.Bolhuis, H., G. Poelarends, H. W. van Veen, B. Poolman, A. J. Driessen, and W. N. Konings. 1995. The lactococcal lmrP gene encodes a proton motive force-dependent drug transporter. J. Biol. Chem. 27026092-26098. [DOI] [PubMed] [Google Scholar]
- 36.Borbat, P. P., K. Surendhran, M. Bortolus, P. Zou, J. H. Freed, and H. S. McHaourab. 2007. Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 5e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borths, E. L., K. P. Locher, A. T. Lee, and D. C. Rees. 2002. The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc. Natl. Acad. Sci. USA 9916642-16647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borths, E. L., B. Poolman, R. N. Hvorup, K. P. Locher, and D. C. Rees. 2005. In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry 4416301-16309. [DOI] [PubMed] [Google Scholar]
- 39.Bos, M. P., V. Robert, and J. Tommassen. 2007. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61191-214. [DOI] [PubMed] [Google Scholar]
- 40.Bos, M. P., B. Tefsen, J. Geurtsen, and J. Tommassen. 2004. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. USA 1019417-9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bos, M. P., and J. Tommassen. 2004. Biogenesis of the gram-negative bacterial outer membrane. Curr. Opin. Microbiol. 7610-616. [DOI] [PubMed] [Google Scholar]
- 42.Bouige, P., D. Laurent, L. Piloyan, and E. Dassa. 2002. Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr. Protein Pept. Sci. 3541-559. [DOI] [PubMed] [Google Scholar]
- 43.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braibant, M., P. Gilot, and J. Content. 2000. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24449-467. [DOI] [PubMed] [Google Scholar]
- 45.Brass, J. M., W. Boos, and R. Hengge. 1981. Reconstitution of maltose transport in malB mutants of Escherichia coli through calcium-induced disruptions of the outer membrane. J. Bacteriol. 14610-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16295-307. [DOI] [PubMed] [Google Scholar]
- 47.Braun, V., S. Mahren, and A. Sauter. 2006. Gene regulation by transmembrane signaling. Biometals 19103-113. [DOI] [PubMed] [Google Scholar]
- 48.Brinkmann, H., and H. Philippe. 1999. Archaea sister group of bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol. Biol. Evol. 16817-825. [DOI] [PubMed] [Google Scholar]
- 49.Bronner, D., B. R. Clarke, and C. Whitfield. 1994. Identification of an ATP-binding cassette transport system required for translocation of lipopolysaccharide O-antigen side-chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol. Microbiol. 14505-519. [DOI] [PubMed] [Google Scholar]
- 50.Brown, M. H., I. T. Paulsen, and R. A. Skurray. 1999. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31394-395. [DOI] [PubMed] [Google Scholar]
- 51.Brune, M., J. L. Hunter, J. E. Corrie, and M. R. Webb. 1994. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 338262-8271. [DOI] [PubMed] [Google Scholar]
- 52.Buchaklian, A. H., A. L. Funk, and C. S. Klug. 2004. Resting state conformation of the MsbA homodimer as studied by site-directed spin labeling. Biochemistry 438600-8606. [DOI] [PubMed] [Google Scholar]
- 53.Buchaklian, A. H., and C. S. Klug. 2006. Characterization of the LSGGQ and H motifs from the Escherichia coli lipid A transporter MsbA. Biochemistry 4512539-12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buchaklian, A. H., and C. S. Klug. 2005. Characterization of the Walker A motif of MsbA using site-directed spin labeling electron paramagnetic resonance spectroscopy. Biochemistry 445503-5509. [DOI] [PubMed] [Google Scholar]
- 55.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 656-63. [DOI] [PubMed] [Google Scholar]
- 56.Buche, A., C. Mendez, and J. A. Salas. 1997. Interaction between ATP, oleandomycin and the OleB ATP-binding cassette transporter of Streptomyces antibioticus involved in oleandomycin secretion. Biochem. J. 321139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buckel, S. D., A. W. Bell, J. K. Mohana Rao, and M. A. Hermodson. 1986. An analysis of the structure of the product of the rbsA gene of Escherichia coli K12. J. Biol. Chem. 2617659-7662. [PubMed] [Google Scholar]
- 58.Camacho, L. R., P. Constant, C. Raynaud, M. A. Laneelle, J. A. Triccas, B. Gicquel, M. Daffe, and C. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 27619845-19854. [DOI] [PubMed] [Google Scholar]
- 59.Campbell, J. D., S. S. Deol, F. M. Ashcroft, I. D. Kerr, and M. S. Sansom. 2004. Nucleotide-dependent conformational changes in HisP: molecular dynamics simulations of an ABC transporter nucleotide-binding domain. Biophys. J. 873703-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campbell, J. D., and M. S. Sansom. 2005. Nucleotide binding to the homodimeric MJ0796 protein: a computational study of a prokaryotic ABC transporter NBD dimer. FEBS Lett. 5794193-4199. [DOI] [PubMed] [Google Scholar]
- 61.Cangelosi, G. A., R. G. Ankenbauer, and E. W. Nester. 1990. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc. Natl. Acad. Sci. USA 876708-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakraburtty, K. 2001. Translational regulation by ABC systems. Res. Microbiol. 152391-399. [DOI] [PubMed] [Google Scholar]
- 63.Chami, M., E. Steinfels, C. Orelle, J. M. Jault, A. Di Pietro, J. L. Rigaud, and S. Marco. 2002. Three-dimensional structure by cryo-electron microscopy of YvcC, an homodimeric ATP-binding cassette transporter from Bacillus subtilis. J. Mol. Biol. 3151075-1085. [DOI] [PubMed] [Google Scholar]
- 64.Chang, G., C. B. Roth, C. L. Reyes, O. Pornillos, Y. J. Chen, and A. P. Chen. 2006. Retraction. Science 3141875. [DOI] [PubMed] [Google Scholar]
- 65.Charbonnel, P., M. Lamarque, J. C. Piard, C. Gilbert, V. Juillard, and D. Atlan. 2003. Diversity of oligopeptide transport specificity in Lactococcus lactis species. A tool to unravel the role of OppA in uptake specificity. J. Biol. Chem. 27814832-14840. [DOI] [PubMed] [Google Scholar]
- 66.Chen, C.-J., J. E. Chin, K. Ueda, D. P. Clark, I. Pastan, M. M. Gottesman, and I. G. Roninson. 1986. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell 47381-389. [DOI] [PubMed] [Google Scholar]
- 67.Chen, H. C., A. J. Newton, and A. Melis. 2005. Role of SulP, a nuclear-encoded chloroplast sulfate permease, in sulfate transport and H2 evolution in Chlamydomonas reinhardtii. Photosynth. Res. 84289-296. [DOI] [PubMed] [Google Scholar]
- 68.Chen, J., G. Lu, J. Lin, A. L. Davidson, and F. A. Quiocho. 2003. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell 12651-661. [DOI] [PubMed] [Google Scholar]
- 69.Chen, J., S. Sharma, F. A. Quiocho, and A. L. Davidson. 2001. Trapping the transition state of an ATP-binding-cassette transporter: evidence for a concerted mechanism of maltose transport. Proc. Natl. Acad. Sci. USA 981525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen, Z. Q., J. S. Dong, A. Ishimura, I. Daar, A. G. Hinnebusch, and M. Dean. 2006. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J. Biol. Chem. 2817452-7457. [DOI] [PubMed] [Google Scholar]
- 71.Chesneau, O., H. Ligeret, N. Hosan-Aghaie, A. Morvan, and E. Dassa. 2005. Molecular analysis of resistance to streptogramin A compounds conferred by the Vga proteins of staphylococci. Antimicrob. Agents Chemother. 49973-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10394-401. [DOI] [PubMed] [Google Scholar]
- 73.Choudhuri, B. S., S. Bhakta, R. Barik, J. Basu, M. Kundu, and P. Chakrabarti. 2002. Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. Biochem. J. 367279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung, Y. J., C. Krueger, D. Metzgar, and M. H. Saier, Jr. 2001. Size comparisons among integral membrane transport protein homologues in bacteria, archaea, and eucarya. J. Bacteriol. 1831012-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cianciotto, N. P., P. Cornelis, and C. Baysse. 2005. Impact of the bacterial type I cytochrome c maturation system on different biological processes. Mol. Microbiol. 561408-1415. [DOI] [PubMed] [Google Scholar]
- 76.Clementz, T., J. J. Bednarski, and C. R. Raetz. 1996. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J. Biol. Chem. 27112095-12102. [DOI] [PubMed] [Google Scholar]
- 77.Cobessi, D., H. Celia, N. Folschweiller, I. J. Schalk, M. A. Abdallah, and F. Pattus. 2005. The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstrom resolution. J. Mol. Biol. 347121-134. [DOI] [PubMed] [Google Scholar]
- 78.Cobessi, D., H. Celia, and F. Pattus. 2005. Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J. Mol. Biol. 352893-904. [DOI] [PubMed] [Google Scholar]
- 79.Cook, G. M., and R. K. Poole. 2000. Oxidase and periplasmic cytochrome assembly in Escherichia coli K-12: CydDC and CcmAB are not required for haem-membrane association. Microbiology 146527-536. [DOI] [PubMed] [Google Scholar]
- 80.Covitz, K.-M. Y., C. H. Panagiotidis, M. Reyes, N. A. Treptow, and H. A. Shuman. 1994. Mutations that alter the transmembrane signalling pathway in an ATP binding cassette (ABC) transporter. EMBO J. 131752-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crouch, M. L., M. Castor, J. E. Karlinsey, T. Kalhorn, and F. C. Fang. 2008. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 67971-983. [DOI] [PubMed] [Google Scholar]
- 82.Cuthbertson, L., M. S. Kimber, and C. Whitfield. 2007. Substrate binding by a bacterial ABC transporter involved in polysaccharide export. Proc. Natl. Acad. Sci. USA 10419529-19534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cuthbertson, L., J. Powers, and C. Whitfield. 2005. The C-terminal domain of the nucleotide-binding domain protein Wzt determines substrate specificity in the ATP-binding cassette transporter for the lipopolysaccharide O-antigens in Escherichia coli serotypes O8 and O9a. J. Biol. Chem. 28030310-30319. [DOI] [PubMed] [Google Scholar]
- 84.Dalmas, O., M. A. Do Cao, M. R. Lugo, F. J. Sharom, A. Di Pietro, and J. M. Jault. 2005. Time-resolved fluorescence resonance energy transfer shows that the bacterial multidrug ABC half-transporter BmrA functions as a homodimer. Biochemistry 444312-4321. [DOI] [PubMed] [Google Scholar]
- 85.Dalmas, O., C. Orelle, A. E. Foucher, C. Geourjon, S. Crouzy, A. Di Pietro, and J. M. Jault. 2005. The Q-loop disengages from the first intracellular loop during the catalytic cycle of the multidrug ABC transporter BmrA. J. Biol. Chem. 28036857-36864. [DOI] [PubMed] [Google Scholar]
- 86.Danese, P. N., and T. J. Silhavy. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 3259-94. [DOI] [PubMed] [Google Scholar]
- 87.Dassa, E., and P. Bouige. 2001. The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152211-229. [DOI] [PubMed] [Google Scholar]
- 88.Dassa, E., M. Hofnung, I. T. Paulsen, and M. H. Saier, Jr. 1999. The Escherichia coli ABC transporters: an update. Mol. Microbiol. 32887-889. [DOI] [PubMed] [Google Scholar]
- 89.Dassa, E., and S. Muir. 1993. Membrane topology of MalG, an inner membrane protein from the maltose transport system of Escherichia coli. Mol. Microbiol. 729-38. [DOI] [PubMed] [Google Scholar]
- 90.Dattelbaum, J. D., L. L. Looger, D. E. Benson, K. M. Sali, R. B. Thompson, and H. W. Hellinga. 2005. Analysis of allosteric signal transduction mechanisms in an engineered fluorescent maltose biosensor. Protein Sci. 14284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Daus, M. L., S. Berendt, S. Wuttge, and E. Schneider. 2007. Maltose binding protein (MalE) interacts with periplasmic loops P2 and P1, respectively, of the MalFG subunits of the maltose ATP binding cassette transporter (MalFGK(2)) from Escherichia coli/Salmonella during the transport cycle. Mol. Microbiol. 661107-1122. [DOI] [PubMed] [Google Scholar]
- 92.Daus, M. L., M. Grote, P. Mueller, M. Doebber, A. Herrmann, H. J. Steinhoff, E. Dassa, and E. Schneider. 2007. ATP-driven MalK dimer closure and reopening and conformational changes of the “EAA” motifs are crucial for function of the maltose ATP-binding cassette transporter (MALFGK2). J. Biol. Chem. 28222387-22396. [DOI] [PubMed] [Google Scholar]
- 93.Daus, M. L., H. Landmesser, A. Schlosser, P. Muller, A. Herrmann, and E. Schneider. 2006. ATP induces conformational changes of periplasmic loop regions of the maltose ATP-binding cassette transporter. J. Biol. Chem. 2813856-3865. [DOI] [PubMed] [Google Scholar]
- 94.Davidson, A. L. 2002. Mechanism of coupling of transport to hydrolysis in bacterial ATP-binding cassette transporters. J. Bacteriol. 1841225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reference deleted.
- 96.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73241-268. [DOI] [PubMed] [Google Scholar]
- 97.Davidson, A. L., S. S. Laghaeian, and D. E. Mannering. 1996. The maltose transport system of Escherichia coli displays positive cooperativity in ATP hydrolysis. J. Biol. Chem. 2714858-4863. [PubMed] [Google Scholar]
- 98.Davidson, A. L., and H. Nikaido. 1990. Overproduction, solubilization and reconstitution of the maltose transport system from Escherichia coli. J. Biol. Chem. 2654254-4260. [PubMed] [Google Scholar]
- 99.Davidson, A. L., and S. Sharma. 1997. Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J. Bacteriol. 1795458-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Davidson, A. L., H. A. Shuman, and H. Nikaido. 1992. Mechanism of maltose transport in Escherichia coli: transmembrane signalling by periplasmic binding proteins. Proc. Natl. Acad. Sci. USA 892360-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264375-382. [DOI] [PubMed] [Google Scholar]
- 102.Dawson, R. J., and K. P. Locher. 2006. Structure of a bacterial multidrug ABC transporter. Nature 443180-185. [DOI] [PubMed] [Google Scholar]
- 103.Dawson, R. J., and K. P. Locher. 2007. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 581935-938. [DOI] [PubMed] [Google Scholar]
- 104.De, E., A. Basle, M. Jaquinod, N. Saint, M. Mallea, G. Molle, and J. M. Pages. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 41189-198. [DOI] [PubMed] [Google Scholar]
- 105.Dean, D. A., A. L. Davidson, and H. Nikaido. 1989. Maltose transport in membrane vesicles of Escherichia coli is linked to ATP hydrolysis. Proc. Natl. Acad. Sci. USA 869134-9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dean, D. A., L. I. Hor, H. A. Shuman, and H. Nikaido. 1992. Interaction between maltose-binding protein and the membrane-associated maltose transporter complex in Escherichia coli. Mol. Microbiol. 62033-2040. [DOI] [PubMed] [Google Scholar]
- 107.Dean, M., A. Rzhetsky, and R. Allikmets. 2001. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 111156-1166. [DOI] [PubMed] [Google Scholar]
- 108.Death, A., L. Notley, and T. Ferenci. 1993. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J. Bacteriol. 1751475-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Debarbieux, L., and C. Wandersman. 2001. Folded HasA inhibits its own secretion through its ABC exporter. EMBO J. 204657-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Decottignies, A., and A. Goffeau. 1997. Complete inventory of the yeast ABC proteins. Nat. Genet. 15137-145. [DOI] [PubMed] [Google Scholar]
- 111.Delepelaire, P. 2004. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694149-161. [DOI] [PubMed] [Google Scholar]
- 112.Delepelaire, P., and C. Wandersman. 1998. The SecB chaperone is involved in the secretion of the Serratia marcescens HasA protein through an ABC transporter. EMBO J. 17936-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.de Lorimier, R. M., J. J. Smith, M. A. Dwyer, L. L. Looger, K. M. Sali, C. D. Paavola, S. S. Rizk, S. Sadigov, D. W. Conrad, L. Loew, and H. W. Hellinga. 2002. Construction of a fluorescent biosensor family. Protein Sci. 112655-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Detmers, F. J., F. C. Lanfermeijer, R. Abele, R. W. Jack, R. Tampe, W. N. Konings, and B. Poolman. 2000. Combinatorial peptide libraries reveal the ligand-binding mechanism of the oligopeptide receptor OppA of Lactococcus lactis. Proc. Natl. Acad. Sci. USA 9712487-12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Deuschle, K., S. Okumoto, M. Fehr, L. L. Looger, L. Kozhukh, and W. B. Frommer. 2005. Construction and optimization of a family of genetically encoded metabolite sensors by semirational protein engineering. Protein Sci. 142304-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dinh, T., I. T. Paulsen, and M. H. Saier, Jr. 1994. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 1763825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dippel, R., and W. Boos. 2005. The maltodextrin system of Escherichia coli: metabolism and transport. J. Bacteriol. 1878322-8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Doerrler, W. T. 2006. Lipid trafficking to the outer membrane of gram-negative bacteria. Mol. Microbiol. 60542-552. [DOI] [PubMed] [Google Scholar]
- 120.Doerrler, W. T., H. S. Gibbons, and C. R. Raetz. 2004. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 27945102-45109. [DOI] [PubMed] [Google Scholar]
- 121.Doerrler, W. T., and C. R. Raetz. 2002. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 27736697-36705. [DOI] [PubMed] [Google Scholar]
- 122.Doerrler, W. T., M. C. Reedy, and C. R. H. Raetz. 2001. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 27611461-11464. [DOI] [PubMed] [Google Scholar]
- 123.Doeven, M. K., R. Abele, R. Tampe, and B. Poolman. 2004. The binding specificity of OppA determines the selectivity of the oligopeptide ATP-binding cassette transporter. J. Biol. Chem. 27932301-32307. [DOI] [PubMed] [Google Scholar]
- 124.Doeven, M. K., J. Kok, and B. Poolman. 2005. Specificity and selectivity determinants of peptide transport in Lactococcus lactis and other microorganisms. Mol. Microbiol. 57640-649. [DOI] [PubMed] [Google Scholar]
- 125.Dong, J., G. Yang, and H. S. McHaourab. 2005. Structural basis of energy transduction in the transport cycle of MsbA. Science 3081023-1028. [DOI] [PubMed] [Google Scholar]
- 126.Dong, J. S., R. Lai, K. Nielsen, C. A. Fekete, H. F. Qiu, and A. G. Hinnebusch. 2004. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 27942157-42168. [DOI] [PubMed] [Google Scholar]
- 127.Doolittle, R. F., M. S. Johnson, I. Husain, B. Van Houten, D. C. Thomas, and A. Sancar. 1986. Domainal evolution of a prokaryotic DNA repair protein and its relationship to active-transport proteins. Nature 323451-452. [DOI] [PubMed] [Google Scholar]
- 128.Doring, K., T. Surrey, P. Nollert, and F. Jahnig. 1999. Effects of ligand binding on the internal dynamics of maltose-binding protein. Eur. J. Biochem. 266477-483. [DOI] [PubMed] [Google Scholar]
- 129.Draper, L. A., R. P. Ross, C. Hill, and P. D. Cotter. 2008. Lantibiotic immunity. Curr. Protein Pept. Sci. 939-49. [DOI] [PubMed] [Google Scholar]
- 130.Duan, X., J. A. Hall, H. Nikaido, and F. A. Quiocho. 2001. Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding. J. Mol. Biol. 3061115-1126. [DOI] [PubMed] [Google Scholar]
- 131.Duan, X., and F. A. Quiocho. 2002. Structural evidence for a dominant role of nonpolar interactions in the binding of a transport/chemosensory receptor to its highly polar ligands. Biochemistry 41706-712. [DOI] [PubMed] [Google Scholar]
- 132.Dubochet, J., A. W. McDowall, B. Menge, E. N. Schmid, and K. G. Lickfeld. 1983. Electron microscopy of frozen-hydrated bacteria. J. Bacteriol. 155381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dumas, F., R. Koebnik, M. Winterhalter, and P. Van Gelder. 2000. Sugar transport through maltoporin of Escherichia coli—role of polar tracks. J. Biol. Chem. 27519747-19751. [DOI] [PubMed] [Google Scholar]
- 134.Duplay, P., S. Szmelcman, H. Bedouelle, and M. Hofnung. 1987. Silent and functional changes in the periplasmic maltose-binding protein of Escherichia coli K12. I. Transport of maltose. J. Mol. Biol. 194663-673. [DOI] [PubMed] [Google Scholar]
- 135.Dwyer, M. A., and H. W. Hellinga. 2004. Periplasmic binding proteins: a versatile superfamily for protein engineering. Curr. Opin. Struct. Biol. 14495-504. [DOI] [PubMed] [Google Scholar]
- 136.Ecker, G. F., K. Pleban, S. Kopp, E. Csaszar, G. J. Poelarends, M. Putman, D. Kaiser, W. N. Konings, and P. Chiba. 2004. A three-dimensional model for the substrate binding domain of the multidrug ATP binding cassette transporter LmrA. Mol. Pharmacol. 661169-1179. [DOI] [PubMed] [Google Scholar]
- 137.Ehrle, R., C. Pick, R. Ulrich, E. Hofmann, and M. Ehrmann. 1996. Characterization of transmembrane domains 6, 7, and 8 of MalF by mutational analysis. J. Bacteriol. 1782255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Elkins, C. A., and K. E. Beenken. 2005. Modeling the tripartite drug efflux pump archetype: structural and functional studies of the macromolecular constituents reveal more than their names imply. J. Chemother. 17581-592. [DOI] [PubMed] [Google Scholar]
- 139.Elkins, C. A., and H. Nikaido. 2003. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J. Bacteriol. 1855349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Entcheva, P., D. A. Phillips, and W. R. Streit. 2002. Functional analysis of Sinorhizobium meliloti genes involved in biotin synthesis and transport. Appl. Environ. Microbiol. 682843-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ernst, R., J. Koch, C. Horn, R. Tampe, and L. Schmitt. 2006. Engineering ATPase activity in the isolated ABC cassette of human TAP1. J. Biol. Chem. 28127471-27480. [DOI] [PubMed] [Google Scholar]
- 142.Eswaran, J., C. Hughes, and V. Koronakis. 2003. Locking TolC entrance helices to prevent protein translocation by the bacterial type I export apparatus. J. Mol. Biol. 327309-315. [DOI] [PubMed] [Google Scholar]
- 143.Faraldo-Gomez, J. D., and M. S. Sansom. 2003. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4105-116. [DOI] [PubMed] [Google Scholar]
- 144.Fath, M. J., and R. Kolter. 1993. ABC transporters—bacterial exporters. Microbiol. Rev. 57995-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Feissner, R. E., C. L. Richard-Fogal, E. R. Frawley, and R. G. Kranz. 2006. ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol. Microbiol. 61219-231. [DOI] [PubMed] [Google Scholar]
- 146.Felder, C. B., R. C. Graul, A. Y. Lee, H. P. Merkle, and W. Sadee. 1999. The Venus flytrap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS PharmSci. 1E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ferenci, T. 1980. The recognition of maltodextrins by Escherichia coli. Eur. J. Biochem. 108613-636. [DOI] [PubMed] [Google Scholar]
- 148.Ferenci, T., W. Boos, M. Schwartz, and S. Szmelcman. 1977. Energy-coupling of the transport system of Escherichia coli dependent on maltose-binding protein. Eur. J. Biochem. 75187-193. [DOI] [PubMed] [Google Scholar]
- 149.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 2951715-1719. [DOI] [PubMed] [Google Scholar]
- 150.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 2822215-2220. [DOI] [PubMed] [Google Scholar]
- 151.Fernandez, E., F. Lombo, C. Mendez, and J. A. Salas. 1996. An ABC transporter is essential for resistance to the antitumor agent mithramycin in the producer Streptomyces argillaceus. Mol. Gen. Genet. 251692-698. [DOI] [PubMed] [Google Scholar]
- 152.Fernandez, L. A., and V. de Lorenzo. 2001. Formation of disulphide bonds during secretion of proteins through the periplasmic-independent type I pathway. Mol. Microbiol. 40332-346. [DOI] [PubMed] [Google Scholar]
- 153.Fernandez-Moreno, M. A., L. Carbo, T. Cuesta, C. Vallin, and F. Malpartida. 1998. A silent ABC transporter isolated from Streptomyces rochei F20 induces multidrug resistance. J. Bacteriol. 1804017-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ferte, J. 2000. Analysis of the tangled relationships between P-glycoprotein-mediated multidrug resistance and the lipid phase of the cell membrane. Eur. J. Biochem. 267277-294. [DOI] [PubMed] [Google Scholar]
- 155.Fetherston, J. D., V. J. Bertolino, and R. D. Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32289-299. [DOI] [PubMed] [Google Scholar]
- 156.Fetsch, E. E., and A. L. Davidson. 2002. Vanadate-catalyzed photocleavage of the signature motif of an ATP-binding cassette (ABC) transporter. Proc. Natl. Acad. Sci. USA 999685-9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fiedler, G., M. Arnold, S. Hannus, and I. Maldener. 1998. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 271193-1202. [DOI] [PubMed] [Google Scholar]
- 158.Flocco, M. M., and S. L. Mowbray. 1994. The 1.9 A X-ray structure of a closed unliganded form of the periplasmic glucose/galactose receptor from Salmonella typhimurium. J. Biol. Chem. 2698931-8936. [PubMed] [Google Scholar]
- 159.Forward, J. A., M. C. Behrendt, N. R. Wyborn, R. Cross, and D. J. Kelly. 1997. TRAP transporters—a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J. Bacteriol. 1795482-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fry, B. N., V. Korolik, J. A. ten Brinke, M. T. Pennings, R. Zalm, B. J. Teunis, P. J. Coloe, and B. A. van der Zeijst. 1998. The lipopolysaccharide biosynthesis locus of Campylobacter jejuni 81116. Microbiology 1442049-2061. [DOI] [PubMed] [Google Scholar]
- 161.Fuellen, G., M. Spitzer, P. Cullen, and S. Lorkowski. 2005. Correspondence of function and phylogeny of ABC proteins based on an automated analysis of 20 model protein data sets. Proteins 61888-899. [DOI] [PubMed] [Google Scholar]
- 162.Fukami-Kobayashi, K., Y. Tateno, and K. Nishikawa. 1999. Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 286279-290. [DOI] [PubMed] [Google Scholar]
- 163.Furuya, K., and C. R. Hutchinson. 1998. The DrrC protein of Streptomyces peucetius, a UvrA-like protein, is a DNA-binding protein whose gene is induced by daunorubicin. FEMS Microbiol. Lett. 168243-249. [DOI] [PubMed] [Google Scholar]
- 164.Gandlur, S. M., L. Wei, J. Levine, J. Russell, and P. Kaur. 2004. Membrane topology of the DrrB protein of the doxorubicin transporter of Streptomyces peucetius. J. Biol. Chem. 27927799-27806. [DOI] [PubMed] [Google Scholar]
- 165.Garcia, O., P. Bouige, C. Forestier, and E. Dassa. 2004. Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J. Mol. Biol. 343249-265. [DOI] [PubMed] [Google Scholar]
- 166.Gatlik-Landwojtowicz, E., P. Aanismaa, and A. Seelig. 2006. Quantification and characterization of P-glycoprotein-substrate interactions. Biochemistry 453020-3032. [DOI] [PubMed] [Google Scholar]
- 167.Gaudet, R., and D. C. Wiley. 2001. Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 204964-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Geourjon, C., C. Orelle, E. Steinfels, C. Blanchet, G. Deleage, A. Di Pietro, and J. M. Jault. 2001. A common mechanism for ATP hydrolysis in ABC transporter and helicase superfamilies. Trends Biochem. Sci. 26539-544. [DOI] [PubMed] [Google Scholar]
- 169.Gerlach, J. H., J. A. Endicott, P. F. Juranka, G. Henderson, F. Sarangi, K. L. Deuchars, and V. Ling. 1986. Homology between P-glycoprotein and bacterial haemolysin transport protein suggests a model for multidrug resistance. Nature 324485-489. [DOI] [PubMed] [Google Scholar]
- 170.Gibbons, S., M. Oluwatuyi, and G. W. Kaatz. 2003. A novel inhibitor of multidrug efflux pumps in Staphylococcus aureus. J. Antimicrob. Chemother. 5113-17. [DOI] [PubMed] [Google Scholar]
- 171.Gilson, E., C. F. Higgins, M. Hofnung, G. Ferro-Luzzi Ames, and H. Nikaido. 1982. Extensive homology between membrane-associated components of histidine and maltose transport systems of Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 2579915-9918. [PubMed] [Google Scholar]
- 172.Goldman, B. S., K. K. Gabbert, and R. G. Kranz. 1996. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J. Bacteriol. 1786348-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Goldman, B. S., K. K. Gabbert, and R. G. Kranz. 1996. Use of heme reporters for studies of cytochrome biosynthesis and heme transport. J. Bacteriol. 1786338-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Goosen, N., and G. F. Moolenaar. 2001. Role of ATP hydrolysis by UvrA and UvrB during nucleotide excision repair. Res. Microbiol. 152401-409. [DOI] [PubMed] [Google Scholar]
- 175.Graham, L. L., R. Harris, W. Villiger, and T. J. Beveridge. 1991. Freeze-substitution of gram-negative eubacteria: general cell morphology and envelope profiles. J. Bacteriol. 1731623-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Greller, G., R. Horlacher, J. DiRuggiero, and W. Boos. 1999. Molecular and biochemical analysis of MalK, the ATP-hydrolyzing subunit of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J. Biol. Chem. 27420259-20264. [DOI] [PubMed] [Google Scholar]
- 177.Greller, G., R. Riek, and W. Boos. 2001. Purification and characterization of the heterologously expressed trehalose/maltose ABC transporter complex of the hyperthermophilic archaeon Thermococcus litoralis. Eur. J. Biochem. 2684011-4018. [DOI] [PubMed] [Google Scholar]
- 178.Grinius, L., G. Dreguniene, E. B. Goldberg, C. H. Liao, and S. J. Projan. 1992. A staphylococcal multidrug resistance gene product is a member of a new protein family. Plasmid 27119-129. [DOI] [PubMed] [Google Scholar]
- 179.Gros, P., J. Croop, and D. Housman. 1986. Mammalian multidrug resistance gene: complete cDNA sequence indicates strong homology to bacterial transport proteins. Cell 47371-380. [DOI] [PubMed] [Google Scholar]
- 180.Guilfoile, P. G., and C. R. Hutchinson. 1991. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc. Natl. Acad. Sci. USA 888553-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guillen-Navarro, K., G. Araiza, A. Garcia-de los Santos, Y. Mora, and M. F. Dunn. 2005. The Rhizobium etli bioMNY operon is involved in biotin transport. FEMS Microbiol. Lett. 250209-219. [DOI] [PubMed] [Google Scholar]
- 182.Guiral, M., O. Viratelle, H. V. Westerhoff, and J. Lankelma. 1994. Cooperative P-glycoprotein mediated daunorubicin transport into DNA-loaded plasma membrane vesicles. FEBS Lett. 346141-145. [DOI] [PubMed] [Google Scholar]
- 183.Hall, J. A., A. K. Ganesan, J. Chen, and H. Nikaido. 1997. Two modes of ligand binding in maltose-binding protein of Escherichia coli. Functional significance in active transport. J. Biol. Chem. 27217615-17622. [DOI] [PubMed] [Google Scholar]
- 184.Hall, J. A., T. E. Thorgeirsson, J. Liu, Y. K. Shin, and H. Nikaido. 1997. Two modes of ligand binding in maltose-binding protein of Escherichia coli. Electron paramagnetic resonance study of ligand-induced global conformational changes by site-directed spin labeling. J. Biol. Chem. 27217610-17614. [DOI] [PubMed] [Google Scholar]
- 185.Hara, T., S. Matsuyama, and H. Tokuda. 2003. Mechanism underlying the inner membrane retention of Escherichia coli lipoproteins caused by Lol avoidance signals. J. Biol. Chem. 27840408-40414. [DOI] [PubMed] [Google Scholar]
- 186.Härle, C., I. Kim, A. Angerer, and V. Braun. 1995. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 141430-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hashimoto, W., J. S. He, Y. Wada, H. Nankai, B. Mikami, and K. Murata. 2005. Proteomics-based identification of outer-membrane proteins responsible for import of macromolecules in Sphingomonas sp A1: alginate-binding flagellin on the cell surface. Biochemistry 4413783-13794. [DOI] [PubMed] [Google Scholar]
- 188.Hashimoto, W., K. Momma, H. Miki, Y. Mishima, E. Kobayashi, O. Miyake, S. Kawai, H. Nankai, B. Mikami, and K. Murata. 1999. Enzymatic and genetic bases on assimilation, depolymerization, and transport of heteropolysaccharides in bacteria. J. Biosci. Bioeng. 87123-136. [DOI] [PubMed] [Google Scholar]
- 189.Havarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16229-240. [DOI] [PubMed] [Google Scholar]
- 190.Hebbeln, P., D. A. Rodionov, A. Alfandega, and T. Eitinger. 2007. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc. Natl. Acad. Sci. USA 1042909-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Henderson, D. P., and S. M. Payne. 1994. Vibrio cholerae iron transport system: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun. 625120-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Herget, M., and R. Tampe. 2007. Intracellular peptide transporters in human compartmentalization of the “peptidome”. Pflugers Arch. 453591-600. [DOI] [PubMed] [Google Scholar]
- 193.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 867-113. [DOI] [PubMed] [Google Scholar]
- 194.Higgins, C. F., and G. F. Ames. 1981. Two periplasmic transport proteins which interact with a common membrane receptor show extensive homology: complete nucleotide sequences. Proc. Natl. Acad. Sci. USA 786038-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Reference deleted.
- 196.Higgins, C. F., I. D. Hiles, G. P. C. Salmond, D. R. Gill, J. A. Downie, I. J. Evans, I. B. Holland, L. Gray, S. D. Buckel, A. W. Bell, and M. A. Hermodson. 1986. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323448-450. [DOI] [PubMed] [Google Scholar]
- 197.Reference deleted.
- 198.Higgins, C. F., I. D. Hiles, K. Whalley, and D. J. Jamieson. 1985. Nucleotide binding by membrane component of bacterial periplasmic binding protein-dependent transport systems. EMBO J. 41033-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Higgins, M. K., E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA 1019994-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hirshberg, M., K. Henrick, L. L. Haire, N. Vasisht, M. Brune, J. E. Corrie, and M. R. Webb. 1998. Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate. Biochemistry 3710381-10385. [DOI] [PubMed] [Google Scholar]
- 201.Hobson, A., R. Weatherwax, and G. F. L. Ames. 1984. ATP-binding sites in the membrane components of the histidine permease, a periplasmic transport system. Proc. Natl. Acad. Sci. USA 817333-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Holland, I. B., H. Benabdelhak, J. Young, A. Pimenta, L. Schmitt, and M. Blight. 2003. Bacterial ABC transporters involved in protein translocation, p. 209-241. In I. B. Holland, S. P. C. Cole, K. Kuchler, and C. F. Higgins (ed.), ABC proteins: from bacteria to man. Academic Press, London, United Kingdom.
- 203.Holland, I. B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293381-399. [DOI] [PubMed] [Google Scholar]
- 204.Holland, I. B., L. Schmitt, and J. Young. 2005. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway. Mol. Membr. Biol. 2229-39. [DOI] [PubMed] [Google Scholar]
- 205.Hollenstein, K., D. C. Frei, and K. P. Locher. 2007. Structure of an ABC transporter in complex with its binding protein. Nature 446213-216. [DOI] [PubMed] [Google Scholar]
- 206.Hopfner, K. P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101789-800. [DOI] [PubMed] [Google Scholar]
- 207.Hor, L. I., and H. A. Shuman. 1993. Genetic analysis of periplasmic binding protein dependent transport in Escherichia coli. Each lobe of maltose-binding protein interacts with a different subunit of the MalFGK2 membrane transport complex. J. Mol. Biol. 233659-670. [DOI] [PubMed] [Google Scholar]
- 208.Hosie, A. H., D. Allaway, M. A. Jones, D. L. Walshaw, A. W. Johnston, and P. S. Poole. 2001. Solute-binding protein-dependent ABC transporters are responsible for solute efflux in addition to solute uptake. Mol. Microbiol. 401449-1459. [DOI] [PubMed] [Google Scholar]
- 209.Hu, Y., S. Rech, R. P. Gunsalus, and D. C. Rees. 1997. Crystal structure of the molybdate binding protein ModA. Nat. Struct. Biol. 4703-707. [DOI] [PubMed] [Google Scholar]
- 210.Huda, N., E. W. Lee, J. Chen, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Molecular cloning and characterization of an ABC multidrug efflux pump, VcaM, in non-O1 Vibrio cholerae. Antimicrob. Agents Chemother. 472413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hughes, A. L. 1994. Evolution of the ATP-binding-cassette transmembrane transporters of vertebrates. Mol. Biol. Evol. 11899-910. [DOI] [PubMed] [Google Scholar]
- 212.Hui, D., C. Morden, F. Zhang, and V. Ling. 2000. Combinatorial analysis of the structural requirements of the Escherichia coli hemolysin signal sequence. J. Biol. Chem. 2752713-2720. [DOI] [PubMed] [Google Scholar]
- 213.Hui, F. M., L. Zhou, and D. A. Morrison. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 15325-31. [DOI] [PubMed] [Google Scholar]
- 214.Huijbregts, R. P., A. I. de Kroon, and B. de Kruijff. 1998. Rapid transmembrane movement of newly synthesized phosphatidylethanolamine across the inner membrane of Escherichia coli. J. Biol. Chem. 27318936-18942. [DOI] [PubMed] [Google Scholar]
- 215.Hunke, S., H. Landmesser, and E. Schneider. 2000. Novel missense mutations that affect the transport function of MalK, the ATP-binding-cassette subunit of the Salmonella enterica serovar Typhimurium maltose transport system. J. Bacteriol. 1821432-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hunke, S., M. Mourez, M. Jehanno, E. Dassa, and E. Schneider. 2000. ATP modulates subunit-subunit interactions in an ATP-binding cassette transporter (MalFGK2) determined by site-directed chemical cross-linking. J. Biol. Chem. 27515526-15534. [DOI] [PubMed] [Google Scholar]
- 217.Hvorup, R. N., B. A. Goetz, M. Niederer, K. Hollenstein, E. Perozo, and K. P. Locher. 2007. Asymmetry in the structure of the ABC transporter binding protein complex BtuCD-BtuF. Science 3171387-1390. [DOI] [PubMed] [Google Scholar]
- 218.Hyde, S. C., P. Emsley, M. J. Hartshorn, M. M. Mimmack, U. Gilaedi, S. R. Pearce, M. P. Gallagher, D. R. Gill, R. E. Hubbard, and C. F. Higgins. 1990. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature 346362-365. [DOI] [PubMed] [Google Scholar]
- 219.Igarashi, Y., K. F. Aoki, H. Mamitsuka, K. Kuma, and M. Kanehisa. 2004. The evolutionary repertoires of the eukaryotic-type ABC transporters in terms of the phylogeny of ATP-binding domains in eukaryotes and prokaryotes. Mol. Biol. Evol. 212149-2160. [DOI] [PubMed] [Google Scholar]
- 220.Ikeno, S., Y. Yamane, Y. Ohishi, N. Kinoshita, M. Hamada, K. S. Tsuchiya, and M. Hori. 2000. ABC transporter genes, kasKLM, responsible for self-resistance of a kasugamycin producer strain. J. Antibiot. 53373-384. [DOI] [PubMed] [Google Scholar]
- 221.Ito, Y., H. Matsuzawa, S. Matsuyama, S. Narita, and H. Tokuda. 2006. Genetic analysis of the mode of interplay between an ATPase subunit and membrane subunits of the lipoprotein-releasing ATP-binding cassette transporter LolCDE. J. Bacteriol. 1882856-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ivetac, A., J. D. Campbell, and M. S. Sansom. 2007. Dynamics and function in a bacterial ABC transporter: simulation studies of the BtuCDF system and its components. Biochemistry 462767-2778. [DOI] [PubMed] [Google Scholar]
- 223.Izadi-Pruneyre, N., N. Wolff, V. Redeker, C. Wandersman, M. Delepierre, and A. Lecroisey. 1999. NMR studies of the C-terminal secretion signal of the haem-binding protein, HasA. Eur. J. Biochem. 261562-568. [DOI] [PubMed] [Google Scholar]
- 224.Jacobs, M. H. J., T. Vanderheide, A. J. M. Driessen, and W. N. Konings. 1996. Glutamate transport in Rhodobacter sphaeroides is mediated by a novel binding protein-dependent secondary transport system. Proc. Natl. Acad. Sci. USA 9312786-12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jacobson, B. L., and F. A. Quiocho. 1988. Sulfate-binding protein dislikes protonated oxyacids. A molecular explanation. J. Mol. Biol. 204783-787. [DOI] [PubMed] [Google Scholar]
- 226.Janas, E., M. Hofacker, M. Chen, S. Gompf, C. van der Does, and R. Tampe. 2003. The ATP hydrolysis cycle of the nucleotide-binding domain of the mitochondrial ATP-binding cassette transporter Mdl1p. J. Biol. Chem. 27826862-26869. [DOI] [PubMed] [Google Scholar]
- 227.Jones, P. M., and A. M. George. 2002. Mechanism of ABC transporters: a molecular dynamics simulation of a well characterized nucleotide-binding subunit. Proc. Natl. Acad. Sci. USA 9912639-12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jones, P. M., and A. M. George. 2007. Nucleotide-dependent allostery within the ABC transporter ATP-binding cassette: a computational study of the MJ0796 dimer. J. Biol. Chem. 28222793-22803. [DOI] [PubMed] [Google Scholar]
- 229.Jones, P. M., and A. M. George. 1999. Subunit interactions in ABC transporters: towards a functional architecture. FEMS Microbiol. Lett. 179187-202. [DOI] [PubMed] [Google Scholar]
- 230.Juliano, R. L., and V. Ling. 1976. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim. Biophys. Acta 455152-162. [DOI] [PubMed] [Google Scholar]
- 231.Kandt, C., Z. Xu, and D. P. Tieleman. 2006. Opening and closing motions in the periplasmic vitamin B12 binding protein BtuF. Biochemistry 4513284-13292. [DOI] [PubMed] [Google Scholar]
- 232.Karcher, A., K. Buttner, B. Martens, R. P. Jansen, and K. P. Hopfner. 2005. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure 13649-659. [DOI] [PubMed] [Google Scholar]
- 233.Karow, M., and C. Georgopoulos. 1993. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 769-79. [DOI] [PubMed] [Google Scholar]
- 234.Karpowich, N., O. Martsinkevich, L. Millen, Y. Yuan, P. L. Dai, K. MacVey, P. J. Thomas, and J. F. Hunt. 2001. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure (Cambridge) 9571-586. [DOI] [PubMed] [Google Scholar]
- 235.Karpowich, N. K., H. H. Huang, P. C. Smith, and J. F. Hunt. 2003. Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding. J. Biol. Chem. 2788429-8434. [DOI] [PubMed] [Google Scholar]
- 236.Kaur, P., D. K. Rao, and S. M. Gandlur. 2005. Biochemical characterization of domains in the membrane subunit DrrB that interact with the ABC subunit DrrA: identification of a conserved motif. Biochemistry 442661-2670. [DOI] [PubMed] [Google Scholar]
- 237.Kaur, P., and J. Russell. 1998. Biochemical coupling between the DrrA and DrrB proteins of the doxorubicin efflux pump of Streptomyces peucetius. J. Biol. Chem. 27317933-17939. [DOI] [PubMed] [Google Scholar]
- 238.Kellermann, O., and S. Szmelcman. 1974. Active transport of maltose in Escherichia coli K12. Involvement of a “periplasmic” maltose binding protein. Eur. J. Biochem. 47139-149. [DOI] [PubMed] [Google Scholar]
- 239.Kelly, D. J., and G. H. Thomas. 2001. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25405-424. [DOI] [PubMed] [Google Scholar]
- 240.Kelly, J., H. Jarrell, L. Millar, L. Tessier, L. M. Fiori, P. C. Lau, B. Allan, and C. M. Szymanski. 2006. Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 1882427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kemner, J. M., X. Liang, and E. W. Nester. 1997. The Agrobacterium tumefaciens virulence gene chvE is part of a putative ABC-type sugar transport operon. J. Bacteriol. 1792452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kempf, B., J. Gade, and E. Bremer. 1997. Lipoprotein from the osmoregulated ABC transport system opuA of Bacillus subtilis—purification of the glycine betaine binding protein and characterization of a functional lipidless mutant. J. Bacteriol. 1796213-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kennedy, K. A., and B. Traxler. 1999. MalK forms a dimer independent of its assembly into the MalFGK2 ATP-binding cassette transporter of Escherichia coli. J. Biol. Chem. 2746259-6264. [DOI] [PubMed] [Google Scholar]
- 244.Kerppola, R. E., and G. F. L. Ames. 1992. Topology of the hydrophobic membrane-bound components of the histidine periplasmic permease. Comparisons with other members of the family. J. Biol. Chem. 2672329-2336. [PubMed] [Google Scholar]
- 245.Khwaja, M., Q. H. Ma, and M. H. Saier. 2005. Topological analysis of integral membrane constituents of prokaryotic ABC efflux systems. Res. Microbiol. 156270-277. [DOI] [PubMed] [Google Scholar]
- 246.Kim, S. H., A. B. Chang, and M. H. Saier, Jr. 2004. Sequence similarity between multidrug resistance efflux pumps of the ABC and RND superfamilies. Microbiology 1502493-2495. [DOI] [PubMed] [Google Scholar]
- 247.Kispal, G., K. Sipos, H. Lange, Z. Fekete, T. Bedekovics, T. Janaky, J. Bassler, D. J. A. Netz, J. Balk, C. Rotte, and R. Lill. 2005. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 24589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Klein, C., and K. D. Entian. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 602793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kobayashi, N., K. Nishino, T. Hirata, and A. Yamaguchi. 2003. Membrane topology of ABC-type macrolide antibiotic exporter MacB in Escherichia coli. FEBS Lett. 546241-246. [DOI] [PubMed] [Google Scholar]
- 250.Kobayashi, N., K. Nishino, and A. Yamaguchi. 2001. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 1835639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37239-253. [DOI] [PubMed] [Google Scholar]
- 252.Kol, M. A., A. van Dalen, A. I. de Kroon, and B. de Kruijff. 2003. Translocation of phospholipids is facilitated by a subset of membrane-spanning proteins of the bacterial cytoplasmic membrane. J. Biol. Chem. 27824586-24593. [DOI] [PubMed] [Google Scholar]
- 253.Koronakis, V., J. Li, E. Koronakis, and K. Stauffer. 1997. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol. Microbiol. 23617-626. [DOI] [PubMed] [Google Scholar]
- 254.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405914-919. [DOI] [PubMed] [Google Scholar]
- 255.Köster, W. 2001. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B-12. Res. Microbiol. 152291-301. [DOI] [PubMed] [Google Scholar]
- 256.Köster, W., and V. Braun. 1990. Iron(III) hydroxamate transport into Escherichia coli: substrate binding to the periplasmic FhuD protein. J. Biol. Chem. 26521407-21410. [PubMed] [Google Scholar]
- 257.Kreimer, D. I., K. P. Chai, and G. F.-L. Ames. 2000. Nonequivalence of the nucleotide-binding subunits of an ABC transporter, the histidine permease, and conformational changes in the membrane complex. Biochemistry 3914183-14195. [DOI] [PubMed] [Google Scholar]
- 258.Kreimer, D. I., H. Malak, J. R. Lakowicz, S. Trakhanov, E. Villar, and V. L. Shnyrov. 2000. Thermodynamics and dynamics of histidine-binding protein, the water-soluble receptor of histidine permease. Implications for the transport of high and low affinity ligands. Eur. J. Biochem. 2674242-4252. [DOI] [PubMed] [Google Scholar]
- 259.Kuan, G., E. Dassa, W. Saurin, M. Hofnung, and M. H. Saier. 1995. Phylogenetic analyses of the ATP-binding constituents of bacterial extracytoplasmic receptor-dependent ABC-type nutrient uptake permeases. Res. Microbiol. 146271-278. [DOI] [PubMed] [Google Scholar]
- 260.Reference deleted.
- 261.Kuhnert, P., B. Heyberger-Meyer, A. P. Burnens, J. Nicolet, and J. Frey. 1997. Detection of RTX toxin genes in gram-negative bacteria with a set of specific probes. Appl. Environ. Microbiol. 632258-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390249-256. [DOI] [PubMed] [Google Scholar]
- 263.Lage, H. 2003. ABC-transporters: implications on drug resistance from microorganisms to human cancers. Int. J. Antimicrob. Agents 22188-199. [DOI] [PubMed] [Google Scholar]
- 264.Lanfermeijer, F. C., F. J. Detmers, W. N. Konings, and B. Poolman. 2000. On the binding mechanism of the peptide receptor of the oligopeptide transport system of Lactococcus lactis. EMBO J. 193649-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Laudenbach, D. E., and A. R. Grossman. 1991. Characterization and mutagenesis of sulfur-regulated genes in a cyanobacterium: evidence for function in sulfate transport. J. Bacteriol. 1732739-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lawson, D. M., C. E. Williams, L. A. Mitchenall, and R. N. Pau. 1998. Ligand size is a major determinant of specificity in periplasmic oxyanion-binding proteins: the 1.2 A resolution crystal structure of Azotobacter vinelandii ModA. Structure 61529-1539. [DOI] [PubMed] [Google Scholar]
- 267.Lazarevic, V., and D. Karamata. 1995. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol. Microbiol. 16345-355. [DOI] [PubMed] [Google Scholar]
- 268.Lebbink, J. H. G., and T. K. Sixma. 2005. Variations on the ABC. Structure 13498-500. [DOI] [PubMed] [Google Scholar]
- 269.Lee, E. W., M. N. Huda, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. EfrAB, an ABC multidrug efflux pump in Enterococcus faecalis. Antimicrob. Agents Chemother. 473733-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lee, S. J., A. Bohm, M. Krug, and W. Boos. 2007. The ABC of binding-protein-dependent transport in archaea. Trends Microbiol. 15389-397. [DOI] [PubMed] [Google Scholar]
- 271.Lee, Y. H., R. K. Deka, M. V. Norgard, J. D. Radolf, and C. A. Hasemann. 1999. Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone. Nat. Struct. Biol. 6628-633. [DOI] [PubMed] [Google Scholar]
- 272.Lemieux, C., C. Otis, and M. Turmel. 2000. Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature 403649-652. [DOI] [PubMed] [Google Scholar]
- 273.Le Roy, F., C. Bisbal, M. Silhol, C. Martinand, B. Lebleu, and T. Salehzada. 2001. The 2-5A/RNase L/RNase L inhibitor (RLI) pathway regulates mitochondrial mRNAs stability in interferon alpha-treated H9 cells. J. Biol. Chem. 27648473-48482. [DOI] [PubMed] [Google Scholar]
- 274.Letoffe, S., P. Delepelaire, and C. Wandersman. 1996. Protein secretion in gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 155804-5811. [PMC free article] [PubMed] [Google Scholar]
- 275.Levdikov, V. M., E. V. Blagova, J. A. Brannigan, L. Wright, A. A. Vagin, and A. J. Wilkinson. 2005. The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J. Mol. Biol. 345879-892. [DOI] [PubMed] [Google Scholar]
- 276.Levina, N., S. Totemeyer, N. R. Stokes, P. Louis, M. A. Jones, and I. R. Booth. 1999. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 181730-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10S122-S129. [DOI] [PubMed] [Google Scholar]
- 278.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64159-204. [DOI] [PubMed] [Google Scholar]
- 279.Linton, K. J., H. N. Cooper, I. S. Hunter, and P. F. Leadlay. 1994. An ABC-transporter from Streptomyces longisporoflavus confers resistance to the polyether-ionophore antibiotic tetronasin. Mol. Microbiol. 11777-785. [DOI] [PubMed] [Google Scholar]
- 280.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 285-13. [DOI] [PubMed] [Google Scholar]
- 281.Liu, C. E., P. Q. Liu, and G. F. L. Ames. 1997. Characterization of the adenosine triphosphatase activity of the periplasmic histidine permease, a traffic ATPase (ABC transporter). J. Biol. Chem. 27221883-21891. [DOI] [PubMed] [Google Scholar]
- 282.Liu, C. E., P. Q. Liu, A. Wolf, E. Lin, and G. F. Ames. 1999. Both lobes of the soluble receptor of the periplasmic histidine permease, an ABC transporter (traffic ATPase), interact with the membrane-bound complex. Effect of different ligands and consequences for the mechanism of action. J. Biol. Chem. 274739-747. [DOI] [PubMed] [Google Scholar]
- 283.Liu, P. Q., C. E. Liu, and G. F. Ames. 1999. Modulation of ATPase activity by physical disengagement of the ATP-binding domains of an ABC transporter, the histidine permease. J. Biol. Chem. 27418310-18318. [DOI] [PubMed] [Google Scholar]
- 284.Liu, Z. Y., M. Jacobs, D. A. Schaff, C. A. McCullen, and A. N. Binns. 2001. ChvD, a chromosomally encoded ATP-binding cassette transporter-homologous protein involved in regulation of virulence gene expression in Agrobacterium tumefaciens. J. Bacteriol. 1833310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 2961091-1098. [DOI] [PubMed] [Google Scholar]
- 286.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95771-778. [DOI] [PubMed] [Google Scholar]
- 287.Lomovskaya, N., S. K. Hong, S. U. Kim, L. Fonstein, K. Furuya, and R. C. Hutchinson. 1996. The Streptomyces peucetius drrC gene encodes a UvrA-like protein involved in daunorubicin resistance and production. J. Bacteriol. 1783238-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Lomovskaya, O., and M. Totrov. 2005. Vacuuming the periplasm. J. Bacteriol. 1871879-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Loo, T. W., M. C. Bartlett, and D. M. Clarke. 2004. The drug-binding pocket of the human multidrug resistance P-glycoprotein is accessible to the aqueous medium. Biochemistry 4312081-12089. [DOI] [PubMed] [Google Scholar]
- 290.Loo, T. W., and D. M. Clarke. 2005. Recent progress in understanding the mechanism of P-glycoprotein-mediated drug efflux. J. Membr. Biol. 206173-185. [DOI] [PubMed] [Google Scholar]
- 291.Loo, T. W., and D. M. Clarke. 1999. The transmembrane domains of the human multidrug resistance P-glycoprotein are sufficient to mediate drug binding and trafficking to the cell surface. J. Biol. Chem. 27424759-24765. [DOI] [PubMed] [Google Scholar]
- 292.Lu, G., J. M. Westbrooks, A. L. Davidson, and J. Chen. 2005. ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation. Proc. Natl. Acad. Sci. USA 10217969-17974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Lubelski, J., A. de Jong, R. van Merkerk, H. Agustiandari, O. P. Kuipers, J. Kok, and A. J. Driessen. 2006. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol. Microbiol. 61771-781. [DOI] [PubMed] [Google Scholar]
- 294.Lubelski, J., W. N. Konings, and A. J. Driessen. 2007. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 71463-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lubelski, J., P. Mazurkiewicz, R. van Merkerk, W. N. Konings, and A. J. Driessen. 2004. ydaG and ydbA of Lactococcus lactis encode a heterodimeric ATP-binding cassette-type multidrug transporter. J. Biol. Chem. 27934449-34455. [DOI] [PubMed] [Google Scholar]
- 296.Lubelski, J., R. van Merkerk, W. N. Konings, and A. J. Driessen. 2006. Nucleotide-binding sites of the heterodimeric LmrCD ABC-multidrug transporter of Lactococcus lactis are asymmetric. Biochemistry 45648-656. [DOI] [PubMed] [Google Scholar]
- 297.Luecke, H., and F. A. Quiocho. 1990. High specificity of a phosphate transport protein determined by hydrogen bonds. Nature 347402-406. [DOI] [PubMed] [Google Scholar]
- 298.Magnusson, U., B. N. Chaudhuri, J. Ko, C. Park, T. A. Jones, and S. L. Mowbray. 2002. Hinge-bending motion of d-allose-binding protein from Escherichia coli: three open conformations. J. Biol. Chem. 27714077-14084. [DOI] [PubMed] [Google Scholar]
- 299.Mannering, D. E., S. Sharma, and A. L. Davidson. 2001. Demonstration of conformational changes associated with activation of the maltose transport complex. J. Biol. Chem. 37612362-12368. [DOI] [PubMed] [Google Scholar]
- 300.Manson, M. D., W. Boos, P. J. Bassford, Jr., and B. A. Rasmussen. 1985. Dependence of maltose transport and chemotaxis on the amount of maltose-binding protein. J. Biol. Chem. 2609727-9733. [PubMed] [Google Scholar]
- 301.Marger, M. D., and M. H. Saier, Jr. 1993. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 1813-20. [DOI] [PubMed] [Google Scholar]
- 302.Margolles, A., M. Putman, H. W. van Veen, and W. N. Konings. 1999. The purified and functionally reconstituted multidrug transporter LmrA of Lactococcus lactis mediates the transbilayer movement of specific fluorescent phospholipids. Biochemistry 3816298-16306. [DOI] [PubMed] [Google Scholar]
- 303.Martineau, P., W. Saurin, M. Hofnung, J. C. Spurlino, and F. A. Quiocho. 1990. Progress in the identification of interaction sites on the periplasmic maltose binding protein from E coli. Biochimie 72397-402. [DOI] [PubMed] [Google Scholar]
- 304.Marvin, J. S., E. E. Corcoran, N. A. Hattangadi, J. V. Zhang, S. A. Gere, and H. W. Hellinga. 1997. The rational design of allosteric interactions in a monomeric protein and its applications to the construction of biosensors. Proc. Natl. Acad. Sci. USA 944366-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Marvin, J. S., and H. W. Hellinga. 2001. Manipulation of ligand binding affinity by exploitation of conformational coupling. Nat. Struct. Biol. 8795-798. [DOI] [PubMed] [Google Scholar]
- 306.Masuda, K., S. Matsuyama, and H. Tokuda. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. USA 997390-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Matsuo, T., J. Chen, Y. Minato, W. Ogawa, T. Mizushima, T. Kuroda, and T. Tsuchiya. 2008. SmdAB, a heterodimeric ABC-type multidrug efflux pump, in Serratia marcescens. J. Bacteriol. 190648-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.McMurry, L., R. E. Petrucci, Jr., and S. B. Levy. 1980. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. USA 773974-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Melis, A., and H. C. Chen. 2005. Chloroplast sulfate transport in green algae—genes, proteins and effects. Photosynth. Res. 86299-307. [DOI] [PubMed] [Google Scholar]
- 310.Mendez, C., and J. A. Salas. 2001. The role of ABC transporters in antibiotic-producing organisms: drug secretion and resistance mechanisms. Res. Microbiol. 152341-350. [DOI] [PubMed] [Google Scholar]
- 311.Meredith, T. C., and R. W. Woodard. 2003. Escherichia coli YrbH is a d-arabinose 5-phosphate isomerase. J. Biol. Chem. 27832771-32777. [DOI] [PubMed] [Google Scholar]
- 312.Merino, G., W. Boos, H. A. Shuman, and E. Bohl. 1995. The inhibition of maltose transport by the unliganded form of the maltose-binding protein of Escherichia coli: experimental findings and mathematical treatment. J. Theor. Biol. 177171-179. [DOI] [PubMed] [Google Scholar]
- 313.Merino, G., and H. A. Shuman. 1998. Truncation of MalF results in lactose transport via the maltose transport system of Escherichia coli. J. Biol. Chem. 2732435-2444. [DOI] [PubMed] [Google Scholar]
- 314.Michiels, J., G. Dirix, J. Vanderleyden, and C. Xi. 2001. Processing and export of peptide pheromones and bacteriocins in gram-negative bacteria. Trends Microbiol. 9164-168. [DOI] [PubMed] [Google Scholar]
- 315.Millet, O., R. P. Hudson, and L. E. Kay. 2003. The energetic cost of domain reorientation in maltose-binding protein as studied by NMR and fluorescence spectroscopy. Proc. Natl. Acad. Sci. USA 10012700-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Mimmack, J. L., M. P. Gallagher, M. P. Hyde, S. R. Pearce, I. R. Booth, and C. F. Higgins. 1989. Energy-coupling to periplasmic binding protein-dependent transport systems: stoichiometry of ATP hydrolysis during transport. Proc. Natl. Acad. Sci. USA 868257-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Miyamoto, A., S. Matsuyama, and H. Tokuda. 2002. Dominant negative mutant of a lipoprotein-specific molecular chaperone, LolA, tightly associates with LolCDE. FEBS Lett. 528193-196. [DOI] [PubMed] [Google Scholar]
- 318.Moody, J. E., L. Millen, D. Binns, J. F. Hunt, and P. J. Thomas. 2002. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J. Biol. Chem. 27721111-21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Mourez, M., M. Hofnung, and E. Dassa. 1997. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J. 163066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Mueller, G. A., W. Y. Choy, D. Yang, J. D. Forman-Kay, R. A. Venters, and L. E. Kay. 2000. Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein. J. Mol. Biol. 300197-212. [DOI] [PubMed] [Google Scholar]
- 321.Muir, M., L. Williams, and T. Ferenci. 1985. Influence of transport energization on the growth yield of Escherichia coli. J. Bacteriol. 1631237-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Murakami, S., R. Nakashima, E. Yamashita, T. Matsumoto, and A. Yamaguchi. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443173-179. [DOI] [PubMed] [Google Scholar]
- 323.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419587-593. [DOI] [PubMed] [Google Scholar]
- 324.Murakami, S., and A. Yamaguchi. 2003. Multidrug-exporting secondary transporters. Curr. Opin. Struct. Biol. 13443-452. [DOI] [PubMed] [Google Scholar]
- 325.Murat, D., P. Bance, I. Callebaut, and E. Dassa. 2006. ATP hydrolysis is essential for the function of the Uup ATP-binding cassette ATPase in precise excision of transposons. J. Biol. Chem. 2816850-6859. [DOI] [PubMed] [Google Scholar]
- 326.Narita, S., K. Kanamaru, S. Matsuyama, and H. Tokuda. 2003. A mutation in the membrane subunit of an ABC transporter LolCDE complex causing outer membrane localization of lipoproteins against their inner membrane-specific signals. Mol. Microbiol. 49167-177. [DOI] [PubMed] [Google Scholar]
- 327.Narita, S., and H. Tokuda. 2006. An ABC transporter mediating the membrane detachment of bacterial lipoproteins depending on their sorting signals. FEBS Lett. 5801164-1170. [DOI] [PubMed] [Google Scholar]
- 328.Navarro, C., L. F. Wu, and M. A. Mandrand-Berthelot. 1993. The nik operon of Escherichia coli encodes a periplasmic binding protein-dependent transport system for nickel. Mol. Microbiol. 91181-1191. [DOI] [PubMed] [Google Scholar]
- 329.Nehme, D., X. Z. Li, R. Elliot, and K. Poole. 2004. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 1862973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Nehme, D., and K. Poole. 2005. Interaction of the MexA and MexB components of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification of MexA extragenic suppressors of a T578I mutation in MexB. Antimicrob. Agents Chemother. 494375-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Neugebauer, H., C. Herrmann, W. Kammer, G. Schwarz, A. Nordheim, and V. Braun. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 1878300-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Neyfakh, A. A., V. E. Bidnenko, and L. B. Chen. 1991. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc. Natl. Acad. Sci. USA 884781-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Nikaido, H., and H. I. Zgurskaya. 2001. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3215-218. [PubMed] [Google Scholar]
- 335.Nikaido, K., and G. F. Ames. 1999. One intact ATP-binding subunit is sufficient to support ATP hydrolysis and translocation in an ABC transporter, the histidine permease. J. Biol. Chem. 27426727-26735. [DOI] [PubMed] [Google Scholar]
- 336.Nishino, K., T. Latifi, and E. A. Groisman. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59126-141. [DOI] [PubMed] [Google Scholar]
- 337.Novotna, G., and J. Janata. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)(LC), from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob. Agents Chemother. 504070-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Novotny, R., C. Schaffer, J. Strauss, and P. Messner. 2004. S-layer glycan-specific loci on the chromosome of Geobacillus stearothermophilus NRS 2004/3a and dTDP-l-rhamnose biosynthesis potential of G. stearothermophilus strains. Microbiology 150953-965. [DOI] [PubMed] [Google Scholar]
- 339.Oh, B. H., G. F. Ames, and S. H. Kim. 1994. Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein. J. Biol. Chem. 26926323-26330. [PubMed] [Google Scholar]
- 340.Ohyama, K., H. Fukuzawa, T. Kohchi, H. Shirai, T. Sano, S. Sano, K. Umesono, Y. Shiki, M. Takeuchi, Z. Chang, S.-I. Aota, H. Inokuchi, and H. Ozeki. 1986. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322572-574. [Google Scholar]
- 341.Olah, G. A., S. Trakhanov, J. Trewhella, and F. A. Quiocho. 1993. Leucine/isoleucine/valine-binding protein contracts upon binding of ligand. J. Biol. Chem. 26816241-16247. [PubMed] [Google Scholar]
- 342.Olano, C., A. M. Rodriguez, C. Mendez, and J. A. Salas. 1995. A second ABC transporter is involved in oleandomycin resistance and its secretion by Streptomyces antibioticus. Mol. Microbiol. 16333-343. [DOI] [PubMed] [Google Scholar]
- 343.Oldham, M. L., D. Khare, F. A. Quiocho, A. L. Davidson, and J. Chen. 2007. Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450515-521. [DOI] [PubMed] [Google Scholar]
- 344.Oloo, E. O., E. Y. Fung, and D. P. Tieleman. 2006. The dynamics of the MgATP-driven closure of MalK, the energy-transducing subunit of the maltose ABC transporter. J. Biol. Chem. 28128397-28407. [DOI] [PubMed] [Google Scholar]
- 345.Oloo, E. O., and D. P. Tieleman. 2004. Conformational transitions induced by the binding of MgATP to the vitamin B12 ATP-binding cassette (ABC) transporter BtuCD. J. Biol. Chem. 27945013-45019. [DOI] [PubMed] [Google Scholar]
- 346.Omote, H., and M. K. Al-Shawi. 2006. Interaction of transported drugs with the lipid bilayer and P-glycoprotein through a solvation exchange mechanism. Biophys. J. 904046-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Orelle, C., O. Dalmas, P. Gros, A. Di Pietro, and J. M. Jault. 2003. The conserved glutamate residue adjacent to the Walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J. Biol. Chem. 27847002-47008. [DOI] [PubMed] [Google Scholar]
- 348.Orelle, C., F. Gubellini, A. Durand, S. Marco, D. Levy, P. Gros, A. Di Pietro, and J. M. Jault. 2008. Conformational change induced by ATP binding in the multidrug ATP-binding cassette transporter BmrA. Biochemistry 472404-2412. [DOI] [PubMed] [Google Scholar]
- 349.Oswald, C., I. B. Holland, and L. Schmitt. 2006. The motor domains of ABC-transporters. What can structures tell us? Naunyn Schmiedebergs Arch. Pharmacol. 372385-399. [DOI] [PubMed] [Google Scholar]
- 350.Otto, M., and F. Gotz. 2001. ABC transporters of staphylococci. Res. Microbiol. 152351-356. [DOI] [PubMed] [Google Scholar]
- 351.Otto, M., A. Peschel, and F. Gotz. 1998. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tu3298. FEMS Microbiol. Lett. 166203-211. [DOI] [PubMed] [Google Scholar]
- 352.Pakotiprapha, D., Y. Inuzuka, B. R. Bowman, G. F. Moolenaar, N. Goosen, D. Jeruzalmi, and G. L. Verdine. 2008. Crystal structure of Bacillus stearothermophilus UvrA provides insight into ATP-modulated dimerization, UvrB interaction, and DNA binding. Mol. Cell 29122-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Park, J. T., D. Raychaudhuri, H. Li, S. Normark, and D. Mengin-Lecreulx. 1998. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-gamma-d-glutamyl-meso-diaminopimelate. J. Bacteriol. 1801215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Patzlaff, J. S., T. van der Heide, and B. Poolman. 2003. The ATP/substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 27829546-29551. [DOI] [PubMed] [Google Scholar]
- 355.Paulsen, I. T., A. M. Beness, and M. H. Saier. 1997. Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology 1432685-2699. [DOI] [PubMed] [Google Scholar]
- 356.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 30175-100. [DOI] [PubMed] [Google Scholar]
- 357.Paulsen, I. T., J. H. Park, P. S. Choi, and M. H. Saier, Jr. 1997. A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol. Lett. 1561-8. [DOI] [PubMed] [Google Scholar]
- 358.Paulsen, I. T., R. A. Skurray, R. Tam, M. H. Saier, Jr., R. J. Turner, J. H. Weiner, E. B. Goldberg, and L. L. Grinius. 1996. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol. 191167-1175. [DOI] [PubMed] [Google Scholar]
- 359.Paulsen, I. T., M. K. Sliwinski, and M. H. Saier, Jr. 1998. Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J. Mol. Biol. 277573-592. [DOI] [PubMed] [Google Scholar]
- 360.Pazzani, C., C. Rosenow, G. J. Boulnois, D. Bronner, K. Jann, and I. S. Roberts. 1993. Molecular analysis of region 1 of the Escherichia coli K5 antigen gene cluster: a region encoding proteins involved in cell surface expression of capsular polysaccharide. J. Bacteriol. 1755978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Petronilli, V., and G. F.-L. Ames. 1991. Binding protein-independent histidine permease mutants. Uncoupling of ATP hydrolysis from transmembrane signaling. J. Biol. Chem. 26616293-16296. [PubMed] [Google Scholar]
- 362.Pflugrath, J. W., and F. A. Quiocho. 1985. Sulphate sequestered in the sulphate-binding protein of Salmonella typhimurium is bound solely by hydrogen bonds. Nature 314257-260. [DOI] [PubMed] [Google Scholar]
- 363.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4629-636. [DOI] [PubMed] [Google Scholar]
- 364.Pigeon, R. P., and R. P. Silver. 1997. Analysis of the G93E mutant allele of KpsM, the membrane component of an ABC transporter involved in polysialic acid translocation in Escherichia coli K1. FEMS Microbiol. Lett. 156217-222. [DOI] [PubMed] [Google Scholar]
- 365.Pimenta, A. L., J. Young, I. B. Holland, and M. A. Blight. 1999. Antibody analysis of the localisation, expression and stability of HlyD, the MFP component of the E. coli haemolysin translocator. Mol. Gen. Genet. 261122-132. [DOI] [PubMed] [Google Scholar]
- 366.Pinkett, H. W., A. T. Lee, P. Lum, K. P. Locher, and D. C. Rees. 2007. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science 315373-377. [DOI] [PubMed] [Google Scholar]
- 367.Pittman, M. S., H. Corker, G. H. Wu, M. B. Binet, A. J. G. Moir, and R. K. Poole. 2002. Cysteine is exported from the Escherichia coli cytoplasm by CydDC, an ATP-binding cassette-type transporter required for cytochrome assembly. J. Biol. Chem. 27749841-49849. [DOI] [PubMed] [Google Scholar]
- 368.Pittman, M. S., H. C. Robinson, and R. K. Poole. 2005. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 28032254-32261. [DOI] [PubMed] [Google Scholar]
- 369.Pleban, K., S. Kopp, E. Csaszar, M. Peer, T. Hrebicek, A. Rizzi, G. F. Ecker, and P. Chiba. 2005. P-glycoprotein substrate binding domains are located at the transmembrane domain/transmembrane domain interfaces: a combined photoaffinity labeling-protein homology modeling approach. Mol. Pharmacol. 67365-374. [DOI] [PubMed] [Google Scholar]
- 370.Poelarends, G. J., and W. N. Konings. 2002. The transmembrane domains of the ABC multidrug transporter LmrA form a cytoplasmic exposed, aqueous chamber within the membrane. J. Biol. Chem. 27742891-42898. [DOI] [PubMed] [Google Scholar]
- 371.Polissi, A., and C. Georgopoulos. 1996. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 201221-1233. [DOI] [PubMed] [Google Scholar]
- 372.Poole, K. 2005. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 5620-51. [DOI] [PubMed] [Google Scholar]
- 373.Poole, K. 2001. Multidrug resistance in gram-negative bacteria. Curr. Opin. Microbiol. 4500-508. [DOI] [PubMed] [Google Scholar]
- 374.Poole, R. K., F. Gibson, and G. H. Wu. 1994. The cydD gene product, component of a heterodimeric ABC transporter, is required for assembly of periplasmic cytochrome-c and of cytochrome-bd in Escherichia coli. FEMS Microbiol. Lett. 117217-224. [DOI] [PubMed] [Google Scholar]
- 375.Poolman, B., J. J. Spitzer, and J. M. Wood. 2004. Bacterial osmosensing: roles of membrane structure and electrostatics in lipid-protein and protein-protein interactions. Biochim. Biophys. Acta 166688-104. [DOI] [PubMed] [Google Scholar]
- 376.Pos, K. M., A. Schiefner, M. A. Seeger, and K. Diederichs. 2004. Crystallographic analysis of AcrB. FEBS Lett. 564333-339. [DOI] [PubMed] [Google Scholar]
- 377.Postle, K., and R. A. Larsen. 2007. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20453-465. [DOI] [PubMed] [Google Scholar]
- 378.Procko, E., I. Ferrin-O'Connell, S. L. Ng, and R. Gaudet. 2006. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol. Cell 2451-62. [DOI] [PubMed] [Google Scholar]
- 379.Prossnitz, E., A. Gee, and G. F. L. Ames. 1989. Reconstitution of the histidine periplasmic transport system in membrane vesicles. Energy coupling and interaction between the binding protein and the membrane complex. J. Biol. Chem. 2645006-5014. [PubMed] [Google Scholar]
- 380.Prossnitz, E., K. Nikaido, S. J. Ulbrich, and G. F.-L. Ames. 1988. Formaldehyde and photoactivatable cross-linking of the periplasmic binding protein to a membrane component of the histidine transport system of Salmonella typhimurium. J. Biol. Chem. 26317917-17920. [PubMed] [Google Scholar]
- 381.Purewal, A. S. 1991. Nucleotide sequence of the ethidium efflux gene from Escherichia coli. FEMS Microbiol. Lett. 66229-231. [DOI] [PubMed] [Google Scholar]
- 382.Putman, M., H. W. Van Veen, J. E. Degener, and W. N. Konings. 2000. Antibiotic resistance: era of the multidrug pump. Mol. Microbiol. 36772-773. [DOI] [PubMed] [Google Scholar]
- 383.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Qu, Q., and F. J. Sharom. 2002. Proximity of bound Hoechst 33342 to the ATPase catalytic sites places the drug binding site of P-glycoprotein within the cytoplasmic membrane leaflet. Biochemistry 414744-4752. [DOI] [PubMed] [Google Scholar]
- 385.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287467-484. [DOI] [PubMed] [Google Scholar]
- 386.Reference deleted.
- 387.Quiocho, F. A. 1996. Atomic basis of the exquisite specificity of phosphate and sulfate transport receptors. Kidney Int. 49943-946. [DOI] [PubMed] [Google Scholar]
- 388.Quiocho, F. A. 1990. Atomic structures of periplasmic binding proteins and the high-affinity active transport systems in bacteria. Philos. Trans. R. Soc. Lond. B 326341-351. [DOI] [PubMed] [Google Scholar]
- 389.Quiocho, F. A., and P. S. Ledvina. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 2017-25. [DOI] [PubMed] [Google Scholar]
- 390.Raetz, C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76295-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Rao, D. K., and P. Kaur. 2008. The Q-loop of DrrA is involved in producing the closed conformation of the nucleotide binding domains and in transduction of conformational changes between DrrA and DrrB. Biochemistry 473038-3050. [DOI] [PubMed] [Google Scholar]
- 393.Ravaud, S., M. A. Do Cao, M. Jidenko, C. Ebel, M. Le Maire, J. M. Jault, A. Di Pietro, R. Haser, and N. Aghajari. 2006. The ABC transporter BmrA from Bacillus subtilis is a functional dimer when in a detergent-solubilized state. Biochem. J. 395345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Reddy, M., and J. Gowrishankar. 2000. Characterization of the uup locus and its role in transposon excisions and tandem repeat deletions in Escherichia coli. J. Bacteriol. 1821978-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Reeves, A. R., J. N. D'Elia, J. Frias, and A. A. Salyers. 1996. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J. Bacteriol. 178823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Ren, D., B. Navarro, H. Xu, L. Yue, Q. Shi, and D. E. Clapham. 2001. A prokaryotic voltage-gated sodium channel. Science 2942372-2375. [DOI] [PubMed] [Google Scholar]
- 397.Reuter, G., T. Janvilisri, H. Venter, S. Shahi, L. Balakrishnan, and H. W. van Veen. 2003. The ATP binding cassette multidrug transporter LmrA and lipid transporter MsbA have overlapping substrate specificities. J. Biol. Chem. 27835193-35198. [DOI] [PubMed] [Google Scholar]
- 398.Reynolds, E., J. I. Ross, and J. H. Cove. 2003. Msr(A) and related macrolide/streptogramin resistance determinants: incomplete transporters? Int. J. Antimicrob. Agents 22228-236. [DOI] [PubMed] [Google Scholar]
- 399.Reynolds, P. E., and P. Courvalin. 2005. Vancomycin resistance in enterococci due to synthesis of precursors terminating in d-alanyl-d-serine. Antimicrob. Agents Chemother. 4921-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Rince, A., A. Dufour, P. Uguen, J. P. Lepennec, and D. Haras. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 634252-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Rocchetta, H. L., and J. S. Lam. 1997. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J. Bacteriol. 1794713-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Rodionov, D. A., P. Hebbeln, M. S. Gelfand, and T. Eitinger. 2006. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2002. Comparative genomics of thiamin biosynthesis in procaryotes—new genes and regulatory mechanisms. J. Biol. Chem. 27748949-48959. [DOI] [PubMed] [Google Scholar]
- 404.Rodriguez, A. M., C. Olano, C. Vilches, C. Mendez, and J. A. Salas. 1993. Streptomyces antibioticus contains at least 3 oleandomycin-resistance determinants, one of which shows similarity with proteins of the ABC-transporter superfamily. Mol. Microbiol. 8571-582. [DOI] [PubMed] [Google Scholar]
- 405.Rodriguez, G. M., and I. Smith. 2006. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol. 188424-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Ross, J. I., E. A. Eady, J. H. Cove, W. J. Cunliffe, S. Baumberg, and J. C. Wootton. 1990. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 41207-1214. [DOI] [PubMed] [Google Scholar]
- 407.Roth, J. R., J. G. Lawrence, M. Rubenfield, S. Kieffer-Higgins, and G. M. Church. 1993. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 1753303-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Rouquette-Loughlin, C. E., J. T. Balthazar, and W. M. Shafer. 2005. Characterization of the MacA-MacB efflux system in Neisseria gonorrhoeae. J. Antimicrob. Chemother. 56856-860. [DOI] [PubMed] [Google Scholar]
- 408a.Ruiz, N., L. S. Gronenberg, D. Kahne, and T. J. Silhavy. 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1055537-5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Russell, R. R. B., J. Aduse-Opoku, I. C. Sutcliffe, L. Tao, and J. J. Ferretti. 1992. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 2674631-4637. [PubMed] [Google Scholar]
- 410.Sack, J. S., M. A. Saper, and F. A. Quiocho. 1989. Periplasmic binding protein structure and function. Refined X-ray structures of the leucine/isoleucine/valine-binding protein and its complex with leucine. J. Mol. Biol. 206171-191. [DOI] [PubMed] [Google Scholar]
- 411.Saier, M. H. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Saier, M. H. 2000. Families of proteins forming transmembrane channels. J. Membr. Biol. 175165-180. [DOI] [PubMed] [Google Scholar]
- 413.Saier, M. H., Jr., R. Tam, A. Reizer, and J. Reizer. 1994. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol. Microbiol. 11841-847. [DOI] [PubMed] [Google Scholar]
- 414.Sakamoto, K., A. Margolles, H. W. van Veen, and W. N. Konings. 2001. Hop resistance in the beer spoilage bacterium Lactobacillus brevis is mediated by the ATP-binding cassette multidrug transporter HorA. J. Bacteriol. 1835371-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Sami, M., K. Suzuki, K. Sakamoto, H. Kadokura, K. Kitamoto, and K. Yoda. 1998. A plasmid pRH45 of Lactobacillus brevis confers hop resistance. J. Gen. Appl. Microbiol. 44361-363. [DOI] [PubMed] [Google Scholar]
- 416.Sanchez, L., W. Pan, M. Vinas, and H. Nikaido. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 1796855-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Sanchez-Fernandez, R., T. G. E. Davies, J. O. D. Coleman, and P. A. Rea. 2001. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 27630231-30244. [DOI] [PubMed] [Google Scholar]
- 418.Sapriel, G., C. Wandersman, and P. Delepelaire. 2002. The N terminus of the HasA protein and the SecB chaperone cooperate in the efficient targeting and secretion of HasA via the ATP-binding cassette transporter. J. Biol. Chem. 2776726-6732. [DOI] [PubMed] [Google Scholar]
- 419.Sauna, Z. E., and S. V. Ambudkar. 2007. About a switch: how P-glycoprotein (ABCB1) harnesses the energy of ATP binding and hydrolysis to do mechanical work. Mol. Cancer Ther. 613-23. [DOI] [PubMed] [Google Scholar]
- 420.Saurin, W., M. Hofnung, and E. Dassa. 1999. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 4822-41. [DOI] [PubMed] [Google Scholar]
- 421.Saurin, W., W. Koster, and E. Dassa. 1994. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol. Microbiol. 12993-1004. [DOI] [PubMed] [Google Scholar]
- 422.Reference deleted.
- 423.Scheffel, F., R. Fleischer, and E. Schneider. 2004. Functional reconstitution of a maltose ATP-binding cassette transporter from the thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius. Biochim. Biophys. Acta 165657-65. [DOI] [PubMed] [Google Scholar]
- 424.Schirmer, T., T. A. Keller, Y. F. Wang, and J. P. Rosenbusch. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science 267512-514. [DOI] [PubMed] [Google Scholar]
- 425.Schmitt, L., H. Benabdelhak, M. A. Blight, I. B. Holland, and M. T. Stubbs. 2003. Crystal structure of the nucleotide-binding domain of the ABC-transporter haemolysin B: identification of a variable region within ABC helical domains. J. Mol. Biol. 330333-342. [DOI] [PubMed] [Google Scholar]
- 426.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 221-20. [DOI] [PubMed] [Google Scholar]
- 427.Schneider, E., S. Wilken, and R. Schmid. 1994. Nucleotide-induced conformational changes of MalK, a bacterial ATP binding cassette transporter protein. J. Biol. Chem. 26920456-20461. [PubMed] [Google Scholar]
- 428.Schoner, B., M. Geistlich, P. Rosteck, Jr., R. N. Rao, E. Seno, P. Reynolds, K. Cox, S. Burgett, and C. Hershberger. 1992. Sequence similarity between macrolide-resistance determinants and ATP-binding transport proteins. Gene 11593-96. [DOI] [PubMed] [Google Scholar]
- 429.Schulein, K., K. Schmid, and R. Benzl. 1991. The sugar-specific outer membrane channel ScrY contains functional characteristics of general diffusion pores and substrate-specific porins. Mol. Microbiol. 52233-2241. [DOI] [PubMed] [Google Scholar]
- 430.Schumacher, M. A., and R. G. Brennan. 2003. Deciphering the molecular basis of multidrug recognition: crystal structures of the Staphylococcus aureus multidrug binding transcription regulator QacR. Res. Microbiol. 15469-77. [DOI] [PubMed] [Google Scholar]
- 431.Schumacher, M. A., and R. G. Brennan. 2002. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 45885-893. [DOI] [PubMed] [Google Scholar]
- 432.Schumacher, M. A., M. C. Miller, and R. G. Brennan. 2004. Structural mechanism of the simultaneous binding of two drugs to a multidrug-binding protein. EMBO J. 232923-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 2942158-2163. [DOI] [PubMed] [Google Scholar]
- 434.Schuurman-Wolters, G. K., and B. Poolman. 2005. Substrate specificity and ionic regulation of GlnPQ from Lactococcus lactis—an ATP-binding cassette transporter with four extracytoplasmic substrate-binding domains. J. Biol. Chem. 28023785-23790. [DOI] [PubMed] [Google Scholar]
- 435.Sebulsky, M. T., B. H. Shilton, C. D. Speziali, and D. E. Heinrichs. 2003. The role of FhuD2 in iron(III)-hydroxamate transport in Staphylococcus aureus. Demonstration that FhuD2 binds iron(III)-hydroxamates but with minimal conformational change and implication of mutations on transport. J. Biol. Chem. 27849890-49900. [DOI] [PubMed] [Google Scholar]
- 436.Seeger, M. A., A. Schiefner, T. Eicher, F. Verrey, K. Diederichs, and K. M. Pos. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 3131295-1298. [DOI] [PubMed] [Google Scholar]
- 437.Seelig, A. 1998. A general pattern for substrate recognition by P-glycoprotein. Eur. J. Biochem. 251252-261. [DOI] [PubMed] [Google Scholar]
- 438.Seelig, A. 2006. Unraveling membrane-mediated substrate-transporter interactions. Biophys. J. 903825-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Seelig, A., X. L. Blatter, and F. Wohnsland. 2000. Substrate recognition by P-glycoprotein and the multidrug resistance-associated protein MRP1: a comparison. Int. J. Clin. Pharmacol. Ther. 38111-121. [DOI] [PubMed] [Google Scholar]
- 440.Senior, A. E., M. K. Al-Shawi, and I. L. Urbatsch. 1995. The catalytic cycle of P-glycoprotein. FEBS Lett. 377285-289. [DOI] [PubMed] [Google Scholar]
- 441.Reference deleted.
- 442.Severi, E., G. Randle, P. Kivlin, K. Whitfield, R. Young, R. Moxon, D. Kelly, D. Hood, and G. H. Thomas. 2005. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 581173-1185. [DOI] [PubMed] [Google Scholar]
- 443.Seydel, A., P. Gounon, and A. P. Pugsley. 1999. Testing the “+2 rule” for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34810-821. [DOI] [PubMed] [Google Scholar]
- 444.Shapiro, A. B., and V. Ling. 1997. Extraction of Hoechst 33342 from the cytoplasmic leaflet of the plasma membrane by P-glycoprotein. Eur. J. Biochem. 250122-129. [DOI] [PubMed] [Google Scholar]
- 445.Shapiro, A. B., and V. Ling. 1997. Positively cooperative sites for drug transport by P-glycoprotein with distinct drug specificities. Eur. J. Biochem. 250130-137. [DOI] [PubMed] [Google Scholar]
- 446.Sharff, A. J., L. E. Rodseth, J. E. Spurlino, and F. A. Quiocho. 1992. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry 3110657-10663. [DOI] [PubMed] [Google Scholar]
- 447.Sharma, S., and A. L. Davidson. 2000. Vanadate-induced trapping of nucleotide by the purified maltose transport complex requires ATP hydrolysis. J. Bacteriol. 1826570-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Sharom, F. J. 2006. Shedding light on drug transport: structure and function of the P-glycoprotein multidrug transporter (ABCB1). Biochem. Cell Biol. 84979-992. [DOI] [PubMed] [Google Scholar]
- 449.Sheps, J. A., S. Ralph, Z. Zhao, D. L. Baillie, and V. Ling. 2004. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 5R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Shilling, R., L. Federici, F. Walas, H. Venter, S. Velamakanni, B. Woebking, L. Balakrishnan, B. Luisi, and H. W. van Veen. 2005. A critical role of a carboxylate in proton conduction by the ATP-binding cassette multidrug transporter LmrA. FASEB J. 191698-1700. [DOI] [PubMed] [Google Scholar]
- 451.Shuman, H. A. 1982. Active transport of maltose in Escherichia coli K-12: role of the periplasmic maltose binding protein and evidence for a substrate recognition site in the cytoplasmic membrane. J. Biol. Chem. 2575455-5461. [PubMed] [Google Scholar]
- 452.Siegers, K., and K. D. Entian. 1995. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 611082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Silver, R. P., W. Aaronson, and W. F. Vann. 1987. Translocation of capsular polysaccharides in pathogenic strains of Escherichia coli requires a 60-kilodalton periplasmic protein. J. Bacteriol. 1695489-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Silver, R. P., K. Prior, C. Nsahlai, and L. F. Wright. 2001. ABC transporters and the export of capsular polysaccharides from gram-negative bacteria. Res. Microbiol. 152357-364. [DOI] [PubMed] [Google Scholar]
- 455.Singh, K. V., G. M. Weinstock, and B. E. Murray. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 461845-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Skrynnikov, N. R., N. K. Goto, D. Yang, W. Y. Choy, J. R. Tolman, G. A. Mueller, and L. E. Kay. 2000. Orienting domains in proteins using dipolar couplings measured by liquid-state NMR: differences in solution and crystal forms of maltodextrin binding protein loaded with beta-cyclodextrin. J. Mol. Biol. 2951265-1273. [DOI] [PubMed] [Google Scholar]
- 457.Smith, P. C., N. Karpowich, L. Millen, J. E. Moody, J. Rosen, P. J. Thomas, and J. F. Hunt. 2002. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Sonne, J., C. Kandt, G. H. Peters, F. Y. Hansen, M. O. Jensen, and D. P. Tieleman. 2007. Simulation of the coupling between nucleotide binding and transmembrane domains in the ABC transporter BtuCD. Biophys. J. 922727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Speiser, D. M., and G. F.-L. Ames. 1991. Salmonella typhimurium histidine periplasmic permease mutations that allow transport in the absence of histidine-binding proteins. J. Bacteriol. 1731444-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Sperandeo, P., R. Cescutti, R. Villa, C. Di Benedetto, D. Candia, G. Deho, and A. Polissi. 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Sperandeo, P., C. Pozzi, G. Deho, and A. Polissi. 2006. Non-essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res. Microbiol. 157547-558. [DOI] [PubMed] [Google Scholar]
- 462.Steinfels, E., C. Orelle, O. Dalmas, F. Penin, B. Miroux, A. Di Pietro, and J. M. Jault. 2002. Highly efficient over-production in E. coli of YvcC, a multidrug-like ATP-binding cassette transporter from Bacillus subtilis. Biochim. Biophys. Acta 15651-5. [DOI] [PubMed] [Google Scholar]
- 463.Steinfels, E., C. Orelle, J. R. Fantino, O. Dalmas, J. L. Rigaud, F. Denizot, A. Di Pietro, and J. M. Jault. 2004. Characterization of YvcC (BmrA), a multidrug ABC transporter constitutively expressed in Bacillus subtilis. Biochemistry 437491-7502. [DOI] [PubMed] [Google Scholar]
- 464.Steinke, A., S. Grau, A. Davidson, E. Hofmann, and M. Ehrmann. 2001. Characterization of transmembrane segments 3, 4, and 5 of MalF by mutational analysis. J. Bacteriol. 183375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 451126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Szmelcman, S., and M. Hofnung. 1975. Maltose transport in Escherichia coli K12: involvement of the bacteriophage λ receptor. J. Bacteriol. 124112-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Szmelcman, S., N. Sassoon, and M. Hofnung. 1997. Residues in the alpha helix 7 of the bacterial maltose binding protein which are important in interactions with the Mal FGK2 complex. Protein Sci. 6628-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Szmelcman, S., M. Schwartz, T. J. Silhavy, and W. Boos. 1976. Maltose transport in Escherichia coli K12. A comparison of transport kinetics in wild type and λ-resistant mutants with the dissociation constants of the maltose-binding protein as measured by fluorescence quenching. Eur. J. Biochem. 6513-19. [DOI] [PubMed] [Google Scholar]
- 469.Takeda, K., H. Miyatake, N. Yokota, S. Matsuyama, H. Tokuda, and K. Miki. 2003. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 223199-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Tamber, S., M. M. Ochs, and R. E. Hancock. 2006. Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J. Bacteriol. 18845-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Tang, C., C. D. Schwieters, and G. M. Clore. 2007. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature 4491078-1082. [DOI] [PubMed] [Google Scholar]
- 473.Taniguchi, N., S. Matsuyama, and H. Tokuda. 2005. Mechanisms underlying energy-independent transfer of lipoproteins from LolA to LolB, which have similar unclosed β-barrel structures. J. Biol. Chem. 28034481-34488. [DOI] [PubMed] [Google Scholar]
- 474.Tao, L., I. C. Sutcliffe, R. R. B. Russell, and J. J. Ferretti. 1993. Transport of sugars, including sucrose, by the MSM transport system of Streptococcus mutans. J. Dent. Res. 721386-1390. [DOI] [PubMed] [Google Scholar]
- 475.Tapia, M. I., M. Mourez, M. Hofnung, and E. Dassa. 1999. Structure-function study of MalF protein by random mutagenesis. J. Bacteriol. 1812267-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Tefsen, B., M. P. Bos, F. Beckers, J. Tommassen, and H. de Cock. 2005. MsbA is not required for phospholipid transport in Neisseria meningitidis. J. Biol. Chem. 28035961-35966. [DOI] [PubMed] [Google Scholar]
- 477.Tefsen, B., J. Geurtsen, F. Beckers, J. Tommassen, and H. de Cock. 2005. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J. Biol. Chem. 2804504-4509. [DOI] [PubMed] [Google Scholar]
- 478.Telmer, P. G., and B. H. Shilton. 2003. Insights into the conformational equilibria of maltose-binding protein by analysis of high affinity mutants. J. Biol. Chem. 27834555-34567. [DOI] [PubMed] [Google Scholar]
- 479.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from E.coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 176487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Thöny-Meyer, L. 2002. Cytochrome c maturation: a complex pathway for a simple task? Biochem. Soc. Trans. 30633-638. [DOI] [PubMed] [Google Scholar]
- 481.Tikhonova, E. B., V. K. Devroy, S. Y. Lau, and H. I. Zgurskaya. 2007. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol. Microbiol. 63895-910. [DOI] [PubMed] [Google Scholar]
- 482.Tombline, G., L. A. Bartholomew, G. A. Tyndall, K. Gimi, I. L. Urbatsch, and A. E. Senior. 2004. Properties of P-glycoprotein with mutations in the “catalytic carboxylate” glutamate residues. J. Biol. Chem. 27946518-46526. [DOI] [PubMed] [Google Scholar]
- 483.Tombline, G., L. A. Bartholomew, I. L. Urbatsch, and A. E. Senior. 2004. Combined mutation of catalytic glutamate residues in the two nucleotide binding domains of P-glycoprotein generates a conformation that binds ATP and ADP tightly. J. Biol. Chem. 27931212-31220. [DOI] [PubMed] [Google Scholar]
- 484.Tombline, G., A. Muharemagic, L. B. White, and A. E. Senior. 2005. Involvement of the “occluded nucleotide conformation” of P-glycoprotein in the catalytic pathway. Biochemistry 4412879-12886. [DOI] [PubMed] [Google Scholar]
- 485.Tomii, K., and M. Kanehisa. 1998. A comparative analysis of ABC transporters in complete microbial genomes. Genome Res. 81048-1059. [DOI] [PubMed] [Google Scholar]
- 486.Trakhanov, S., N. K. Vyas, H. Luecke, D. M. Kristensen, J. Ma, and F. A. Quiocho. 2005. Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity. Biochemistry 446597-6608. [DOI] [PubMed] [Google Scholar]
- 487.Treptow, N. A., and H. A. Shuman. 1988. Allele-specific malE mutations that restore interactions between maltose-binding protein and the inner-membrane components of the maltose transport system. J. Mol. Biol. 202809-822. [DOI] [PubMed] [Google Scholar]
- 488.Treptow, N. A., and H. A. Shuman. 1985. Genetic evidence for substrate and periplasmic-binding-protein recognition by the MalF and MalG proteins, cytoplasmic membrane components of the Escherichia coli maltose transport system. J. Bacteriol. 163654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Trias, J., and H. Nikaido. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 26515680-15684. [PubMed] [Google Scholar]
- 490.Trias, J., E. Y. Rosenberg, and H. Nikaido. 1988. Specificity of the glucose channel formed by protein D1 of Pseudomonas aeruginosa. Biochim. Biophys. Acta 938493-496. [DOI] [PubMed] [Google Scholar]
- 491.Tsuda, H., Y. Yamashita, Y. Shibata, Y. Nakano, and T. Koga. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 463756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Turmel, M., C. Otis, and C. Lemieux. 1999. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc. Natl. Acad. Sci. USA 9610248-10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Urbatsch, I. L., M. Julien, I. Carrier, M. E. Rousseau, R. Cayrol, and P. Gros. 2000. Mutational analysis of conserved carboxylate residues in the nucleotide binding sites of P-glycoprotein. Biochemistry 3914138-14149. [DOI] [PubMed] [Google Scholar]
- 494.Urbatsch, I. L., B. Sankaran, S. Bhagat, and A. E. Senior. 1995. Both P-glycoprotein nucleotide-binding sites are catalytically active. J. Biol. Chem. 27026956-26962. [DOI] [PubMed] [Google Scholar]
- 495.Urbatsch, I. L., B. Sankaran, J. Weber, and A. E. Senior. 1995. P-glycoprotein is stably inhibited by vanadate-induced trapping of nucleotide at a single catalytic site. J. Biol. Chem. 27019383-19390. [DOI] [PubMed] [Google Scholar]
- 496.van den Berg van Saparoea, H. B., J. Lubelski, R. van Merkerk, P. S. Mazurkiewicz, and A. J. Driessen. 2005. Proton motive force-dependent Hoechst 33342 transport by the ABC transporter LmrA of Lactococcus lactis. Biochemistry 4416931-16938. [DOI] [PubMed] [Google Scholar]
- 497.van der Heide, T., and B. Poolman. 2002. ABC transporters: one, two or four extracytoplasmic substrate-binding sites? EMBO Rep. 3938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Reference deleted.
- 499.van der Meer, J. R., J. Polman, M. M. Beerthuyzen, R. J. Siezen, O. P. Kuipers, and W. M. De Vos. 1993. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 1752578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.van Veen, H. W., R. Callaghan, L. Soceneantu, A. Sardini, W. N. Konings, and C. F. Higgins. 1998. A bacterial antibiotic-resistance gene that complements the human multidrug-resistance P-glycoprotein gene. Nature 391291-295. [DOI] [PubMed] [Google Scholar]
- 501.van Veen, H. W., C. F. Higgins, and W. N. Konings. 2001. Molecular basis of multidrug transport by ATP-binding cassette transporters: a proposed two-cylinder engine model. J. Mol. Microbiol. Biotechnol. 3185-192. [PubMed] [Google Scholar]
- 502.van Veen, H. W., C. F. Higgins, and W. N. Konings. 2001. Multidrug transport by ATP binding cassette transporters: a proposed two-cylinder engine mechanism. Res. Microbiol. 152365-374. [DOI] [PubMed] [Google Scholar]
- 503.van Veen, H. W., A. Margolles, M. Muller, C. F. Higgins, and W. N. Konings. 2000. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J. 192503-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.van Veen, H. W., K. Venema, H. Bolhuis, I. Oussenko, J. Kok, B. Poolman, A. J. Driessen, and W. N. Konings. 1996. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc. Natl. Acad. Sci. USA 9310668-10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Vazquez de Aldana, C. R., M. J. Marton, and A. G. Hinnebusch. 1995. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2 alpha kinase GCN2 in amino acid-starved cells. EMBO J. 143184-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Venter, H., R. A. Shilling, S. Velamakanni, L. Balakrishnan, and H. W. Van Veen. 2003. An ABC transporter with a secondary-active multidrug translocator domain. Nature 426866-870. [DOI] [PubMed] [Google Scholar]
- 507.Venter, H., S. Velamakanni, L. Balakrishnan, and H. W. van Veen. 2008. On the energy-dependence of Hoechst 33342 transport by the ABC transporter LmrA. Biochem. Pharmacol. 75866-874. [DOI] [PubMed] [Google Scholar]
- 508.Verdon, G., S. V. Albers, B. W. Dijkstra, A. J. Driessen, and A. M. Thunnissen. 2003. Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations. J. Mol. Biol. 330343-358. [DOI] [PubMed] [Google Scholar]
- 509.Verdon, G., S. V. Albers, N. van Oosterwijk, B. W. Dijkstra, A. J. M. Driessen, and A. Thunnissen. 2003. Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus. J. Mol. Biol. 334255-267. [DOI] [PubMed] [Google Scholar]
- 510.Vigano, C., A. Margolles, H. W. van Veen, W. N. Konings, and J. M. Ruysschaert. 2000. Secondary and tertiary structure changes of reconstituted LmrA induced by nucleotide binding or hydrolysis. A Fourier transform attenuated total reflection infrared spectroscopy and tryptophan fluorescence quenching analysis. J. Biol. Chem. 27510962-10967. [DOI] [PubMed] [Google Scholar]
- 511.Vyas, N. K., M. N. Vyas, and F. A. Quiocho. 2003. Crystal structure of M tuberculosis ABC phosphate transport receptor: specificity and charge compensation dominated by ion-dipole interactions. Structure 11765-774. [DOI] [PubMed] [Google Scholar]
- 512.Wacker, M., D. Linton, P. G. Hitchen, M. Nita-Lazar, S. M. Haslam, S. J. North, M. Panico, H. R. Morris, A. Dell, B. W. Wren, and M. Aebi. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 2981790-1793. [DOI] [PubMed] [Google Scholar]
- 513.Wakasugi, T., T. Nagai, M. Kapoor, M. Sugita, M. Ito, S. Ito, J. Tsudzuki, K. Nakashima, T. Tsudzuki, Y. Suzuki, A. Hamada, T. Ohta, A. Inamura, K. Yoshinaga, and M. Sugiura. 1997. Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division. Proc. Natl. Acad. Sci. USA 945967-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Walsh, F. M., and S. G. Amyes. 2004. Microbiology and drug resistance mechanisms of fully resistant pathogens. Curr. Opin. Microbiol. 7439-444. [DOI] [PubMed] [Google Scholar]
- 515.Wanders, R. J. A., W. F. Visser, C. W. T. van Roermund, S. Kemp, and H. R. Waterham. 2007. The peroxisomal ABC transporter family. Pflugers Arch. 453719-734. [DOI] [PubMed] [Google Scholar]
- 516.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58611-647. [DOI] [PubMed] [Google Scholar]
- 517.Wandersman, C., M. Schwartz, and T. Ferenci. 1979. Escherichia coli mutants impaired in maltodextrin transport. J. Bacteriol. 1401-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Ward, A., C. L. Reyes, J. Yu, C. B. Roth, and G. Chang. 2007. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl. Acad. Sci. USA 10419005-19010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Weiner, J. H., C. E. Furlong, and L. A. Heppel. 1971. A binding protein for l-glutamine and its relation to active transport in E. coli. Arch. Biochem. Biophys. 142715-717. [DOI] [PubMed] [Google Scholar]
- 520.Weng, J., J. Ma, K. Fan, and W. Wang. 2008. The conformational coupling and translocation mechanism of vitamin B12 ATP-binding cassette transporter BtuCD. Biophys. J. 94612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Whitfield, C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 7539-68. [DOI] [PubMed] [Google Scholar]
- 522.Whitfield, C. 1995. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 3178-185. [DOI] [PubMed] [Google Scholar]
- 523.Wiener, M. C. 2005. TonB-dependent outer membrane transport: going for baroque? Curr. Opin. Struct. Biol. 15394-400. [DOI] [PubMed] [Google Scholar]
- 524.Wilkinson, A. J., and K. H. G. Verschueren. 2003. Crystal structures of periplasmic solute-binding proteins in ABC transport complexes illuminate their function, p. 187-207. In I. B. Holland, S. P. C. Cole, K. Kuchler, and C. F. Higgins (ed.), ABC proteins: from bacteria to man. Academic Press, London, United Kingdom.
- 525.Winans, S. C., R. A. Kerstetter, and E. W. Nester. 1988. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J. Bacteriol. 1704047-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Woebking, B., G. Reuter, R. A. Shilling, S. Velamakanni, S. Shahi, H. Venter, L. Balakrishnan, and H. W. van Veen. 2005. Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA. J. Bacteriol. 1876363-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Wolf, A., K. C. Lee, J. F. Kirsch, and G. F. L. Ames. 1996. Ligand-dependent conformational plasticity of the periplasmic histidine-binding protein HisJ. Involvement in transport specificity. J. Biol. Chem. 27121243-21250. [DOI] [PubMed] [Google Scholar]
- 528.Wolf, A., E. W. Shaw, B. H. Oh, H. De Bondt, A. K. Joshi, and G. F. Ames. 1995. Structure/function analysis of the periplasmic histidine-binding protein. Mutations decreasing ligand binding alter the properties of the conformational change and of the closed form. J. Biol. Chem. 27016097-16106. [DOI] [PubMed] [Google Scholar]
- 529.Wolff, N., G. Sapriel, C. Bodenreider, A. Chaffotte, and P. Delepelaire. 2003. Antifolding activity of the SecB chaperone is essential for secretion of HasA, a quickly folding ABC pathway substrate. J. Biol. Chem. 27838247-38253. [DOI] [PubMed] [Google Scholar]
- 530.Wolschendorf, F., M. Mahfoud, and M. Niederweis. 2007. Porins are required for uptake of phosphates by Mycobacterium smegmatis. J. Bacteriol. 1892435-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Wright, G. D. 2005. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Deliv. Rev. 571451-1470. [DOI] [PubMed] [Google Scholar]
- 532.Wu, J., and R. W. Woodard. 2003. Escherichia coli YrbI is 3-deoxy-d-manno-octulosonate 8-phosphate phosphatase. J. Biol. Chem. 27818117-18123. [DOI] [PubMed] [Google Scholar]
- 533.Wu, T., A. C. McCandlish, L. S. Gronenberg, S. S. Chng, T. J. Silhavy, and D. Kahne. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 10311754-11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Xu, Y., K. V. Singh, X. Qin, B. E. Murray, and G. M. Weinstock. 2000. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect. Immun. 68815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2212-218. [DOI] [PubMed] [Google Scholar]
- 536.Yamaguchi, K., F. Yu, and M. Inouye. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53423-432. [DOI] [PubMed] [Google Scholar]
- 537.Yarmus, M., A. Mett, and R. Shapira. 2000. Cloning and expression of the genes involved in the production of and immunity against the bacteriocin lacticin RM. Biochim. Biophys. Acta 1490279-290. [DOI] [PubMed] [Google Scholar]
- 538.Yarunin, A., V. G. Panse, E. Petfalski, C. Dez, D. Tollervey, and E. C. Hurt. 2005. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 24580-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Yasuhisa, K., M. Shin-ya, M. Michinori, and U. Kazumitsu. 2007. Mechanism of multidrug recognition by MDR1/ABCB1. Cancer Sci. 981303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Ye, J., A. R. Osborne, M. Groll, and T. A. Rapoport. 2004. RecA-like motor ATPases—lessons from structures. Biochim. Biophys. Acta 16591-18. [DOI] [PubMed] [Google Scholar]
- 541.Yerushalmi, H., M. Lebendiker, and S. Schuldiner. 1995. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 2706856-6863. [DOI] [PubMed] [Google Scholar]
- 542.Young, J., and I. B. Holland. 1999. ABC transporters: bacterial exporters—revisited five years on. Biochim. Biophys. Acta 1461177-200. [DOI] [PubMed] [Google Scholar]
- 543.Yu, E. W., J. R. Aires, and H. Nikaido. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 1855657-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300976-980. [DOI] [PubMed] [Google Scholar]
- 545.Zaitseva, J., S. Jenewein, T. Jumpertz, I. B. Holland, and L. Schmitt. 2005. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 241901-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Zaitseva, J., S. Jenewein, A. Wiedenmann, H. Benabdelhak, I. B. Holland, and L. Schmitt. 2005. Functional characterization and ATP-induced dimerization of the isolated ABC-domain of the haemolysin B transporter. Biochemistry 449680-9690. [DOI] [PubMed] [Google Scholar]
- 547.Zaitseva, J., C. Oswald, T. Jumpertz, S. Jenewein, A. Wiedenmann, I. B. Holland, and L. Schmitt. 2006. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. EMBO J. 253432-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Zgurskaya, H. I., and H. Nikaido. 1999. AcrA is a highly asymmetric protein capable of spanning the periplasm. J. Mol. Biol. 285409-420. [DOI] [PubMed] [Google Scholar]
- 549.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 1824264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37219-225. [DOI] [PubMed] [Google Scholar]
- 551.Zhang, F., J. A. Sheps, and V. Ling. 1993. Complementation of transport-deficient mutants of Escherichia coli alpha-hemolysin by second-site mutations in the transporter hemolysin B. J. Biol. Chem. 26819889-19895. [PubMed] [Google Scholar]
- 552.Zhou, Z. M., K. A. White, A. Polissi, C. Georgopoulos, and C. R. H. Raetz. 1998. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 27312466-12475. [DOI] [PubMed] [Google Scholar]
- 553.Zolghadr, B., S. Weber, Z. Szabo, A. J. Driessen, and S. V. Albers. 2007. Identification of a system required for the functional surface localization of sugar binding proteins with class III signal peptides in Sulfolobus solfataricus. Mol. Microbiol. 64795-806. [DOI] [PubMed] [Google Scholar]
- 554.Zou, J. Y., M. M. Flocco, and S. L. Mowbray. 1993. The 1.7 A refined X-ray structure of the periplasmic glucose/galactose receptor from Salmonella typhimurium. J. Mol. Biol. 233739-752. [DOI] [PubMed] [Google Scholar]