Abstract
Linezolid resistance has dominantly been mediated by mutations in 23S rRNA or ribosomal protein L4 genes. Recently, cfr has demonstrated the ability to produce a phenotype of resistance to not only oxazolidinones, but also other antimicrobial classes (phenicols, lincosamides, pleuromutilins, and streptogramin A). We describe the first detection of cfr-mediated linezolid resistance in Staphylococcus aureus and Staphylococcus epidermidis recovered from human infection cases monitored during the 2007 LEADER Program.
Linezolid, the first oxazolidinone class agent used in clinical practice, has demonstrated potent antimicrobial activity against gram-positive pathogens, including methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and Streptococcus spp. (3). According to the LEADER Program (5), nearly all S. aureus strains (>99.9%) and coagulase-negative staphylococci (98.4%) isolated in the United States were susceptible to linezolid. Furthermore, similar results (99.8% susceptibility) were observed when testing a global collection of gram-positive isolates evaluated by the ZAAPS Program in the same year (5, 6). Linezolid resistance has appeared only sporadically since its introduction in 2000, and it is usually mediated by the presence of mutations in one or more alleles of the target 23S rRNA gene (4, 11). However, some linezolid-resistant isolates fail to display these mutations, indicating the presence of other resistance mechanisms.
Previously, the cfr gene was described as a chloramphenicol resistance mechanism in Staphylococcus sciuri (14). The cfr-encoded product, a methyltransferase, provides posttranscriptional methylation of the 23S rRNA at position A2503. This methylation affects the binding of at least four antimicrobial classes (phenicols, lincosamides, pleuromutilins, and streptogramin A), leading to a multidrug-resistant phenotype (10). This gene has been detected in Staphylococcus spp. of animal origin in Europe (7, 8, 10, 14). One recent report described the detection of cfr in a Staphylococcus aureus isolate recovered from the respiratory tract of an infected patient in Colombia (16).
The LEADER Program evaluates the activity of linezolid and numerous comparator agents against gram-positive clinical isolates recovered from more than 50 medical centers within the United States. During the 2007 LEADER Program, linezolid-resistant S. aureus (004-737X) and Staphylococcus epidermidis (426-3147L) were forwarded to JMI Laboratories (North Liberty, IA) and tested for susceptibility by the Clinical and Laboratory Standards Institute (CLSI) reference broth microdilution method (1). The S. aureus strain was isolated from a 45-year-old paraplegic female patient residing in a nursing home. She was admitted to a hospital in Ohio showing symptoms of urinary tract infection, pneumonia, and sepsis, which required mechanical ventilation. Candida spp. were recovered from a blood culture, and the patient received an antifungal agent plus ciprofloxacin therapy. Pseudomonas spp. were also recovered from a urine culture. S. aureus (004-737X), Klebsiella pneumoniae, and Pseudomonas alcaligenes were recovered from a bronchio-alveolar lavage specimen. The patient subsequently received azithromycin, vancomycin, linezolid, and piperacillin-tazobactam and remained hospitalized for 1 month secondary to respiratory failure. S. epidermidis (426-3147L) was isolated from a 79-year-old female living in a long-term care facility. Between July and September 2007, she was admitted to a hospital in Arizona and returned to the long-term care facility multiple times before being placed in hospital care. S. epidermidis was recovered from a blood culture 3 days after the second hospital admission. She received vancomycin, cefepime, and ampicillin/sulbactam. No linezolid use could be documented.
Both isolates (004-737X and 426-3147L) displayed linezolid-nonsusceptible phenotypes (MICs of 8 and >256 μg/ml, respectively), which were confirmed by the Etest (AB Biodisk, Solna, Sweden) and the disk diffusion methods (2), with results at 8 and >256 μg/ml or 19 and 6 mm, respectively. Additionally, the isolates showed resistance to chloramphenicol, clindamycin, quinupristin-dalfopristin, retapamulin, oxacillin, ciprofloxacin, erythromycin (S. aureus only), tetracycline, and trimethoprim-sulfamethoxazole but remained susceptible to vancomycin (Table 1). These results led to screenings for the G2576T mutation in the 23S rRNA genes (11) and the previously described cfr gene (8). The G2576T mutation was not present, but a positive PCR result was obtained using cfr-specific primers, which was confirmed in both isolates by sequencing. S. aureus 004-737X was further analyzed for the characterization of SCCmec types and the presence of PVL genes (lukF-PV and lukS-PV) (12) and was subjected to pulsed-field gel electrophoresis (PFGE). The PFGE pattern was compared to those of contemporary community-acquired and hospital-associated methicillin-resistant S. aureus clones prevalent in the United States (15). Additionally, both isolates were screened for erythromycin resistance determinants, as previously described (9). Characterization of SCCmec types (I through VI) of S. aureus isolate 004-737X was unsuccessful, and the reaction for the presence of PVL genes was negative. The isolate showed a unique PFGE pattern compared to those of the predominant U.S. clones, and erythromycin resistance in the isolate 004-737X was mediated by ermA, while the isolate 426-3147L showed negative results for the most common ermA, ermB, ermC, and mefA resistance genes. This latter result does not exclude the possibility that the isolate 426-3147L harbored other ribosomal methylation or efflux pump genes, which could explain the decreased susceptibility (4 μg/ml) to erythromycin.
TABLE 1.
Antimicrobial susceptibility profiles of cfr-harboring S. aureus (004-737X) and S. epidermidis (426-3147L) isolates
| Antimicrobial agent | MIC (μg/ml)
|
|
|---|---|---|
| 004-737X | 426-3147L | |
| Linezolid | 8 | >256 |
| Chloramphenicol | >256 | >256 |
| Clindamycin | >256 | >256 |
| Quinupristin-dalfopristin | 8 | 4 |
| Retapamulin | 32 | >32 |
| Oxacillin | >2 | >2 |
| Ciprofloxacin | >32 | >32 |
| Erythromycin | >256a | 4b |
| Mupirocin | 0.25 | 64 |
| Tetracycline | >8 | >8 |
| Trimethoprim-sulfamethoxazole | >2 | >2 |
| Vancomycin | 0.5 | 2 |
Erythromycin resistance is mediated by ermA.
No macrolide resistance mechanism was detected.
Plasmid DNA was extracted using the plasmid DNA midi kit (Qiagen GmbH, Hilden, Germany), separated on 1% agarose gel in Tris-acetate-EDTA buffer on a Criterion sub-cell GT system (Bio-Rad, Hercules, CA), and transferred onto a nylon membrane by Southern blotting (13). A labeled cfr probe was used for hybridization, which was revealed with a nonradioactive DIG-High Prime DNA labeling and detection kit (Roche Diagnostics GmbH, Mannheim, Germany). Plasmid sizes were determined by comparison with standard plasmid DNAs extracted from Escherichia coli NCTC 50192 and NCTC 50193. Analysis of the plasmid content of isolates 004-737X and 426-3147L revealed the presence of two plasmids in each isolate (550 and 55 kb, and 175 and 75 kb, respectively). Experiments showed that the 55- and 175-kb plasmid DNAs from isolates 004-737X and 426-3147L, respectively, hybridized with the cfr-specific probe (data not shown).
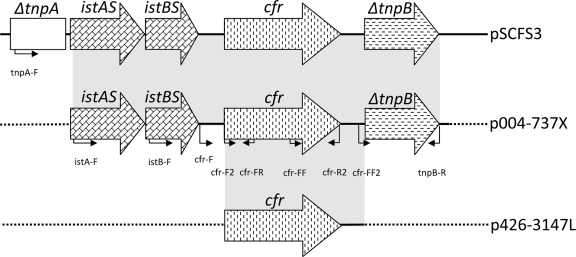
Surrounding cfr DNA sequences were accessed by primer walking. Downstream of the cfr gene, the presence of ΔtnpB was noted in the S. aureus isolate, which was identical to the structure described for the pSCFS3 plasmid found in an S. aureus isolate collected from the respiratory tract of a swine (7) (AM086211) (Fig. 1). The DNA sequence upstream of the cfr gene in the S. aureus isolate showed the presence of istAS and istBS genes, which were also identical to those of the pSCFS3 plasmid, suggesting that these insertion sequences may be involved in the mobilization of the cfr gene (7). However, a PCR using a primer targeting ΔtnpA (tnpA-F), which was located further upstream of the cfr gene on the pSCFS3 plasmid, yielded a negative result, suggesting that the upstream region of cfr on this isolate significantly differed from that of the pSCFS3 plasmid. PCRs performed with the primers cfr-F and cfr-R2 or cfr-F2 and tnpB-R for the 426-3147L isolate yielded negative results, also indicating additional distinct DNA sequences upstream and downstream of the cfr gene on this isolate. Further analysis of the cfr genetic context in the 426-3147L isolate is ongoing.
FIG. 1.
Schematic representation of cfr surrounding DNA sequences in S. aureus (004-737X) and S. epidermidis (426-3147L) isolates. The genetic context of pSCFS3 is also shown for comparison purposes (AM086211). Genes are indicated by boxes, and the arrows indicate their transcriptional orientations. Small arrows indicate primers targeting regions and their respective orientations. Dashes indicate unknown DNA sequences. The background shading indicates identity to the pSCFS3 DNA sequence.
Linezolid resistance, as described in numerous earlier reports, has been mediated by mutations in 23S rRNA or other ribosomal protein genes, implying the slow dissemination of resistance by these mechanisms (10). However, the detection of a plasmid-borne cfr-mediated linezolid resistance gene in staphylococci recovered from human specimens in the United States adds a new dimension to the threat against the clinical utility of several antimicrobial classes, including the oxazolidinones.
Although S. aureus 004-737X did not belong to one of the prevalent clones in the United States (15) and cfr-carrying Staphylococcus sp. isolates appear rare (10), these data require continued active resistance surveillance programs (such as LEADER and ZAAPS). This must be combined with effective infection control strategies in case further spread of this resistance mechanism is observed by those programs. The dissemination of the cfr-mediated resistance genes among staphylococcal clinical isolates is especially worrisome given the potential for rapid simultaneous increases in resistance rates for several antimicrobial classes.
Nucleotide sequence accession number.
The nucleotide sequences of the cfr gene from S. aureus 004-737X have been deposited in the GenBank database under the accession number EU598691.
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7, 7th edition. CLSI, Wayne, PA.
- 2.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. M100-S17. CLSI, Wayne, PA.
- 3.Diekema, D. J., and R. N. Jones. 2001. Oxazolidinone antibiotics. Lancet 358:1975-1982. [DOI] [PubMed] [Google Scholar]
- 4.Farrell, D. J., I. Morrissey, S. Bakker, S. Buckridge, and D. Felmingham. 2004. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob. Agents Chemother. 48:3169-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones, R. N., T. R. Fritsche, H. S. Sader, and J. E. Ross. 2007. LEADER surveillance program results for 2006: an activity and spectrum analysis of linezolid using clinical isolates from the United States (50 medical centers). Diagn. Microbiol. Infect. Dis. 59:309-317. [DOI] [PubMed] [Google Scholar]
- 6.Jones, R. N., T. R. Fritsche, H. S. Sader, and J. E. Ross. 2007. Zyvox Annual Appraisal of Potency and Spectrum Program results for 2006: an activity and spectrum analysis of linezolid using clinical isolates from 16 countries. Diagn. Microbiol. Infect. Dis. 59:199-209. [DOI] [PubMed] [Google Scholar]
- 7.Kehrenberg, C., F. M. Aarestrup, and S. Schwarz. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kehrenberg, C., and S. Schwarz. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lina, G., A. Quaglia, M. E. Reverdy, R. Leclercq, F. Vandenesch, and J. Etienne. 1999. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long, K. S., J. Poehlsgaard, C. Kehrenberg, S. Schwarz, and B. Vester. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meka, V. G., S. K. Pillai, G. Sakoulas, C. Wennersten, L. Venkataraman, P. C. DeGirolami, G. M. Eliopoulos, R. C. Moellering, Jr., and H. S. Gold. 2004. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J. Infect. Dis. 190:311-317. [DOI] [PubMed] [Google Scholar]
- 12.Mendes, R. E., H. S. Sader, L. Deshpande, and R. N. Jones. 2008. Antimicrobial activity of tigecycline against community-acquired methicillin-resistant Staphylococcus aureus isolates recovered from North American medical centers. Diagn. Microbiol. Infect. Dis. 60:433-436. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., P. MacCallum, and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Schwarz, S., C. Werckenthin, and C. Kehrenberg. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toh, S. M., L. Xiong, C. A. Arias, M. V. Villegas, K. Lolans, J. Quinn, and A. S. Mankin. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]