Abstract
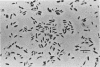
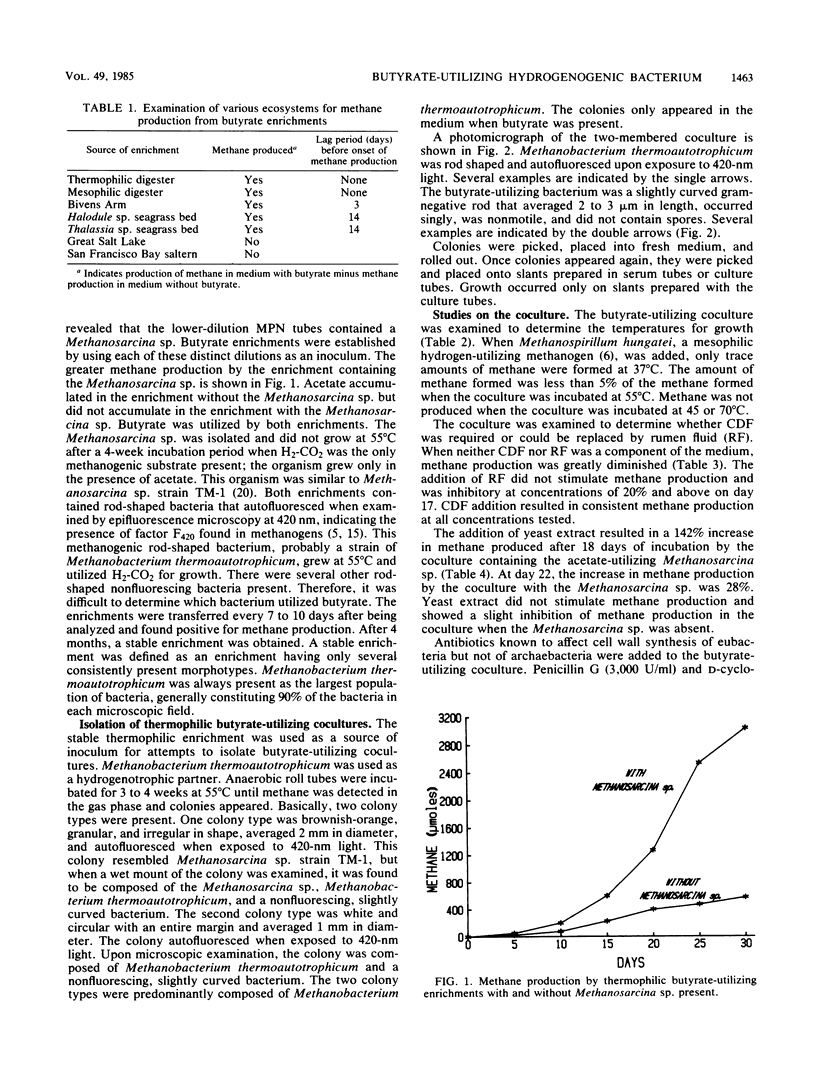
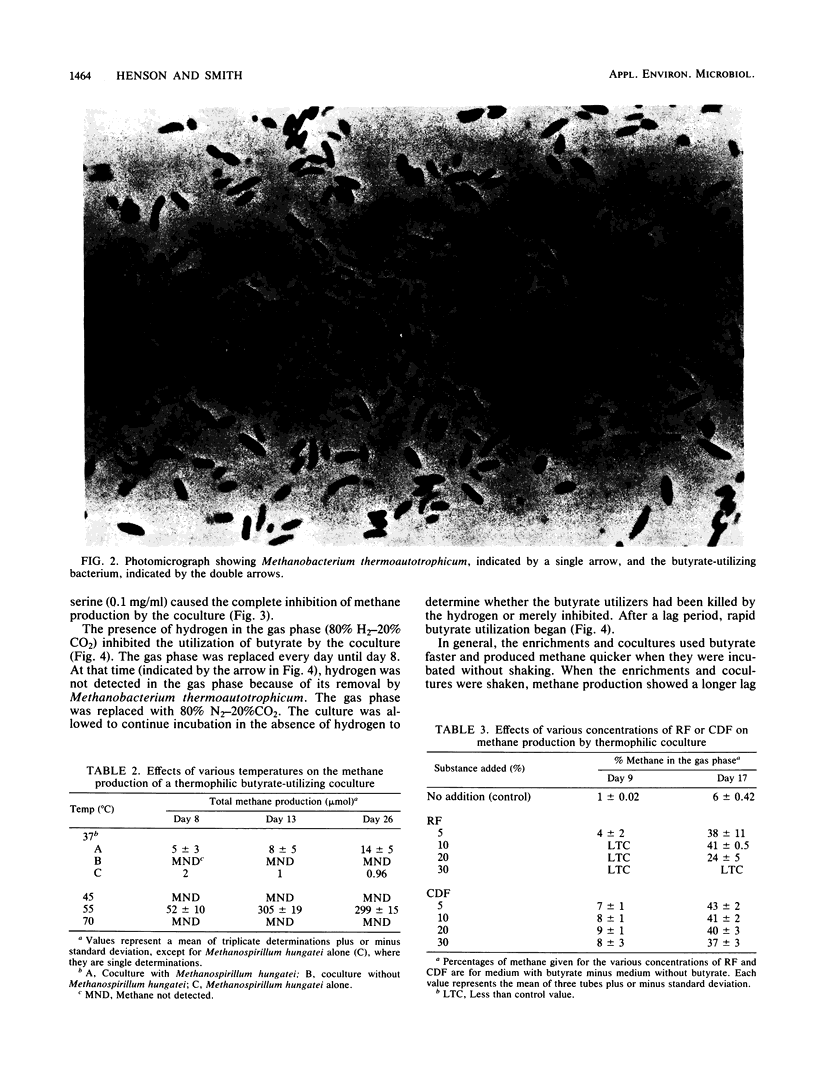
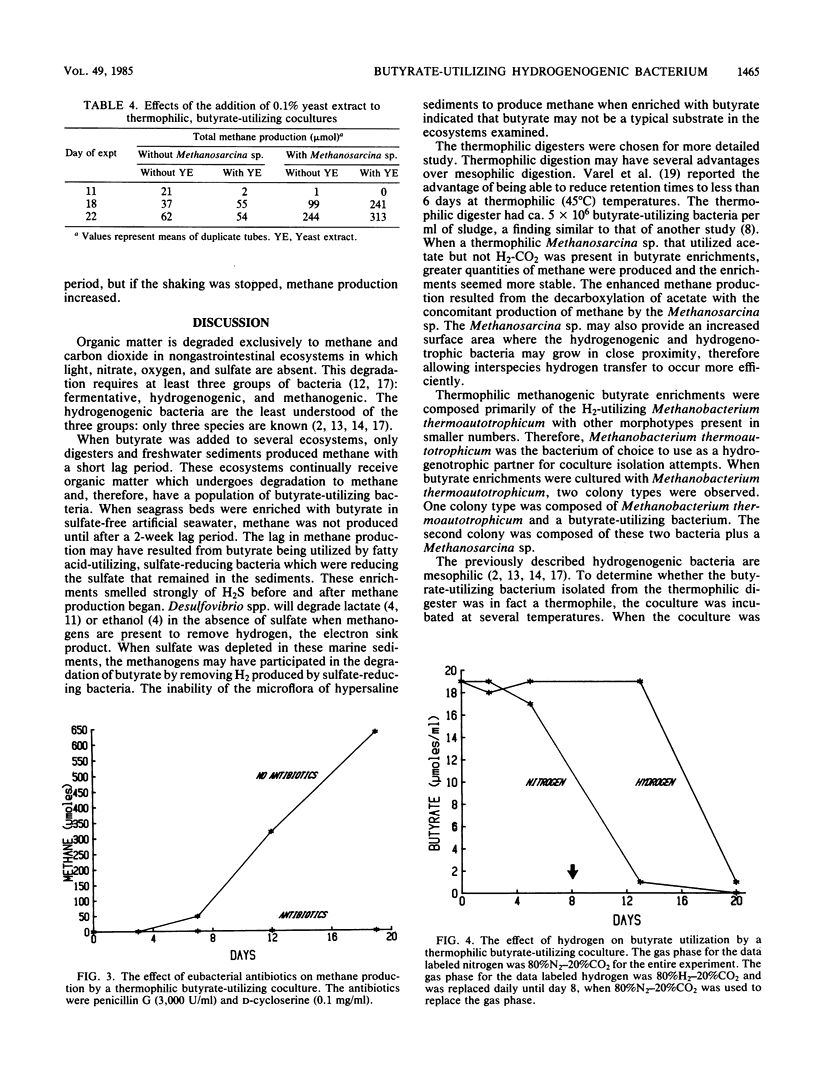
Sludge from a thermophilic, 55°C digester produced methane without a lag period when enriched with butyrate. The sludge was found by most-probable-number enumeration to have ca. 5 × 106 butyrate-utilizing bacteria per ml. A thermophilic butyrate-utilizing bacterium was isolated in coculture with Methanobacterium thermoautotrophicum. This bacterium was a gram-negative, slightly curved rod, occurred singly, was nonmotile, and did not appear to produce spores. When this coculture was incubated with Methanospirillum hungatei at 37°C, the quantity of methane produced was less than 5% of the methane produced when the coculture was incubated at 55°C, the routine incubation temperature. The coculture required clarified digester fluid. The addition of yeast extract to medium containing 5% clarified digester fluid stimulated methane production when a Methanosarcina sp. was present. Hydrogen in the gas phase prevented butyrate utilization. However, when the hydrogen was removed, butyrate utilization began. Penicillin G and d-cycloserine caused the complete inhibition of butyrate utilization by the coculture. The ability of various ecosystems to convert butyrate to methane was studied. Marine sediments enriched with butyrate required a 2-week incubation period before methanogenesis began. Hypersaline sediments did not produce methane after 3 months when enriched with butyrate.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone D. R., Bryant M. P. Propionate-Degrading Bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from Methanogenic Ecosystems. Appl Environ Microbiol. 1980 Sep;40(3):626–632. doi: 10.1128/aem.40.3.626-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M. P., Campbell L. L., Reddy C. A., Crabill M. R. Growth of desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl Environ Microbiol. 1977 May;33(5):1162–1169. doi: 10.1128/aem.33.5.1162-1169.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddema H. J., Vogels G. D. Improved identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1978 Nov;36(5):752–754. doi: 10.1128/aem.36.5.752-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie R. I., Bryant M. P. Metabolic Activity of Fatty Acid-Oxidizing Bacteria and the Contribution of Acetate, Propionate, Butyrate, and CO(2) to Methanogenesis in Cattle Waste at 40 and 60 degrees C. Appl Environ Microbiol. 1981 Jun;41(6):1363–1373. doi: 10.1128/aem.41.6.1363-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M. J., Bryant M. P. Anaerobic Degradation of Lactate by Syntrophic Associations of Methanosarcina barkeri and Desulfovibrio Species and Effect of H(2) on Acetate Degradation. Appl Environ Microbiol. 1981 Feb;41(2):346–354. doi: 10.1128/aem.41.2.346-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M. J., Bryant M. P., Hespell R. B., Costerton J. W. Syntrophomonas wolfei gen. nov. sp. nov., an Anaerobic, Syntrophic, Fatty Acid-Oxidizing Bacterium. Appl Environ Microbiol. 1981 Apr;41(4):1029–1039. doi: 10.1128/aem.41.4.1029-1039.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink R. W., Dugan P. R. Tentative identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 Mar;33(3):713–717. doi: 10.1128/aem.33.3.713-717.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivard C. J., Henson J. M., Thomas M. V., Smith P. H. Isolation and Characterization of Methanomicrobium paynteri sp. nov., a Mesophilic Methanogen Isolated from Marine Sediments. Appl Environ Microbiol. 1983 Aug;46(2):484–490. doi: 10.1128/aem.46.2.484-490.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton D. R., Tiedje J. M. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic Acid. Appl Environ Microbiol. 1984 Oct;48(4):840–848. doi: 10.1128/aem.48.4.840-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varel V. H., Hashimoto A. G., Chen Y. R. Effect of temperature and retention time on methane production from beef cattle waste. Appl Environ Microbiol. 1980 Aug;40(2):217–222. doi: 10.1128/aem.40.2.217-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder S. H., Mah R. A. Isolation and Characterization of a Thermophilic Strain of Methanosarcina Unable to Use H(2)-CO(2) for Methanogenesis. Appl Environ Microbiol. 1979 Nov;38(5):996–1008. doi: 10.1128/aem.38.5.996-1008.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]