Abstract
Inhibitors of DNA methyltransferase, typified by 5-aza-2′-deoxycytidine (5-Aza-CdR), induce the expression of genes transcriptionally down-regulated by de novo methylation in tumor cells. We utilized gene expression microarrays to examine the effects of 5-Aza-CdR treatment in HT29 colon adenocarcinoma cells. This analysis revealed the induction of a set of genes that implicated IFN signaling in the HT29 cellular response to 5-Aza-CdR. Subsequent investigations revealed that the induction of this gene set correlates with the induction of signal transducer and activator of transcription (STAT) 1, 2, and 3 genes and their activation by endogenous IFN-α. These observations implicate the induction of the IFN-response pathway as a major cellular response to 5-Aza-CdR and suggests that the expression of STATs 1, 2, and 3 can be regulated by DNA methylation. Consistent with STAT’s limiting cell responsiveness to IFN, we found that 5-Aza-CdR treatment sensitized HT29 cells to growth inhibition by exogenous IFN-α2a, indicating that 5-Aza-CdR should be investigated as a potentiator of IFN responsiveness in certain IFN-resistant tumors.
DNA cytosine methyltransferase I (DNA MeTase) recognizes hemimethylated CpG dinucleotides in mammalian DNA and catalyzes the transfer of methyl groups to cytosine residues in newly synthesized DNA (1). The methylation of cytosines within CpG islands located in core promoter regions can negatively regulate the transcription of the adjacent genes. The basis for this negative regulation may involve recruitment of histone deacetylases to methylated CpG islands (1). Holliday first suggested a relationship between abnormal DNA methylation and cancer (2). Subsequently, a number of methylation-silenced tumor suppressor genes, including p16Ink4a, retinoblastoma, estrogen receptor, hMLH1, and E-cadherin, have been identified in cancer cells in vitro and in vivo (3–8). It is becoming clear that epigenetic processes constitute a significant factor in the formation of cancer (9). In this regard, DNA methylation abnormalities have been implicated in colon cancers in both mouse and human tumor model systems (6, 10–12).
5-Aza-2′-deoxycytidine (5-Aza-CdR) inhibits DNA methylation and often is used in vitro to induce the reexpression of genes putatively silenced by promoter methylation (8). 5-Aza-CdR is substituted for cytosine during replication and is recognized by DNA MeTase (13). Attempted transfer of methyl groups to 5-Aza-CdR, however, covalently traps the enzyme to newly synthesized DNA (14, 15). This sequestration ultimately depletes cellular stores of DNA MeTase and results in widespread genomic hypomethylation. Clinical trials have demonstrated promise in the use of 5-Aza-CdR (decitabine) for treating leukemia, and current trials are evaluating 5-Aza-CdR in the treatment of lung and prostate cancers (16–19). It is plausible that the antitumor activity of 5-Aza-CdR results from the induction of methylation-regulated tumor-suppressive pathways.
The identification of methylation-silenced genes is offering new insights into tumor development and may reveal the potential for inhibiting DNA methylation as a cancer treatment (20). In this regard, a number of strategies have been used to uncover methylation-regulated genes, including candidate gene analysis, representational difference analysis, restriction landmark genome scanning, methylation-sensitive, arbitrarily primed PCR, and methylated DNA-binding protein affinity chromatography (21–24). Another strategy, gene expression microarrays, is particularly suited for identifying candidate, methylation-silenced genes and for assessing the downstream, cellular consequences of reactivating these genes. Microarray technology permits the systematic examination of thousands of gene expression changes simultaneously and has been used to follow the transcriptional changes that accompany disease development and cellular responses to environmental stimuli (25–29).
In view of the clinical interest in 5-Aza-CdR and our incomplete understanding of the cellular consequences of inhibiting DNA MeTase, we have utilized gene expression microarrays to probe the effects of treating colon tumor cells with 5-Aza-CdR. Here, we show that 5-Aza-CdR inhibits the growth of HT29 colon carcinoma cells and that this growth inhibition parallels the transcriptional induction of IFN-responsive genes. Subsequent analysis revealed induction of signal transducers and activators of transcription 1, 2, and 3 (STATs 1, 2, and 3), elements central to IFN signaling. Given the established growth-inhibitory properties of IFNs, our data offer a new model for understanding the cellular consequences of inhibiting DNA MeTase.
Materials and Methods
Cell Culture and Drug Treatments.
HT29 adenocarcinoma cells (American Type Culture Collection) were cultured at 37°C in 5% CO2 by using McCoy’s medium supplemented with 10% FBS (GIBCO). For treatments with 5-Aza-CdR, cells were exposed to 500 nM 5-Aza-CdR (Sigma) 24 hr after passage in complete culture medium. Control cultures were treated in parallel with vehicle (PBS). Twenty-four hours after drug addition, culture medium was replaced with drug-free medium. Control and 5-Aza-CdR-treated cells were subcultured at equal densities at 1 and 5 days after the initial treatment, and proliferation was measured at the subsequent time point by using a Coulter counter.
In other experiments, HT29 cells (control or pretreated with 500 nM 5-Aza-CdR) were exposed to human recombinant IFN-α2a (a gift from Roche) at 1 × 105 units/ml or human recombinant IFN-γ (GIBCO) at 5 × 102 units/ml. RNA was harvested for microarray expression analysis at 10, 24, and 96 hr. IFN concentrations were established by measuring growth inhibition in HT29 cells after treatment and approximating the IC50 for each IFN type (data not shown).
Construction of Microarrays.
The cDNA clones on the microarray were obtained from Research Genetics (Huntsville, AL) and Genome Systems (St. Louis). Transformants were grown overnight at 37°C in 96-well microtiter dishes containing 0.2 ml/well Terrific Broth supplemented with ampicillin. Cultures were transferred to a Millipore multiscreen, 96-well glass-fiber filtration plate (MAFB NOB), and growth medium was voided. Twenty-five microliters of 25 mM Tris⋅HCl, pH 8/10 mM EDTA/50 μl of 0.2 M NaOH/1% SDS/160 μl of 0.7 M potassium acetate, pH 4.8/5.3 M guanidine hydrochloride was added to each well of the glass filtration plate. Cell lysates were drawn through the glass filters under vacuum, and filter-bound DNA was washed four times with 200 μl of 80% ethanol. Plasmid DNAs were eluted by centrifugation after the addition of 65 μl of distilled H2O. Samples were collected in a 96-well microtiter dish during centrifugation.
PCR amplifications (30 cycles, 52°C annealing) were performed in 100-μl reaction volumes in a 96-well format by using 2 μl of purified plasmid as template and vector-specific primers (typically T7 and T3). PCR products were combined with 200 μl of binding solution (150 mM potassium acetate, pH 4.8/5.3 M guanidine hydrochloride) in a Millipore multiscreen glass-fiber filtration plate. Vacuum was applied to void the binding solution, and bound PCR product was washed four times with 200 μl of 80% ethanol. Products were eluted in 65 μl of distilled H2O. PCR product size ranged from 300 bp to 2.0 kb, with 1.0 kb as a typical length. DNA was prepared for spotting by diluting the purified PCR products in DMSO at a final concentration of 20–45 ng/μl.
Microarray slides were produced by using a Generation III Microarray Spotter (Molecular Dynamics). Each microarray contained 4,608 minimally redundant cDNAs spotted in duplicate on 3-aminoproply-trimethoxy silane-coated (Sigma) slides and UV crosslinked in a Stratalinker (Stratagene).
Generation of Microarray Probes, Microarray Hybridizations, and Scanning.
Total RNA was isolated by using Trizol reagent (GIBCO) and poly(A) RNA was selected by using an Oligotex Kit (Qiagen). First-strand cDNA probes were generated by incorporation of Cy3-dCTP or Cy5-dCTP (Amersham Pharmacia) during reverse transcription of purified mRNA (1 μg) with SuperScript II (GIBCO). After synthesis, RNA/cDNA hybrids were denatured and the mRNA was hydrolyzed with NaOH. Single-stranded cDNA probes were transferred to a Millipore glass-fiber filtration plate containing two volumes of 150 mM potassium acetate, pH 4.8, and 5.3 M guanidine hydrochloride. The mixture was voided by vacuum, and bound cDNA was washed four times with 80% ethanol. Probes were eluted by the addition of 50 μl of distilled H2O, recovered by vacuum concentration, and reconstituted in 30 μl of 5× SSC/0.1% SDS/0.1 μg/ml salmon sperm DNA/50% formamide. After denaturation at 94°C, the hybridization mixture was deposited onto an arrayed slide under a coverslip.
Hybridizations were performed overnight at 42°C in a humidified chamber. After hybridization, slides were washed for 10 min in 1× SSC/0.2% SDS and then for 20 min in 0.1× SSC/0.2% SDS. Slides were rinsed briefly in distilled water and dried with compressed air, and the fluorescent hybridization signatures were captured by using the “Avalanche” dual-laser confocal scanner (Molecular Dynamics). Fluorescent intensities were quantified by using arrayvision 4.0 (Imaging Research, St. Catherine’s, ON, Canada).
Northern Blotting and Reverse Transcription–PCR.
Five micrograms of total RNA was fractionated through formaldehyde-containing agarose gels and transferred onto nylon membranes (Amersham Pharmacia). Hybridizations with 32P-labeled probes were carried out by using Rapid-hyb buffer (Amersham Pharmacia). Reverse transcription–PCR of type I (α, β) and II (γ) IFN genes was carried out on cDNAs prepared from vehicle-treated and 500 nM 5-Aza-CdR-treated HT29 cells 9 days after treatment. The primers used for amplification of IFN-α are within the coding region and are capable of amplifying each member of the IFN-α gene cluster.
Cell Fractionations and Western Blotting.
Nuclear and cytoplasmic fractions were prepared as described previously (30). Protein extracts (50 μg) were fractionated through 10% SDS/PAGE gels (Novex) and blotted onto poly(vinylidene difluoride) membranes (Amersham Pharmacia). Antibody to DNA methyltransferase I was a kind gift from Moshe Szyf (McGill University, Montreal, Canada). STATs 1, 2, and 3 antibodies were purchased from Transduction Laboratories (Lexington, KY). Final protein detection employed a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (GIBCO) and chemiluminescence (NEN Renaissance).
Results
5-Aza-CdR Treatment Inhibits the Proliferation of HT29 Cells.
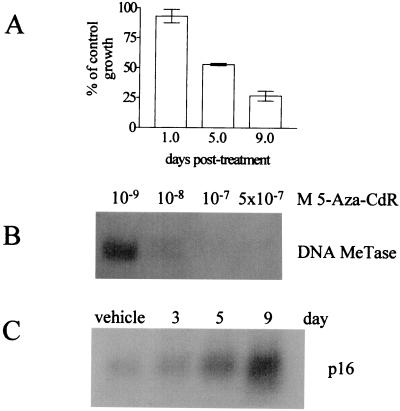
HT29 colon adenocarcinoma cells are p53- and APC-deficient and mismatch repair-proficient. Treatment of these cells with 500 nM 5-Aza-CdR for 24 hr caused a time-dependent, 3-fold inhibition of proliferation (Fig. 1A). As determined by flow cytometric analysis of propidium iodide-stained cells, apoptosis failed to account for the reduced cell numbers in response to 5-Aza-CdR. Rather, growth inhibition was characterized by an increased proportion of cells in G1 (data not shown). Treatment with 5-Aza-CdR depleted HT29 cells of soluble, nuclear DNA MeTase I as determined by Western analyses of nuclear protein extracts (Fig. 1B). This depletion corresponded with the reexpression of a known methylation-silenced gene, p16 (8) (Fig. 1C). The kinetics of growth inhibition, depletion of DNA MeTase I, and the reactivation of p16 were consistent with the mechanistic properties of 5-Aza-CdR and verified our HT29 cell model system. Although the induction of p16 may contribute to the growth inhibition seen in response to 5-Aza-CdR (31), we hypothesized that the genomewide nature of 5-Aza-CdR-induced hypomethylation was likely to affect other growth inhibitory pathways.
Figure 1.
5-Aza-CdR inhibits HT29 cell proliferation and sequesters DNA MeTase I. (A) HT29 cells were treated with vehicle or 500 nM 5-Aza-CdR for 24 hr. After this treatment, the drug was removed and cell proliferation was measured by directly counting cells at the indicated time points (see Materials and Methods). Data are presented as mean count ± 1 SD, (n = 3). (B) HT29 cells were treated with the indicated concentrations of 5-Aza-CdR for 24 hr, and the presence of DNA MeTase I (200 kDa) was assessed in nuclear protein extracts by immunoblotting. Sequestration of DNA MeTase I by 500 nM 5-Aza-CdR continued for 4 days after treatment (data not shown) (C) The expression of p16 in HT29 cells at time points after treatment with 500 nM 5-Aza-CdR was measured by Northern blot analysis.
5-Aza-CdR-Treatment Induces the Expression of IFN-Responsive Genes in HT29 Cells.
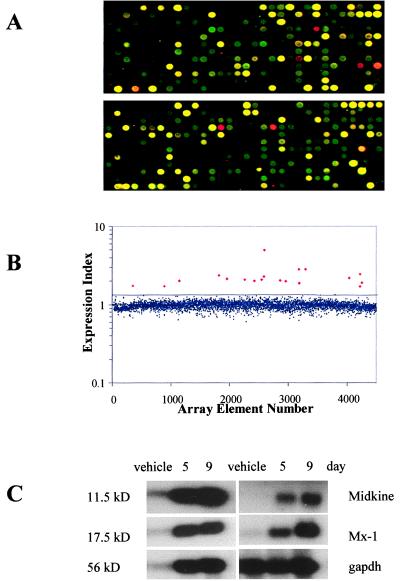
To investigate the molecular mechanisms involved in 5-Aza-CdR-induced growth inhibition in HT29 cells, we constructed and utilized high-density cDNA microarrays to analyze gene expression changes coincident with 5-Aza-CdR treatment. Our array was composed of 4,608 randomly selected, minimally redundant cDNAs from the Unigene set (32). Labeled cDNA probes were prepared from vehicle-treated and 500 nM 5-Aza-CdR-treated HT29 cells 9 days after the initial drug exposure, a time that coincided with maximal growth inhibition (Fig. 1A). First-strand cDNAs were reverse-transcribed from mRNA samples in the presence of Cy-3dCTP (vehicle-treated) or Cy-5dCTP (5-Aza-CdR-treated). After labeling, the two probes were hybridized simultaneously to the microarray slide (Fig. 2A). Subsequent analysis revealed up-regulation of 19 genes by greater than 2 SD above the mean expression ratio for the entire gene set (Fig. 2B). We confirmed the induction of these genes with Northern analyses (Fig. 2C) and their identity by DNA sequencing (Table 1). We noted that 10 of 19 genes induced by 5-Aza-CdR were established IFN-response genes (Table 1) (27, 34–36). Because IFNs are established cell growth inhibitors (37–39), the stimulation of IFN-responsive genes in 5-Aza-CdR-treated HT29 cells presented an attractive hypothesis to explain the coincident growth inhibition (Fig. 1A).
Figure 2.
Microarray analysis of gene expression changes in HT29 cells after 5-Aza-CdR treatment. (A) A cDNA microarray containing 4,608 target genes was constructed from a set of minimally redundant expressed sequence tags (ESTs). The microarray was hybridized with cDNAs prepared from vehicle-treated [Cy3-dCTP-labeled (green)] and 500 nM 5-Aza-CdR-treated HT29 cells [Cy5-dCTP (red)] 9 days after treatment. Two representative 12 × 32 gene grids (of 12) are displayed. (B) The fluorescent signal from the hybridized microarray slide was detected, quantified, and plotted as a ratio (Cy-5 signal/Cy-3 signal) for each array element. The average expression ratio for all genes on the array was normalized to 1.0 and had a SD of 0.177. The black line indicates a trend line 2 SD above the mean expression ratio for all genes on the microarray. The small, blue diamonds are genes below this cutoff; the large, red diamonds are genes above the cutoff. (C) Microarray expression data were confirmed by Northern blot analysis. Induction of five representative transcripts (see Table 1) 5 and 9 days after 5-Aza-CdR treatment is shown, along with glyceraldehyde-3-phosphate dehydrogenase (gapdh), an RNA-loading control. 11.5 kD, IFN-inducible protein 27; 17.5 kD, IFN-induced 17-kDa protein; 56 kD, IFN-induced protein 56.
Table 1.
Genes up-regulated by 5-Aza-CdR treatment of HT29 cells*
| 5-Aza-CdR-induced gene† | Unigene number or image ID | Regulation by IFN-α‡ | Regulation by IFN-γ‡ |
|---|---|---|---|
| Human mRNA for Stac | Hs. 56045 | + | + |
| IFN-induced protein 56 | Hs. 20315 | + | − |
| IFNα-inducible protein 27 | Hs. 2867 | + | + |
| IFN-induced 17-kDa protein | IID 149319 | + | + |
| EST | Hs. 6166 | + | − |
| EST | Hs. 165240 | + | + |
| Myxovirus resistance gene 2 | Hs. 926 | + | − |
| Purinergic receptor P2Y5 | Hs. 189999 | + | + |
| EST | Hs. 109309 | + | − |
| CpG island DNA fragment | No match§ | + | + |
| TGF-β superfamily member MIC-1 | Hs. 116577 | + | + |
| IFN-induced protein IFI-6-16 | Hs. 21205 | + | − |
| EST | Hs. 47783 | + | + |
| MHC class I | Hs. 77961 | + | + |
| Midkine | Hs. 82045 | − | + |
| Myxovirus resistance gene 1 | Hs. 76391 | + | − |
| 2′-5′-Oligoadenylate synthetase 3 | No match¶ | + | + |
| Nuclear antigen SP100 | Hs. 77617 | + | − |
| IFN-inducible protein 10 | Hs. 2248 | − | + |
Bold type indicates previously identified IFN-responsive genes.
Genes induced in HT29 cells by treatment with 500 nM 5-Aza-CdR at day 9 that were up-regulated by greater than 2 SD above the mean expression ratio for all genes on the microarray (see Fig. 2B), listed in descending order.
† The identity of each gene was verified by DNA sequencing and blast analysis (33).
‡ The expression of the genes induced by 5-Aza-CdR (column 1) was measured by microarray analysis after treatment of HT29 cells with 1 × 105 units/ml IFN-α2a or 5 × 102 units/ml IFN-γ for 10, 24, and 96 hr. Expression was scored as induced if the gene was up-regulated at any time point.
§ Identical to accession numbers Z61029 and Z61030 (100% identity over 115 bases) in the nonredundant Genbank database.
¶ Identical to accession number NM_006187 (100% identity over 366 bases) in the nonredundant Genbank database.
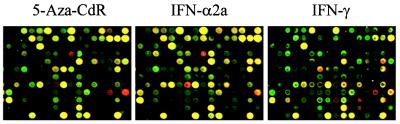
To determine whether these 10 genes are regulated by IFN in HT29 cells, and to assess whether the other 9 genes also are responsive to IFN, we conducted microarray experiments on HT29 cells treated for 10, 24, or 96 hr with either IFN-α or IFN-γ (Fig. 3). Interestingly, each of the 19 genes regulated by 5-Aza-CdR were also induced by either IFN-α or IFN-γ (Table 1). Comparison of the induced genes revealed a significantly greater overlap between 5-Aza-CdR- and IFN-α-induced genes than between 5-Aza-CdR- and IFN-γ-induced genes (17/19 vs. 12/19 genes, respectively).
Figure 3.
Microarray expression profiling of 5-Aza-CdR and IFN-treated HT29 cells. HT29 cells were treated with 500 nM 5-Aza-CdR, 1 × 105 units/ml IFN-α2a, or 5 × 102 units/ml IFN-γ. RNA was harvested 9 days (5-Aza-CdR) or 4 days (96 hr) (IFN-α or -γ) after treatment and used to generate probes for microarray analysis. Shown in the figure is a representative section of the microarray after hybridization with Cy-5-labeled cDNAs from 5-Aza-CdR-, IFN-α-, or IFN-γ-treated cells and Cy-3-labeled cDNAs from control cells. Four genes up-regulated by 5-Aza-CdR treatment are on the displayed grid. They are IFN-α-inducible protein 6–16 (row 4, column 9), expressed sequence tag (EST) Hs.109309 (8, 7), EST Hs.165240 (9, 14), and human mRNA for Stac (9, 16).
5-Aza-CdR Treatment Induces the Nuclear Accumulation and the Expression of STATs 1, 2, and 3.
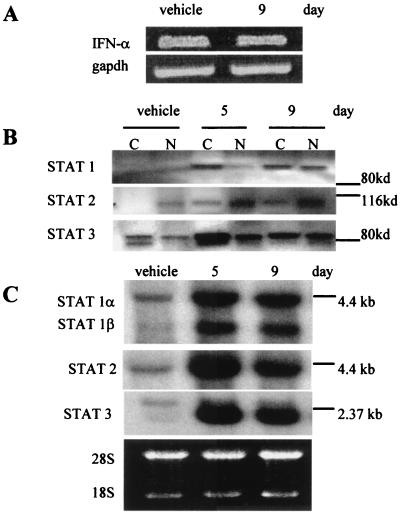
A simple explanation for the activation of IFN-responsive genes by 5-Aza-CdR is that the drug stimulated the synthesis and release of IFNs. Consistent with this possibility is the observation that the expression of IFN-γ can be regulated by DNA methylation (40–42). We, therefore, measured the mRNA levels for IFNs-α, -β, and -γ in HT29 after treatment with 500 nM 5-Aza-CdR. Reverse transcription–PCR analysis detected only IFN-α, and its mRNA level remained unchanged after 5-Aza-CdR treatment (Fig. 4A). We also were unable to detect any increase in IFN-α (or the presence of IFN-γ) by Western blot analysis of protein extracts from 5-Aza-CdR-treated HT29 cells at 2, 5, or 7 days after treatment (data not shown). These observations eliminated increased levels of IFNs as an explanation for the induction of IFN-responsive genes by 5-Aza-CdR. In addition, the transfer of medium harvested from 5-Aza-CdR-treated cells at 9 days after treatment onto control HT29 cell cultures did not inhibit cell growth. This indicates that the growth inhibition observed in 5-Aza-CdR-treated cells does not result from an increase in secreted IFN protein or of other growth inhibitory cytokines. These data caused us to investigate the IFN-signaling pathway to account for the induction of IFN-responsive genes by 5-Aza-CdR.
Figure 4.
5-Aza-CdR treatment activates STATs 1, 2, and 3 in HT29 cells. (A) The expression level of IFN-α in HT29 cells before and after 500 nM 5-Aza-CdR treatment was measured by reverse transcription–PCR along with gapdh to confirm equivalent cDNA input. (B) STAT transcription factor levels were measured by Western blotting. Cytoplasmic (C) and nuclear (N) cell extracts were prepared from HT29 cells after treatment with vehicle or 500 nM 5-Aza-CdR. A poly(vinylidene difluoride) membrane harboring the protein extracts was probed sequentially with mAbs specific to STATs 1, 2, and 3. In each case, the antibodies recognized proteins of the appropriate molecular weight for each STAT. Molecular mass markers are indicated. (C) The expression of STAT 1, 2, and 3 genes was measured by Northern blotting. RNA was isolated from HT29 cells after treatment with vehicle or 500 nM 5-Aza-CdR. The locations of molecular mass markers are indicated. Ethidium bromide staining confirmed equal RNA loading (28S, 18S rRNAs).
Because STAT transcription factors are effectors of IFN signaling (43), we next examined whether 5-Aza-CdR treatment caused them to accumulate in the nuclei of HT29 cells. To address this, we performed Western blot analyses on fractionated HT29 cells by using antibodies specific for STATs 1, 2, 3, 4, 5, and 6. We observed a time-dependent increase of STATs 1, 2, and 3 in the nuclei of HT29 cells after treatment with 500 nM 5-Aza-CdR (Fig. 4B). In contrast, STATs 4, 5, and 6 did not accumulate in the nuclei after 5-Aza-CdR treatment (data not shown). We also noted an increase in the total cellular levels of STATs 1, 2, and 3 after 5-Aza-CdR treatment (Fig. 4B). This novel observation raised the possibility that inhibition of DNA MeTase induced the expression of STATs 1, 2, and 3.
Because cDNAs corresponding to STATs 1, 2, and 3 were not on the microarray, we next performed Northern blot analyses on RNAs from 5-Aza-CdR-treated HT29 cells by using probes specific for STATs 1, 2, and 3. Fig. 4C illustrates the time-dependent up-regulation of STATs 1, 2, and 3 mRNA levels after 5-Aza-CdR treatment. This induction correlated temporally with growth inhibition in response to 5-Aza-CdR and implicates the transcriptional activation of STATs 1, 2, and 3 in the response of HT29 cells to 5-Aza-CdR. We also found that the STAT genes were expressed above control levels for at least 17 days (5 cell passages) after treatment with 5-Aza-CdR (data not shown).
5-Aza-CdR Treatment Sensitizes HT29 Cells to Exogenous IFN-α2a.
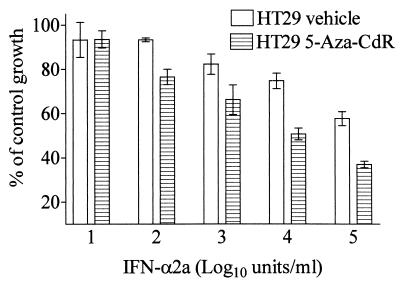
The above data suggest that STATs 1, 2, and 3 limit the response of HT29 cells to IFNs. With this in mind, we hypothesized that 5-Aza-CdR treatment could potentiate the response of HT29 cells to IFN-α. To test this hypothesis, we exposed control and 5-Aza-CdR-treated HT29 cells to various concentrations of IFN-α2a and measured growth rates. We found that 5-Aza-CdR increased the responsiveness of HT29 cells to growth inhibition mediated by IFN-α2a (Fig. 5). This effect corresponded to at least a 5-fold increase in the potency (IC50 of 2 × 105 units IFN/ml for control cells vs. IC50 of 4 × 104 units IFN/ml for 5-Aza-CdR-treated cells) of IFN-α for inhibiting HT29 cell growth. It is important to note that the increased responsiveness was observed despite the high level of growth inhibition elicited by 5-Aza-CdR treatment alone (Fig. 1A).
Figure 5.
5-Aza-CdR treatment increases the responsiveness of HT29 cells to growth inhibition mediated by exogenous IFN-α2A. HT29 cells were treated with 500 nM 5-Aza-CdR or vehicle (PBS). Ten days after removal of the drug, triplicate wells were treated with a concentration curve of IFN-α2a. Four days later, cell proliferation was measured by using a Coulter counter. Percentage of control growth was calculated by dividing the mean cell count at each IFN concentration by the mean cell count of untreated control cells (either HT29 or 5-Aza-CdR-treated HT29 cells, respectively). Data are presented as mean ± 1 SD, (n = 3). Similar results were obtained in four independent experiments.
Discussion
Transcriptional silencing of tumor-suppressor genes by CpG methylation may contribute to the development of human carcinomas. A model wherein methylation-induced gene silencing accompanies tumor development raises the potential for drug-induced reactivation of methylation-silenced tumor-suppressor genes as a therapeutic strategy. In this context, pharmacological inhibition of DNA MeTase by 5-Aza-CdR inhibits the growth of bladder, colon, and melanoma tumor cell lines, whereas control human fibroblasts are unaffected (31). Also, consistent with this model, a number of methylation-silenced tumor-suppressor genes have been identified by candidate gene approaches in tumor cells (3–5, 7, 11, 12, 44, 45). Among these, Bender et al. have demonstrated induction of p16 in a number of tumor cells that are growth-inhibited after 5-Aza-CdR treatment and that this induction correlates with the methylation status of the p16 promoter (31). However, it is reasonable to assume that the pharmacology of 5-Aza-CdR extends beyond p16-mediated growth arrest in that tumor cells in which p16 is not induced by 5-Aza-CdR are also growth-inhibited (31).
Our observation that 5-Aza-CdR inhibits HT29 cell growth parallels the results seen in other tumor cell lines (Fig. 1) (31) and validated them as a model system for microarray expression analysis. However, the results of our microarray analysis led to a new hypothesis for explaining the growth-inhibitory properties of 5-Aza-CdR in tumor cells in vitro and, perhaps, the efficacy of this compound in vivo. Our data indicate that STAT 1, 2, and 3 expression is induced by 5-Aza-CdR, that these proteins accumulate in the nucleus of 5-Aza-CdR-treated cells, and that these phenomenon parallel 5-Aza-CdR-induced growth inhibition. These data suggest that the presence of STAT proteins in tumor cells can dictate responsiveness to certain chemotherapeutics and raise the possibility that STATs 1, 2, and 3 are methylation-silenced tumor suppressors.
Our microarray approach started with an unbiased look at HT29 cell responses to 5-Aza-CdR and led us, indirectly, to the IFN-signaling pathway as a potential tumor-suppressive pathway. e saw that the genes responding most robustly to 5-Aza-CdR treatment in HT29 cells were also responsive to IFN treatment. This suggested the activation of the IFN-signaling pathway as a major cellular response to 5-Aza-CdR. The induction of IFN-responsive genes presents an attractive hypothesis for explaining 5-Aza-CdR-mediated growth inhibition in that IFNs are established growth-inhibitory cytokines (37, 39). However, it was unlikely that each of these IFN-responsive genes were regulated directly by promoter methylation. As an alternative, the microarray data pointed us to the up-regulation of STATs 1, 2, and 3, which are required to mediate the growth-inhibitory effects of IFN-γ (STAT 1) and IFN-α (STATs 1, 2, and 3) (43, 46, 47).
The up-regulation of STATs in response to 5-Aza-CdR may be explained in at least two ways. One explanation is that STAT genes are directly silenced by de novo methylation in tumor cells. In support of this model, the 5′ regions of STAT 1, 2, and 3 cDNAs contain likely CpG island targets for methylation (48) (see also GenBank accession no. L29277 for STAT 3). A second explanation is that induction of STATs 1, 2, and 3 by 5-Aza-CdR is the result of the epigenetic activation of another, upstream regulator of STAT expression. We do not believe it is likely that the stimulation of STATs and the IFN-induced gene set is due to nonspecific cellular toxicity or growth arrest because microarray experiments performed in our laboratory with agents such as TNF, TRAIL, FasL, and TGF-β have not revealed the induction of a similar gene set as that seen with 5-Aza-CdR and IFN-α and, to a lesser extent, with IFN-γ (data not shown). Whatever model accounts for the increased expression of STATs, it is unlikely that the simple up-regulation of these genes also results in their activation and nuclear accumulation. Rather, our data support a scenario in which endogenous IFN-α is responsible for activating STATs 1, 2, and 3. Several lines of evidence support this explanation. First, our analysis indicates the presence of IFN-α in control and 5-Aza-CdR-treated HT29 cells whereas IFN-β and -γ were undetectable under either condition. Second, our microarray analysis showed substantial overlap in genes induced by 5-Aza-CdR and those induced by the direct addition of IFN-α. Finally, our observation that STATs 1, 2, and 3 each accumulated in HT29 cell nuclei follows a number of studies demonstrating that IFN-α stimulation leads to activation of STAT1/2 or STAT1/3 heterodimers (43, 49, 50).
Our observation that 5-Aza-CdR stimulates expression of STATs 1, 2, and 3 holds important clinical implications. First, the expression of STAT 1 in certain metastatic melanoma and gastric adenocarcinoma cell lines is greatly depressed and correlates with a reduced level of responsiveness of these tumors to IFN-α (51–53). In the clinic, metastatic melanomas often fail to respond to IFN-α (54). Dampening of the IFN-response pathway by methylation silencing of STATs or other signaling components could account for lack of responsiveness of certain melanomas to IFN-α. Further, the activation of STATs by 5-Aza-CdR treatment raises the possibility that this drug could sensitize resistant tumor cells to IFN. As an initial test of this hypothesis, we examined the sensitivity of HT29 cells to the growth-inhibitory effects of IFN-α before and after treatment with 5-Aza-CdR. We saw that 5-Aza-CdR treatment increased the responsiveness of HT29 cells to IFN-α-mediated growth inhibition This result offers a plausible new line of investigation on the combination of 5-Aza-CdR and IFNs for the treatment of certain IFN-resistant tumors.
In conclusion, our work shows the value of microarray expression analyses in analyzing the mechanistic actions of pharmaceutical agents. Two previous studies have utilized microarrays to examine the specificity of drug actions in yeast. Gray et al. examined the transcriptional perturbations elicited by structural analogs of cyclin-dependent kinases inhibitors (28). In another study, Marton et al. compared the transcriptional profiles resulting from cyclosporin A and FK506 treatment of yeast mutant strains defective in calcineurin and immunophilin genes (29). These studies illustrate that microarrays can be used to examine drug-target specificity and potential secondary drug effects. We have extended these approaches by presenting a microarray-based evaluation of a clinically relevant compound in a human cell line. Although the explicit mechanistic basis for inhibition of DNA MeTase by 5-Aza-CdR is known, our study provides new, testable hypotheses that may explain the consequences of inhibiting DNA methylation in a clinical setting. Further pharmacological studies that utilize microarrays are likely to reveal new lines of investigation, both in vitro and in vivo, that more focused experimental approaches may overlook.
Acknowledgments
We thank Ann Jones for cell culture expertise. We thank the DNA Sequencing Facility, DNA/Peptide Synthesis Facility, and Flow Cytometry Facilities at the University of Utah. This work was supported by grants from the Huntsman Cancer Foundation.
Abbreviations
- 5-Aza-CdR
5-aza-2′-deoxycytidine
- DNA MeTase
DNA cytosine methyltransferase
- STAT
signal transducer and activator of transcription
- EST
expressed sequence tag
References
- 1.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 2.Holliday R. Br J Cancer. 1979;40:513–522. doi: 10.1038/bjc.1979.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Issa J P, Ottaviano Y L, Celano P, Hamilton S R, Davidson N E, Baylin S B. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 4.Ottaviano Y L, Issa J P, Parl F F, Smith H S, Baylin S B, Davidson N E. Cancer Res. 1994;54:2552–2555. [PubMed] [Google Scholar]
- 5.Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Proc Natl Acad Sci USA. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman J G, Umar A, Polyak K, Graff J R, Ahuja N, Issa J P, Markowitz S, Willson J K, Hamilton S R, Kinzler K W, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Zulueta M, Bender C M, Yang A S, Nguyen T, Beart R W, Van Tornout J M, Jones P A. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 8.Bender C M, Zingg J M, Jones P A. Pharm Res. 1998;15:175–187. doi: 10.1023/a:1011946030404. [DOI] [PubMed] [Google Scholar]
- 9.Bird A P. Cancer Surv. 1996;28:87–101. [PubMed] [Google Scholar]
- 10.Laird P W, Jackson-Grusby L, Fazeli A, Dickinson S L, Jung W E, Li E, Weinberg R A, Jaenisch R. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 11.Veigl M L, Kasturi L, Olechnowicz J, Ma A H, Lutterbaugh J D, Periyasamy S, Li G M, Drummond J, Modrich P L, Sedwick W D, et al. Proc Natl Acad Sci USA. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cote S, Sinnett D, Momparler R L. Anticancer Drugs. 1998;9:743–750. doi: 10.1097/00001813-199810000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Momparler R L. Pharmacol Ther. 1985;30:287–299. doi: 10.1016/0163-7258(85)90053-1. [DOI] [PubMed] [Google Scholar]
- 14.Juttermann R, Li E, Jaenisch R. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson A T, Vertino P M, Spitzner J R, Baylin S B, Muller M T, Davidson N E. J Biol Chem. 1997;272:32260–32266. doi: 10.1074/jbc.272.51.32260. [DOI] [PubMed] [Google Scholar]
- 16.Kantarjian H M, O’Brien S M, Estey E, Giralt S, Beran M, Rios M B, Keating M, de Vos D, Talpaz M. Leukemia. 1997;11, Suppl 1:S35–S36. [PubMed] [Google Scholar]
- 17.Momparler R L, Rivard G E, Gyger M. Pharmacol Ther. 1985;30:277–286. doi: 10.1016/0163-7258(85)90052-x. [DOI] [PubMed] [Google Scholar]
- 18.Momparler R L, Bouffard D Y, Momparler L F, Dionne J, Belanger K, Ayoub J. Anticancer Drugs. 1997;8:358–368. doi: 10.1097/00001813-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Thibault A, Figg W D, Bergan R C, Lush R M, Myers C E, Tompkins A, Reed E, Samid D. Tumori. 1998;84:87–89. doi: 10.1177/030089169808400120. [DOI] [PubMed] [Google Scholar]
- 20.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 21.Ushijima T, Morimura K, Hosoya Y, Okonogi H, Tatematsu M, Sugimura T, Nagao M. Proc Natl Acad Sci USA. 1997;94:2284–2289. doi: 10.1073/pnas.94.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatada I, Sugama T, Mukai T. Nucleic Acids Res. 1993;21:5577–5582. doi: 10.1093/nar/21.24.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalgo M L, Liang G, Spruck C H, III, Zingg J M, Rideout W M, III, Jones P A. Cancer Res. 1997;57:594–599. [PubMed] [Google Scholar]
- 24.Shiraishi M, Chuu Y H, Sekiya T. Proc Natl Acad Sci USA. 1999;96:2913–2918. doi: 10.1073/pnas.96.6.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 26.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 27.Der S D, Zhou A, Williams B R, Silverman R H. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray N S, Wodicka L, Thunnissen A M, Norman T C, Kwon S, Espinoza F H, Morgan D O, Barnes G, LeClerc S, Meijer L, et al. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 29.Marton M J, DeRisi J L, Bennett H A, Iyer V R, Meyer M R, Roberts C J, Stoughton R, Burchard J, Slade D, Dai H, et al. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Wu P, Siegel M I, Egan R W, Billah M M. J Biol Chem. 1995;270:9558–9563. doi: 10.1074/jbc.270.16.9558. [DOI] [PubMed] [Google Scholar]
- 31.Bender C M, Pao M M, Jones P A. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 32.Miller G S, Fuchs R. Comput Appl Biosci. 1997;13:81–87. doi: 10.1093/bioinformatics/13.1.81. [DOI] [PubMed] [Google Scholar]
- 33.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stadler M, Chelbi-Alix M K, Koken M H, Venturini L, Lee C, Saib A, Quignon F, Pelicano L, Guillemin M C, Schindler C, et al. Oncogene. 1995;11:2565–2573. [PubMed] [Google Scholar]
- 35.Rasmussen U B, Wolf C, Mattei M G, Chenard M P, Bellocq J P, Chambon P, Rio M C, Basset P. Cancer Res. 1993;53:4096–4101. [PubMed] [Google Scholar]
- 36.Luster A D, Jhanwar S C, Chaganti R S, Kersey J H, Ravetch J V. Proc Natl Acad Sci USA. 1987;84:2868–2871. doi: 10.1073/pnas.84.9.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeffer L M, Dinarello C A, Herberman R B, Williams B R, Borden E C, Bordens R, Walter M R, Nagabhushan T L, Trotta P P, Pestka S. Cancer Res. 1998;58:2489–2499. [PubMed] [Google Scholar]
- 38.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 39.Nathan C. In: Inflammation: Basic Principles and Clinical Correlates. 2nd Ed. Gallin J I, Goldstein I M, editors. New York: Raven; 1992. pp. 265–289. [Google Scholar]
- 40.Fukunaga R, Matsuyama M, Okamura H, Nagata K, Nagata S, Sokawa Y. Nucleic Acids Res. 1986;14:4421–4436. doi: 10.1093/nar/14.11.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young H A, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard J R, Penix L, Wilson C B, Melvin A J, McGurn M E, et al. J Immunol. 1994;153:3603–3610. [PubMed] [Google Scholar]
- 42.Mikovits J A, Young H A, Vertino P, Issa J P, Pitha P M, Turcoski-Corrales S, Taub D D, Petrow C L, Baylin S B, Ruscetti F W. Mol Cell Biol. 1998;18:5166–5177. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 44.Belinsky S A, Nikula K J, Palmisano W A, Michels R, Saccomanno G, Gabrielson E, Baylin S B, Herman J G. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lapidus R G, Ferguson A T, Ottaviano Y L, Parl F F, Smith H S, Weitzman S A, Baylin S B, Issa J P J, Davidson N E. Clin Cancer Res. 1996;2:805–810. [PubMed] [Google Scholar]
- 46.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ihle J N, Thierfelder W, Teglund S, Stravapodis D, Wang D, Feng J, Parganas E. Ann N Y Acad Sci. 1998;865:1–9. doi: 10.1111/j.1749-6632.1998.tb11157.x. [DOI] [PubMed] [Google Scholar]
- 48.Yan R, Qureshi S, Zhong Z, Wen Z, Darnell J E., Jr Nucleic Acids Res. 1995;23:459–463. doi: 10.1093/nar/23.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C H, Murti A, Pfeffer L M. Proc Natl Acad Sci USA. 1998;95:5568–5572. doi: 10.1073/pnas.95.10.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stancato L F, David M, Carter-Su C, Larner A C, Pratt W B. J Biol Chem. 1996;271:4134–4137. doi: 10.1074/jbc.271.8.4134. [DOI] [PubMed] [Google Scholar]
- 51.Abril E, Real L M, Serrano A, Jimenez P, Garcia A, Canton J, Trigo I, Garrido F, Ruiz-Cabello F. Cancer Immunol Immunother. 1998;47:113–120. doi: 10.1007/s002620050511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun W H, Pabon C, Alsayed Y, Huang P P, Jandeska S, Uddin S, Platanias L C, Rosen S T. Blood. 1998;91:570–576. [PubMed] [Google Scholar]
- 53.Wong L H, Krauer K G, Hatzinisiriou I, Estcourt M J, Hersey P, Tam N D, Edmondson S, Devenish R J, Ralph S J. J Biol Chem. 1997;272:28779–28785. doi: 10.1074/jbc.272.45.28779. [DOI] [PubMed] [Google Scholar]
- 54.Punt C J. Melanoma Res. 1998;8:95–104. doi: 10.1097/00008390-199804000-00001. [DOI] [PubMed] [Google Scholar]