Abstract
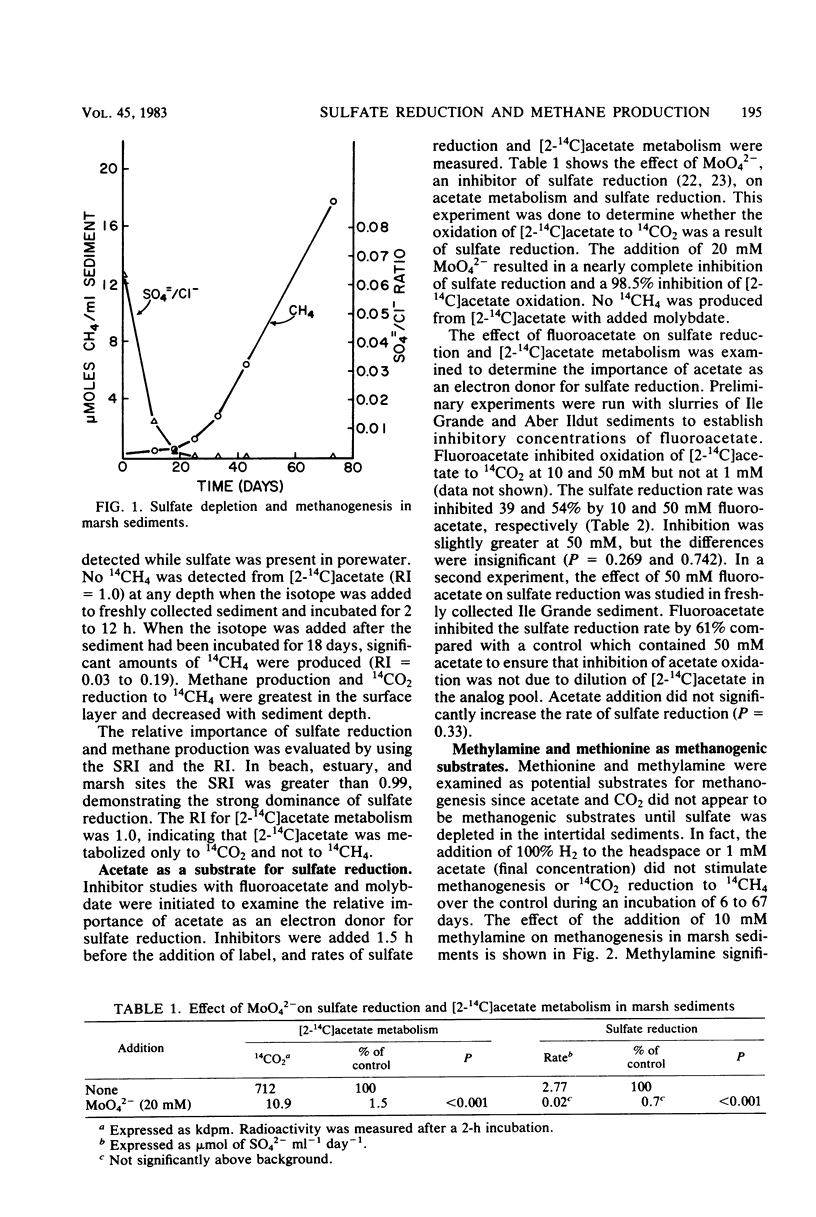
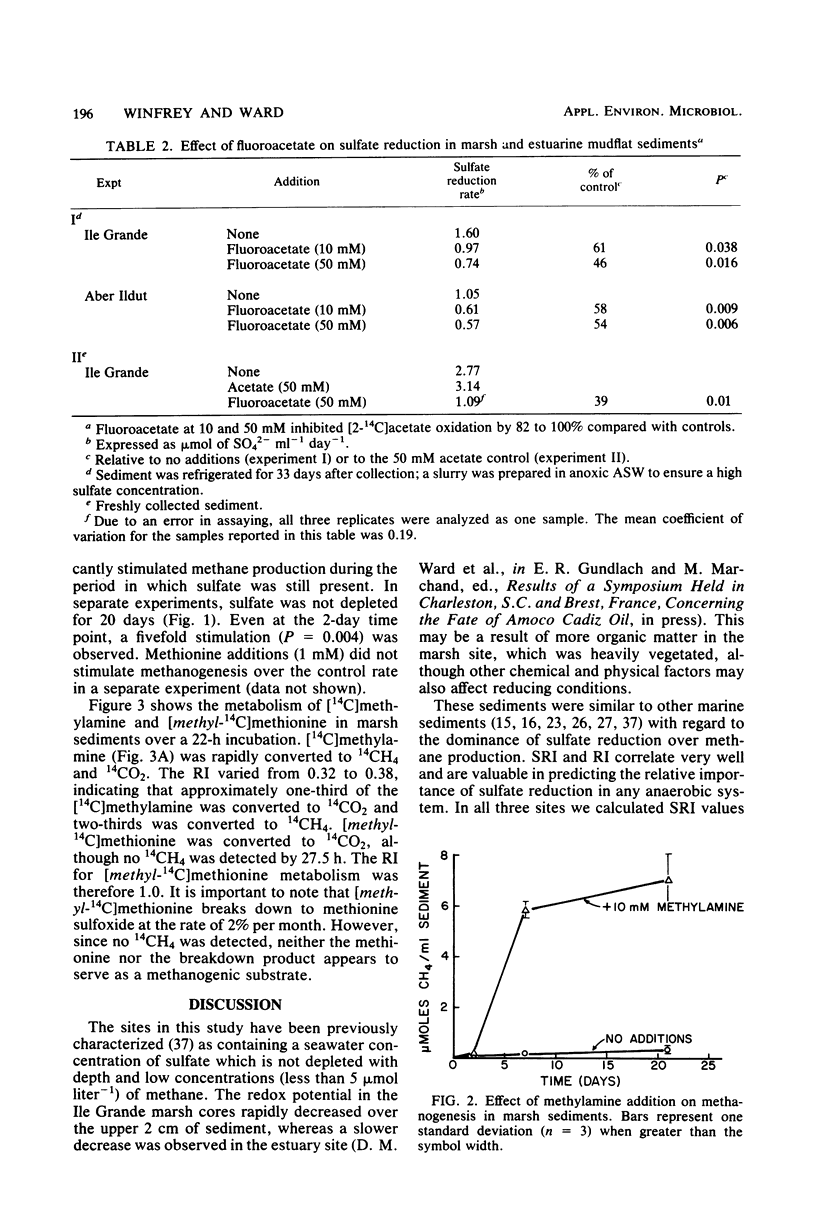
The activity of and potential substrates for methane-producing bacteria and sulfate-reducing bacteria were examined in marsh, estuary, and beach intertidal sediments. Slow rates of methane production were detected in all sediments, although rates of sulfate reduction were 100- to 1,000-fold higher. After sulfate was depleted in sediments, the rates of methane production sharply increased. The addition of methylamine stimulated methanogenesis in the presence of sulfate, and [14C]methylamine was rapidly converted to 14CH4 and 14CO2 in freshly collected marsh sediment. Acetate, hydrogen, or methionine additions did not stimulate methanogenesis. [methyl-14C]methionine and [2-14C]acetate were converted to 14CO2 and not to 14CH4 in fresh sediment. No reduction of 14CO2 to 14CH4 occurred in fresh sediment. Molybdate, an inhibitor of sulfate reduction, inhibited [2-14C]acetate metabolism by 98.5%. Fluoracetate, an inhibitor of acetate metabolism, inhibited sulfate reduction by 61%. These results suggest that acetate is a major electron donor for sulfate reduction in marine sediments. In the presence of high concentrations of sulfate, methane may be derived from novel substrates such as methylamine.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abram J. W., Nedwell D. B. Hydrogen as a substrate for methanogenesis and sulphate reduction in anaerobic saltmarsh sediment. Arch Microbiol. 1978 Apr 27;117(1):93–97. doi: 10.1007/BF00689357. [DOI] [PubMed] [Google Scholar]
- Abram J. W., Nedwell D. B. Inhibition of methanogenesis by sulphate reducing bacteria competing for transferred hydrogen. Arch Microbiol. 1978 Apr 27;117(1):89–92. doi: 10.1007/BF00689356. [DOI] [PubMed] [Google Scholar]
- Banat I. M., Lindström E. B., Nedwell D. B., Balba M. T. Evidence for coexistence of two distinct functional groups of sulfate-reducing bacteria in salt marsh sediment. Appl Environ Microbiol. 1981 Dec;42(6):985–992. doi: 10.1128/aem.42.6.985-992.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappenberg T. E. Interrelations between sulfate-reducing and methane-producing bacteria in bottom deposits of a fresh-water lake. II. Inhibition experiments. Antonie Van Leeuwenhoek. 1974;40(2):297–306. doi: 10.1007/BF00394388. [DOI] [PubMed] [Google Scholar]
- Cappenberg T. E., Prins R. A. Interrelations between sulfate-reducing and methane-producing bacteria in bottom deposits of a fresh-water lake. 3. Experiments with 14C-labeled substrates. Antonie Van Leeuwenhoek. 1974;40(3):457–469. doi: 10.1007/BF00399358. [DOI] [PubMed] [Google Scholar]
- Laanbroek H. J., Pfennig N. Oxidation of short-chain fatty acids by sulfate-reducing bacteria in freshwater and in marine sediments. Arch Microbiol. 1981 Jan;128(3):330–335. doi: 10.1007/BF00422540. [DOI] [PubMed] [Google Scholar]
- Mah R. A., Ward D. M., Baresi L., Glass T. L. Biogenesis of methane. Annu Rev Microbiol. 1977;31:309–341. doi: 10.1146/annurev.mi.31.100177.001521. [DOI] [PubMed] [Google Scholar]
- Martens C. S., Berner R. A. Methane production in the interstitial waters of sulfate-depleted marine sediments. Science. 1974 Sep 27;185(4157):1167–1169. doi: 10.1126/science.185.4157.1167. [DOI] [PubMed] [Google Scholar]
- Mountfort D. O., Asher R. A., Mays E. L., Tiedje J. M. Carbon and electron flow in mud and sandflat intertidal sediments at delaware inlet, nelson, new zealand. Appl Environ Microbiol. 1980 Apr;39(4):686–694. doi: 10.1128/aem.39.4.686-694.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R., Zeikus J. G. Rapid method for the radioisotopic analysis of gaseous end products of anaerobic metabolism. Appl Microbiol. 1974 Aug;28(2):258–261. doi: 10.1128/am.28.2.258-261.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland R. S. Methane production in shallow-water, tropical marine sediments. Appl Microbiol. 1975 Oct;30(4):602–608. doi: 10.1128/am.30.4.602-608.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig N., Biebl H. Desulfuromonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. Arch Microbiol. 1976 Oct 11;110(1):3–12. doi: 10.1007/BF00416962. [DOI] [PubMed] [Google Scholar]
- Sansone F. J., Martens C. S. Methane production from acetate and associated methane fluxes from anoxic coastal sediments. Science. 1981 Feb 13;211(4483):707–709. doi: 10.1126/science.211.4483.707. [DOI] [PubMed] [Google Scholar]
- Senior E., Lindström E. B., Banat I. M., Nedwell D. B. Sulfate reduction and methanogenesis in the sediment of a saltmarsh on the East coast of the United kingdom. Appl Environ Microbiol. 1982 May;43(5):987–996. doi: 10.1128/aem.43.5.987-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. L., Klug M. J. Electron donors utilized by sulfate-reducing bacteria in eutrophic lake sediments. Appl Environ Microbiol. 1981 Jul;42(1):116–121. doi: 10.1128/aem.42.1.116-121.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen J., Christensen D., Jørgensen B. B. Volatile Fatty acids and hydrogen as substrates for sulfate-reducing bacteria in anaerobic marine sediment. Appl Environ Microbiol. 1981 Jul;42(1):5–11. doi: 10.1128/aem.42.1.5-11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. M., Olson G. J. Terminal processes in the anaerobic degradation of an algal-bacterial mat in a high-sulfate hot spring. Appl Environ Microbiol. 1980 Jul;40(1):67–74. doi: 10.1128/aem.40.1.67-74.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. M. Thermophilic methanogenesis in a hot-spring algal-bacterial mat (71 to 30 degrees C). Appl Environ Microbiol. 1978 Jun;35(6):1019–1026. doi: 10.1128/aem.35.6.1019-1026.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer P. J., Zeikus J. G. One carbon metabolism in methanogenic bacteria. Cellular characterization and growth of Methanosarcina barkeri. Arch Microbiol. 1978 Oct 4;119(1):49–57. doi: 10.1007/BF00407927. [DOI] [PubMed] [Google Scholar]
- Widdel F., Pfennig N. A new anaerobic, sporing, acetate-oxidizing, sulfate-reducing bacterium, Desulfotomaculum (emend.) acetoxidans. Arch Microbiol. 1977 Feb 4;112(1):119–122. doi: 10.1007/BF00446665. [DOI] [PubMed] [Google Scholar]
- Widdel F., Pfennig N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov., sp. nov. Arch Microbiol. 1981 Jul;129(5):395–400. doi: 10.1007/BF00406470. [DOI] [PubMed] [Google Scholar]
- Winfrey M. R., Zeikus J. G. Anaerobic metabolism of immediate methane precursors in Lake Mendota. Appl Environ Microbiol. 1979 Feb;37(2):244–253. doi: 10.1128/aem.37.2.244-253.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfrey M. R., Zeikus J. G. Effect of sulfate on carbon and electron flow during microbial methanogenesis in freshwater sediments. Appl Environ Microbiol. 1977 Feb;33(2):275–281. doi: 10.1128/aem.33.2.275-281.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfrey M. R., Zeikus J. G. Microbial methanogenesis and acetate metabolism in a meromictic lake. Appl Environ Microbiol. 1979 Feb;37(2):213–221. doi: 10.1128/aem.37.2.213-221.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder S. H., Brock T. D. Methane, carbon dioxide, and hydrogen sulfide production from the terminal methiol group of methionine by anaerobic lake sediments. Appl Environ Microbiol. 1978 Feb;35(2):344–352. doi: 10.1128/aem.35.2.344-352.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]


