Abstract
The Hfq protein is recognized as a global regulatory molecule that facilitates certain RNA-RNA interactions in bacteria. BLAST analysis identified a 630-nucleotide open reading frame in the genome of Moraxella catarrhalis ATCC 43617 that was highly conserved among M. catarrhalis strains and which encoded a predicted protein with significant homology to the Hfq protein of Escherichia coli. This protein, containing 210 amino acids, was more than twice as large as the Hfq proteins previously described for other bacteria. The C-terminal half of the M. catarrhalis Hfq protein was very hydrophilic and contained two different types of amino acid repeats. A mutation in the M. catarrhalis hfq gene affected both the growth rate of this organism and its sensitivity to at least two different types of stress in vitro. Provision of the wild-type M. catarrhalis hfq gene in trans eliminated these phenotypic differences in the hfq mutant. This M. catarrhalis hfq mutant exhibited altered expression of some cell envelope proteins relative to the wild-type parent strain and also had a growth advantage in a continuous flow biofilm system. The presence of the wild-type M. catarrhalis hfq gene in trans in an E. coli hfq mutant fully reversed the modest growth deficiency of this E. coli mutant and partially reversed the stress sensitivity of this E. coli mutant to methyl viologen. The use of an electrophoretic mobility shift assay showed that this M. catarrhalis Hfq protein could bind RNA derived from a gene whose expression was altered in the M. catarrhalis hfq mutant.
Moraxella catarrhalis is an unencapsulated gram-negative coccobacillus that was regarded for many years as a harmless commensal microorganism that could be isolated from the human nasopharynx. However, within the last few decades, M. catarrhalis has been shown to be a pathogen that is capable of producing disease in both adults and children (27, 62). In infants and very young children, this bacterium is an important cause of otitis media (27, 40, 62). This organism can also cause infectious exacerbations of chronic obstructive pulmonary disease (39, 43, 52) and may be responsible for 2 to 4 million occurrences of this type each year in the United States (43).
Although there are some data, derived primarily from immune response studies, about which surface antigens of this organism are expressed during growth in vivo (2, 34, 41, 42), the molecular bases for expression of most if not all of these gene products remain to be determined. To date, studies on gene expression by M. catarrhalis have involved mainly investigations of phase-variable genes, including uspA1 (28), uspA2H (65), and hag (37, 45), and genes encoding proteins involved in restriction and modification (51), and there has been one recent study which showed that temperature can affect expression of the UspA1 protein (25). However, with the exception of the mutant analysis done by Furano and Campagnari (18) working with the M. catarrhalis fur gene, no global regulators of M. catarrhalis have been identified and studied in detail in this microorganism.
Hfq, or host factor 1, is a global regulatory protein that has been widely characterized in many bacterial species in recent years (for reviews, see references 11, 19, and 61). The Hfq protein forms a hexameric ring-shaped structure similar to that of the eukaryotic splicing proteins of the Sm family that have RNA-binding activities (50). Several functions have been ascribed to the Hfq protein, starting with its role in the replication of the RNA phage Qβ (17), to facilitating bacterial cell responses to environmental stresses (for reviews, see references 21, 22, 30, and 57). Most of the phenotypes linked to the Hfq protein were shown to be the result of its ability to function as an RNA chaperone that allows RNA-RNA interactions involving mRNA and small RNA molecules (sRNA) (20, 30, 38, 56). These sRNAs typically affect the expression of their cognate mRNAs by altering either their stability or translation (58). This fact has allowed identification of additional bacterial sRNA molecules by their binding to Hfq (68).
We describe here the M. catarrhalis hfq gene and its encoded protein product, which is more than twice the size of the Hfq proteins of other bacterial species studied to date. In addition, an isogenic M. catarrhalis hfq deletion mutant was constructed and characterized in several in vitro systems to determine whether the M. catarrhalis Hfq protein was involved in regulating the stress response of this bacterium. Finally, purified recombinant M. catarrhalis Hfq protein was shown to bind in vitro to the RNA transcribed from one particular M. catarrhalis gene. The abundance of transcript from this latter gene was significantly increased in the M. catarrhalis hfq mutant, making this gene a possible target for control by the Hfq protein and an sRNA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
M. catarrhalis O35E (3) was used as the wild-type M. catarrhalis strain in the present study. M. catarrhalis strains were grown in brain heart infusion (BHI) broth (Becton Dickinson, Sparks, MD) with aeration or on BHI solidified with 1.5% agar at 37°C in an atmosphere of 95% air and 5% CO2. Autoagglutination of M. catarrhalis strains was measured as described previously (65). When appropriate, BHI medium was supplemented with kanamycin (15 μg/ml) or spectinomycin (15 μg/ml). Mueller-Hinton (MH) broth (Becton Dickinson) was used for some experiments as noted. The Escherichia coli strain MC4100 and its hfq mutant GS081 have been described (67). E. coli strains DH5α (48) and TOP10 (Invitrogen, Carlsbad, CA) were used for cloning purposes. All E. coli strains were grown by using Luria-Bertani (LB) medium as described previously (48). When necessary, LB medium was supplemented with ampicillin (100 μg/ml), kanamycin (30 μg/ml), or spectinomycin (100 μg/ml). The M. catarrhalis plasmid cloning vector pWW115 (64) was used for complementation analysis with M. catarrhalis mutants. Plasmid pACYC177 was obtained from New England Biolabs (Beverly, MA).
Western blot analysis and protein identification.
Whole-cell lysates (WCL) were prepared as described previously (7). Outer membrane vesicles were prepared from M. catarrhalis by using the method described by Murphy and Loeb (44). For Western blot analysis, proteins in samples were resolved by using 15% (wt/vol) polyacrylamide separating gels for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes. The Hfq antiserum (described below) was used at a dilution of 1:1,000 in phosphate-buffered saline-Tween containing 10% (wt/vol) dried milk and incubated with the membrane either for 2 h at room temperature or overnight at 4°C. Horseradish peroxidase-conjugated goat anti-mouse antibody (Jackson Immunoresearch, West Grove, PA) was used as the secondary antibody. The antigen-antibody complexes were detected by using Western Lightning Chemiluminescence Reagent Plus (New England Nuclear, Boston, MA). For the identification of proteins that were differentially expressed by the hfq mutant, proteins in either WCL or outer membrane vesicles were resolved by SDS-PAGE, transferred to polyvinylidene difluoride, and then stained with Coomassie blue. The selected protein bands were excised from the membranes and subjected to N-terminal amino acid sequence analysis by means of Edman degradation in the Protein Chemistry Technology Center at the University of Texas Southwestern Medical Center at Dallas.
DNA and RNA isolation.
Chromosomal DNA from agar plate-grown M. catarrhalis cells was isolated by using a DNA Easy kit (Invitrogen). Plasmid DNA from either M. catarrhalis or E. coli was isolated by using a miniprep spin kit (Qiagen). RNA was isolated from M. catarrhalis strains grown to early stationary phase (optical density at 600 nm [OD600] = 1.5) by using an RNeasy Midi kit (Qiagen).
Construction of M. catarrhalis hfq mutants.
Overlapping extension PCR (26) was used to replace most of the hfq-like region of the M. catarrhalis O35E hfq open reading frame (ORF) with a promoterless kanamycin resistance cartridge (35). Briefly, with M. catarrhalis O35E chromosomal DNA as the template, the oligonucleotide primers AA133 (5′-GTAAGCCGTTAAGCCATTGGCAAT-3′) and AA135 (5′-CCTAGTTAGTCACCCGGTTTGTCCTTTTGACATAAGTTTCTCCAA-3′) were used to amplify the region (designated amplicon 133-135) immediately upstream of codon 7 in the hfq ORF, and the primers AA136 (5′-GGAATAATGCGACCCGTGTATAAGCATG CCATTTCAACGGTTGTA-3′) and AA137 (5′-CGCCCGAATTTGAGATGGCAA-3′) were used to amplify the region (designated amplicon 136-137) immediately downstream of codon 53 in the hfq ORF. The primers AA111 (5′-GGGTGACTAACTAGGAGGAATAAAT-3′) and AA116 (5′-GGGTCGCATTATTCCCTCCAGGTA-3′) were used to amplify the promoterless kanamycin resistance (kan) cartridge from pUC18K3 (35). The primers AA133 and AA116 were used together with amplicon 133-135 and the kan cartridge as templates to obtain a PCR product that contained both the upstream region and the kan cartridge, whereas the primers AA111 and AA137 were used with amplicon 136-137 and the kan cartridge to produce a different PCR product that contained the kan cartridge and the downstream region. These two PCR products were then used together as templates for another round of PCR using the primers AA133 and AA137. After verification of the nucleotide sequence of the final PCR product, it was used to transform wild-type M. catarrhalis O35E. One kanamycin-resistant transformant was selected for further characterization and was designated O35E.hfq::kan. A similar approach was used to obtain a PCR product that lacked almost all of the M. catarrhalis hfq ORF but contained the relevant flanking DNA. The primers used for this overlapping extension PCR series were the primers AA133 and AA172 (5′-GGTTTGTCCTTTTGACATAAGTTTCTC-3′) and the primers AA171 (5′-ATGTCAAAAGG ACAAACCGATTTTGAAAATCAATCCTGATTTGGT-3′) and AA137. The final PCR product was used to transform O35E.hfq::kan and the transformant colonies were screened for the loss of kanamycin resistance. One kanamycin-sensitive transformant was chosen for further characterization and was designated O35EΔhfq; this mutant contained an in-frame deletion of almost the entire hfq ORF. Nucleotide sequence analysis showed two other nucleotide changes near this deletion in this mutant: one in the intergenic region between the mia and hfq ORFs and the other within the remainder of the hfq ORF. There were no nucleotide changes in the kpsF gene located immediately downstream from the hfq gene.
Complementation of the M. catarrhalis hfq mutant.
The wild-type M. catarrhalis O35E hfq gene was PCR amplified by using the primers AA160 (5′-ATGGATCCGGCGTCTTCTCATTTACATTGCTT-3′; the BamHI site is underlined) and AA159 (5′-TTAGGAGCTCTGAGTTAG AAGGTATCACCC-3′; the SacI site is underlined) with O35E chromosomal DNA as a template. After digestion with both BamHI and SacI, the PCR product was ligated with BamHI- and SacI-digested pWW115 (64), and this ligation mixture was used to transform O35E. One spectinomycin-resistant transformant was selected for further characterization, and its plasmid was designated pAA200. This plasmid was used to transform the O35EΔhfq mutant to obtain the recombinant strain O35EΔhfq(pAA200). Plasmid pWW115 was used to transform O35EΔhfq to obtain a negative control strain for complementation analysis.
Complementation of an E. coli hfq mutant with the M. catarrhalis Hfq protein.
The primers AA160 and AA293 (5′-AGCTGCAGTGAGTTAGAAGGTATCACCC-3′; the PstI site is underlined) were used to amplify the M. catarrhalis hfq gene using O35E chromosomal DNA as a template. The PCR product was then digested using BamHI and PstI and ligated to pACYC177 that had been digested with the same restriction enzymes. The ligation mixture was used to transform the E. coli hfq mutant GS081 (67). Kanamycin-resistant transformants were screened for the expression of the M. catarrhalis Hfq protein by Western blot analysis, and one transformant was selected for further characterization; its plasmid was designated pAA105. As a negative control, an irrelevant piece of DNA from the 5′ region of the M. catarrhalis O35E uspA2 gene was amplified using the primers AA16 (5′-CGCGGATCCTGAAAACCATGAAACTT CTCCC-3′; the BamHI site is underlined) and AA294 (5′-ACCTGCAGTTGGGTAGCGATTTTGGT GGTGAC CGA-3′; the PstI site is underlined), ligated into the BamHI/PstI sites of pACYC177, and transformed into E. coli GS081. The resultant control plasmid was designated pAA105B.
Methyl viologen sensitivity assay.
Bacterial cells grown overnight on solidified media were suspended in 5 ml of BHI medium to a final OD600 of 0.2. A 500-μl portion of this suspension was mixed with an equal volume of BHI broth and then serially diluted. A 10-μl aliquot of each dilution was spotted onto BHI agar with or without 40 μM methyl viologen (Sigma, St. Louis, MO). The spots were allowed to dry, and then the plates were incubated overnight.
Salt sensitivity assay.
Bacterial cells were grown, diluted, and spotted as described immediately above except that the cells were spotted onto BHI agar plates that contained either the normal concentration of NaCl in BHI (i.e., 1× NaCl; 5 mg/ml) or 8× NaCl (i.e., 40 mg/ml). The spots were allowed to dry, and then the plates were incubated for 24 and 48 h, and the growth of the bacteria was examined at each time point.
Cloning and purification of the M. catarrhalis Hfq protein.
The primer pair AA138 (5′-AGGGATCCGCACAGACAGACAAACCAAC-3′; the BamHI site is underlined) and AA163 (5′-GT GGTGGTGGTGGTGGTGGGATTGATTTTCAAAATCGTCATCATA-3′) and the primer pair AA152 (5′-CACCACCACCACCACCACTGATTTGGTCATCATCTTGACTTATCC-3′) and AA156 (5′-CAAAACGCTTAATGTCAGTTT-3′) were used in overlapping extension PCR to introduce nucleotides encoding six histidine residues (His) at the C terminus of the M. catarrhalis O35E Hfq protein. The final PCR product was ligated to the pCR2.1 TOPO vector (Invitrogen) and transformed into E. coli TOP10 cells. Kanamycin-resistant transformants were screened for His tag expression, and one transformant was selected for further characterization; its plasmid was designated pAA201. To purify the His-tagged Hfq protein, recombinant E. coli cells grown overnight in broth were harvested, suspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]) containing lysozyme (1 μg/ml), and incubated on ice for 30 min. The cells were then subjected to sonication using a Branson Sonifier 450 sonicator (Proquip, Inc., Macedonia, OH). Unbroken cells and debris were removed by centrifugation at 26,000 × g for 20 min at 4°C. The supernatant was then mixed with NiNTA agarose beads (Qiagen, Valencia, CA) and incubated with agitation at 4°C for 1 h. The suspension was then loaded into a chromatography column and washed four times (5× bed volume) with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole [pH 8.0]). The His-tagged Hfq protein was eluted using elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole [pH 8.0]). Selected fractions were pooled, and dialyzed against storage buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 50 mM NH4Cl, and 20% glycerol (vol/vol). The concentration of purified, recombinant Hfq protein was determined by using the Bradford protein assay (Bio-Rad), and then portions of the protein were stored at −70°C for later use.
Production of polyclonal antibody against the recombinant M. catarrhalis Hfq protein.
The recombinant Hfq protein was used to immunize mice to obtain polyclonal antibody against this protein. Briefly, 100 μg of the protein was mixed with TiterMax (Sigma) and injected intraperitoneally into BALB/c mice. Four weeks later, the mice were boosted with 30 μg of the purified protein in TiterMax. After two more weeks, the mice were euthanized, and blood was collected for serum preparation.
RT-PCR analysis.
RNA that had been isolated from M. catarrhalis O35E grown into the early stationary phase of growth was digested with DNase I (MessageClean kit; GenHunter Corp, Nashville, TN) to remove any DNA contamination. Oligonucleotide primers were designed to amplify the regions between the hfq gene and the two flanking genes so that each primer would bind to a sequence within the ORF of each gene. The reverse transcription (RT) reaction was carried out by using MultiScribe reverse transcriptase (Applied Biosystems, Foster City, CA) followed by PCR amplification. As controls, the reaction was also done using either DNA alone as the template or with RNA template in the absence of reverse transcriptase.
Real-time RT-PCR.
Primers for real-time RT-PCR were designed by using Primer Express software (Applied Biosystems). The primer pair AA173 (5′-CAATGGGCGAAAGCCTGAT-3′) and AA174 (5′-GTGCTTTACAACCAAAAGGCCT-3′) was specific for 16S RNA, and the primer pair AA305 (5′-TGTTGAGACGGTGGTTGACAAT-3′) and AA306 (5′-GCCGTACATTTCCTTGGCAA-3′) was specific for M. catarrhalis ORF 1068 (hereafter referred to as ORF 1068). The reaction was performed as described previously (6). The data analysis was carried out by using the 7500 System SDS software v.13 (Applied Biosystems), applying the relative quantification ΔΔCT method. The level of the ORF 1068 message was normalized according to the level of the 16S RNA, and the normalized wild-type message level was used as the calibrator. The Student t test was used for statistical analysis of these data.
Competitive index-based biofilm experiments.
The streptomycin-resistant mutant of the wild-type strain O35E (O35E-Smr) (7) and the hfq mutant O35E.hfq::kan were grown separately in MH broth to a density equivalent to 108 CFU/ml, and then equal volumes of each culture were mixed in a 1:1 ratio. Serial dilutions of this inoculum mixture were plated on BHI agar plates containing the appropriate antibiotic to determine the relative percentages of each strain in the input mixture. A 3-ml portion of the mixture was used to inoculate 200 ml of MH broth that was then allowed to grow overnight at 37°C with aeration. A second 3-ml portion was used to inoculate a sterile Sorbarod cellulose filter that was inserted into a short piece of silicone tubing as described previously (45). After inoculation, sterile MH broth was allowed to drip along the length of the silicone tubing onto this Sorbarod filter as described previously (45). Cells present in the resultant biofilm were collected the next day by scraping the silicone tubing, starting at a point approximately 0.25 in. above the Sorbarod filter and continuing in the direction of the top of the tubing. The harvested cells from the biofilm, as well as the cells grown in liquid medium, were serially diluted and plated on agar media containing the appropriate antibiotics to determine the relative percentage of each strain in the output mixture. These data were subjected to repeated-measures analysis and pairwise comparisons were performed by using the Tukey HSD procedure at the 0.05 significance level (SAS 9.13; SAS, Inc., Cary, NC).
Preparation of a radiolabeled RNA probe.
The primers AA373 (5′-GTCAAATTTCACTATGCATTATCTT-3′) and AA281 (5′-TGACTGTGTCTGTATCGCC-3′) were used to amplify an ∼300-bp fragment that contained the putative transcriptional start point of ORF 1068 and part of the 5′ region of this ORF. This PCR fragment was used as a template for another PCR using the primers AA281 and AA374 (5′-CAGAGATGCATAATACGACTCACTATAGGGAGAGTCAAATTTCACTATGCATTATCTT-3′; the T7 core promoter sequence is underlined). The resultant PCR product AA373-AA281 was used in an vitro transcription reaction using a T7 MAXIscript kit (Ambion, Austin, TX) in the presence of [α-32P]UTP for 1 h at 37°C according to the manufacturer's protocol. The DNA template was then digested with Turbo DNase (Ambion), and the RNA product was partially purified by using an RNeasy MinElute cleanup kit (Qiagen).
EMSA.
The radiolabeled RNA probe described immediately above was used in an electrophoretic mobility shift assay (EMSA) with the purified recombinant M. catarrhalis Hfq protein. Briefly, equivalent amounts of RNA fragment (ca. 20,000 cpm) were incubated with increasing concentrations of purified Hfq protein in a 25-μl reaction volume in a binding buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 80 mM NaCl, 10% (vol/vol) glycerol, and 0.01% (vol/vol) dodecyl maltoside for 30 min at 37°C. Each reaction tube also contained E. coli tRNA (100 ng/μl; Roche) to minimize nonspecific binding. The reaction mixtures were then loaded onto a native 6% polyacrylamide gel and run in 0.5× Tris-borate-EDTA at room temperature at 200 V for 2 h. The gel was then exposed to a storage phosphor intensifying screen (GE Healthcare, Piscataway, NJ) and scanned by using a Storm 820 scanner (GE Healthcare). The image was analyzed by using ImageQuant v.5.2 software (Molecular Dynamics, Sunnyvale, CA). To determine whether binding was specific for the RNA fragment used in this experiment, the binding reaction was also carried out in the presence of 25 ng of the same RNA fragment that had not been radiolabeled.
RESULTS
Identification of an Hfq-like protein encoded by M. catarrhalis.
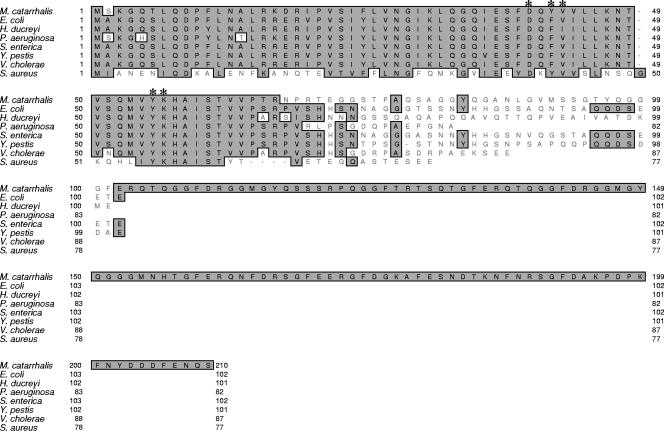
The use of the nucleotide sequence of the E. coli hfq gene in a BLAST search against the M. catarrhalis ATCC 43617 genome (GenBank accession numbers AX067426 to AX067466) identified a Hfq-like protein encoded by a 630-nucleotide (nt) ORF. The predicted encoded protein contained 210 amino acids (Fig. 1), and nucleotide sequence analysis of the hfq ORF from five additional M. catarrhalis strains (O35E, FIN2344, V1118, 7169, and O12E) revealed that this ORF was identical among all six strains (data not shown). Amino acid alignment of the predicted M. catarrhalis Hfq protein with Hfq proteins from other well-studied bacterial species (Fig. 1) showed high degrees of identity and similarity between these latter proteins and the N-terminal one-third of the M. catarrhalis Hfq protein. For example, all of the amino acids predicted to form the RNA-binding pocket in different Hfq proteins (5, 49, 50, 69) are conserved in the M. catarrhalis Hfq protein (Fig. 1).
FIG. 1.
Comparison of the deduced amino acid sequence of the M. catarrhalis Hfq protein with the sequences of Hfq proteins from other bacteria. The deduced amino acid sequence of the M. catarrhalis ATCC 43617 Hfq protein (top sequence) was aligned to the amino acid sequences of seven other bacterial Hfq proteins. The asterisks indicate the amino acids predicted to form the nucleotide-binding pocket in other Hfq proteins (5, 49, 50, 69). This figure was generated by using the CLUSTAL W alignment program in MacVector (version 6.5).
In contrast to these other Hfq proteins that typically contained between 77 and 99 amino acids, the M. catarrhalis Hfq protein contained more than 100 additional amino acids; these were present in the C-terminal half of this macromolecule (Fig. 1). Hydrophilicity analysis of the amino acid sequence of the E. coli Hfq protein indicated that the C-terminal domain containing approximately 30 amino acids was hydrophilic (data not shown). Analysis of the much larger number of additional amino acids in the C-terminal half of the M. catarrhalis Hfq protein indicated that this region is highly hydrophilic in nature but is not predicted to form a regular secondary structure (data not shown). The amino acid sequence GF appears nine times in these 142 amino acids in the M. catarrhalis Hfq protein, and the sequence GFERQTQGGFDRGGMGYQ appears twice. When the complete amino acid sequence of the M. catarrhalis Hfq protein was used in a BLAST search against the nonredundant protein databases, the predicted proteins with the highest level of identity (E values [31] of 2 × 10−30) were found in Psychrobacter species. These three different Psychrobacter proteins (accession numbers ZP_01272067.1, YP_580756.1, and YP_264208.1) each possessed predicted Hfq proteins that were very similar in size to the M. catarrhalis Hfq protein, containing 183 or 201 amino acids, but no description of these Psychrobacter proteins has been published to date.
Genetic organization of the M. catarrhalis hfq chromosomal locus.
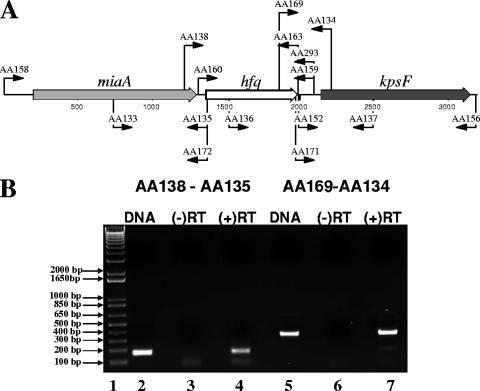
Upon examination of the hfq chromosomal locus in M. catarrhalis strain ATCC 43617, we found that the hfq gene was preceded by an miaA gene which encodes a predicted delta 2-isopentenylpyrophosphate transferase (12) (Fig. 2A). Immediately downstream of the M. catarrhalis hfq gene, we found an ORF with homology to kpsF, which encodes a predicted arabinose-5-phosphate isomerase (60).
FIG. 2.
Genetic organization of the M. catarrhalis hfq chromosomal locus. (A) Schematic representation of the M. catarrhalis hfq chromosomal locus in M. catarrhalis ATCC 43617, including the two genes flanking the hfq ORF. The relative positions of the different oligonucleotide primers used for PCR and RT-PCR are indicated by the arrows. (B) Photograph of an agarose gel showing the results of a RT-PCR experiment involving the hfq ORF and the two flanking genes. The primer pair used in each experiment is indicated on the top of the gel. Lane 1 contains DNA size markers. Lanes 2 and 5 contain samples from RT-PCRs where DNA was used as the template. Lanes 3 and 6 contain samples from RT-PCRs where RNA was used as the template but no reverse transcriptase was added [(−)RT]. Lanes 4 and 7 contain samples from RT-PCRs where RNA was used as the template and reverse transcriptase was added [(+)RT].
RT-PCR analysis was performed with RNA from M. catarrhalis O35E to determine whether the M. catarrhalis hfq gene was transcriptionally linked to either of the two flanking genes. Primer AA135, which binds to the 5′ end of the hfq gene on the negative strand (Fig. 2A), was used in a reverse transcriptase reaction, followed by a PCR using the primer AA138, which binds to the 3′ end of the miaA gene (Fig. 2A). This RT-PCR yielded a ∼200-bp PCR product (Fig. 2B, lane 4) similar to the one obtained using O35E chromosomal DNA as a template (Fig. 2B, lane 2), and no product was obtained in the PCR in which the reverse transcriptase enzyme was not added (Fig. 2B, lane 3). These results indicated that M. catarrhalis hfq and miaA genes are linked transcriptionally. Primer AA134, which binds to the 5′ end of the kpsF gene on the negative strand (Fig. 2A), was used in an RT-PCR with primer AA169, which binds to the 3′ end of the hfq gene (Fig. 2A). The results obtained with these two primers (Fig. 2B, lanes 5 to 7) indicated that the hfq and kpsF genes are transcriptionally linked in M. catarrhalis O35E.
Construction and complementation of M. catarrhalis hfq mutants.
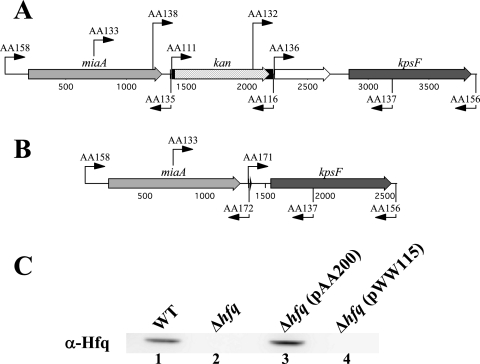
Overlapping extension PCR, followed by transformation and allelic exchange, was used to construct two different M. catarrhalis hfq mutants. The first mutant, O35E.hfq::kan, had a promoterless kan cartridge in place of most of the hfq-like region (i.e., nt 19 to 159) in the hfq ORF (Fig. 3A). This mutant was then used to construct the deletion mutant O35EΔhfq in which the region from nt 16 to 612, comprising almost the entire hfq ORF, was deleted in-frame (Fig. 3B) as described in Materials and Methods. The M. catarrhalis cloning vector pWW115 carrying the wild-type hfq gene from M. catarrhalis O35E was used to complement the O35EΔhfq mutant, yielding the recombinant M. catarrhalis strain O35EΔhfq(pAA200). The recombinant M. catarrhalis strain O35EΔhfq(pWW115) was used as a negative control. When subjected to Western blot analysis with mouse polyclonal antibody raised against the recombinant M. catarrhalis Hfq protein, both the wild-type O35E strain and the complemented O35EΔhfq mutant (Fig. 3C, lanes 1 and 3, respectively) expressed an antibody-reactive band of approximately 23 kDa, which is very similar to the predicted mass of the M. catarrhalis Hfq protein. This same antibody-reactive band was absent from both the hfq deletion mutant O35EΔhfq and the negative control strain O35EΔhfq(pWW115) (Fig. 3C, lanes 2 and 4, respectively).
FIG. 3.
Construction of M. catarrhalis hfq mutants. (A and B) Schematic representation of the M. catarrhalis chromosomal locus containing the hfq gene and flanking regions in the hfq mutant O35E.hfq::kan (A) and the hfq deletion mutant O35EΔhfq (B). The relative positions of the different primers used for PCR are indicated by the arrows. (C) Western blot analysis using mouse polyclonal antibody raised against the recombinant M. catarrhalis O35E Hfq protein to probe WCL derived from the wild-type O35E strain (WT, lane 1), the O35EΔhfq mutant (Δhfq, lane 2), the O35EΔhfq mutant containing the pAA200 plasmid with the wild-type O35E hfq gene [Δhfq(pAA200), lane 3], and this same mutant containing only the plasmid vector [Δhfq(pWW115), lane 4].
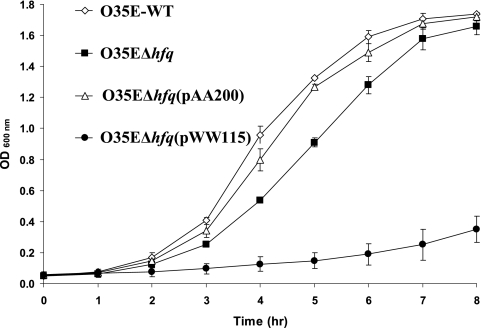
The M. catarrhalis hfq mutant is growth deficient.
A phenotypic feature common to the hfq mutants of some other bacterial species is a growth deficiency in vitro (15, 53, 59). Upon examination of the growth characteristics of the M. catarrhalis hfq deletion mutant, it was observed that this mutant (Fig. 4, solid squares) exhibited a modest growth deficiency compared to the wild-type parent strain O35E (Fig. 4, open diamonds). This deficiency involved both a prolongation of the lag phase and a very slight decrease in final culture density relative to that obtained with the wild-type parent strain. The hfq mutant did not autoagglutinate more rapidly than the wild-type parent strain (data not shown). The observed growth deficiency of the O35EΔhfq mutant was substantially reversed by the introduction of a wild-type M. catarrhalis hfq gene in trans (Fig. 4, open triangles). However, for reasons that remain unclear, the negative control strain O35EΔhfq(pWW115) (Fig. 4, solid circles) showed a greater growth deficiency than that observed with the hfq deletion mutant.
FIG. 4.
Growth profiles of wild-type, mutant, and complemented mutant strains of M. catarrhalis. Cells of the wild-type strain O35E (⋄), the hfq deletion mutant O35EΔhfq (▪), the complemented hfq deletion mutant O35EΔhfq(pAA200) (▵), and the negative control O35EΔhfq(pWW115) (•) were suspended in BHI broth to an OD600 of 1.0 and then diluted 1:20. These suspensions were allowed to grow with aeration at 37°C, and the growth was monitored by measuring the absorbance at 600 nm every hour.
The M. catarrhalis hfq mutant exhibits increased sensitivity to stress.
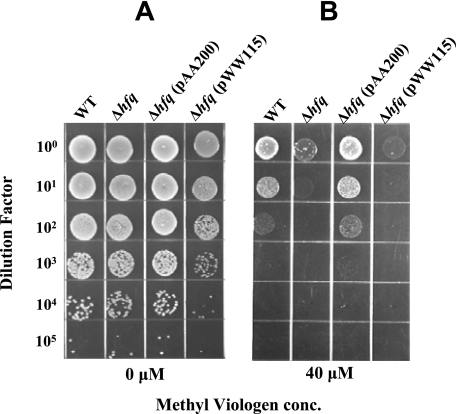
Methyl viologen (paraquat) is a redox-cycling agent that causes cytotoxicity by producing a cytosolic superoxide flux (23). No difference was apparent in the growth of the wild-type parent strain O35E and the hfq deletion mutant when they were spotted onto BHI agar (Fig. 5A), but the hfq deletion mutant was ∼100-fold more sensitive to 40 μM methyl viologen than was the wild-type parent strain (Fig. 5B). The increase in susceptibility to methyl viologen observed with the O35EΔhfq mutant was reversed by complementing this mutant with a wild-type M. catarrhalis hfq gene provided in trans, whereas the same mutant containing only the plasmid vector pWW115 was as sensitive to methyl viologen as the hfq deletion mutant (Fig. 5B).
FIG. 5.
Effect of the hfq mutation on the sensitivity of M. catarrhalis to methyl viologen. Bacterial cells of each strain were suspended to the same OD600 and then serially diluted and spotted onto BHI agar (A) and BHI agar containing 40 μM methyl viologen (B). Four strains were tested: wild-type O35E strain (WT), the O35EΔhfq mutant (Δhfq), the O35EΔhfq mutant containing the pAA200 plasmid with the wild-type O35E hfq gene [Δhfq(pAA200)], and this same mutant containing only the plasmid vector [Δhfq(pWW115)]. The plates were dried and incubated overnight, and the resultant growth was photographed.
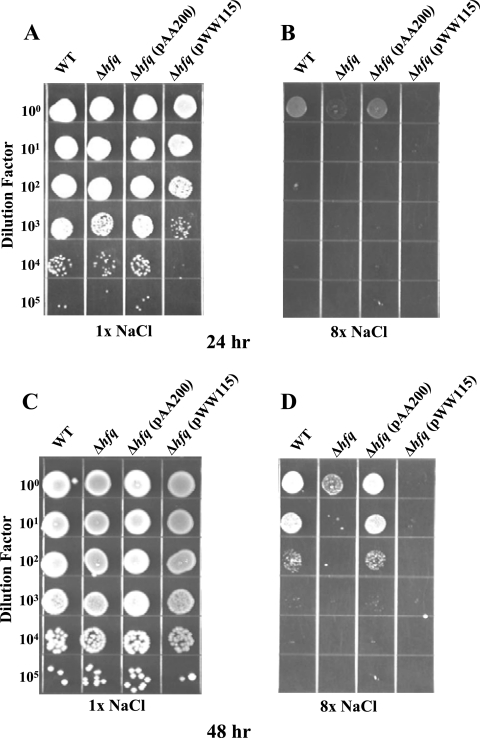
Both the wild-type strain O35E and the O35EΔhfq mutant were inhibited by a high salt concentration (compare Fig. 6A and B). However, the O35EΔhfq mutant was more sensitive to inhibition by a high concentration of salt. The difference in growth of the wild-type strain and the hfq mutant in the presence of a high salt concentration was modest after 24 h (Fig. 6B), whereas the difference became much more apparent after 48 h (Fig. 6D). In the presence of a high salt concentration, the complemented mutant O35EΔhfq(pAA200) grew to the same extent as the wild-type parent strain (Fig. 6B and D) and the negative control strain O35EΔhfq(pWW115) grew to lesser extent than the O35EΔhfq mutant (Fig. 6B and 6D).
FIG. 6.
Effect of the hfq mutation on the sensitivity of M. catarrhalis to increased salt concentration. Bacterial cells of the same four strains described in Fig. 5 were suspended to the same OD600 and then serially diluted and plated onto BHI agar containing 1× NaCl (5 mg/ml; A and C) and onto BHI agar containing 8× NaCl (40 mg/ml; B and D) and incubated for 24 h (A and B) and 48 h (C and D) at 37°C.
Effect of the hfq mutation on protein expression by M. catarrhalis.
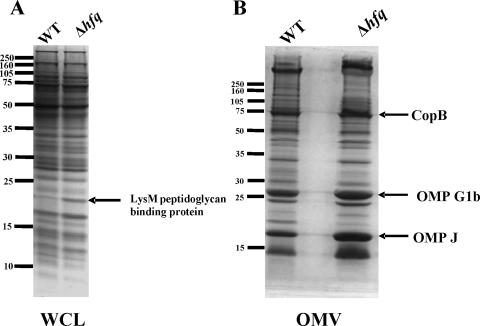
Comparison of the protein profiles of WCL of the wild-type O35E parent strain and the hfq deletion mutant (Fig. 7A) revealed a band of ∼18 kDa that was more highly expressed in the mutant. N-terminal sequence analysis of the protein contained in this band yielded the amino acid sequence GVFEFAKDIG. When used to search the M. catarrhalis ATCC 43617 genome (GenBank accession numbers AX067426 to AX067466), this sequence most closely matched that of a predicted M. catarrhalis protein that contains a LysM peptidoglycan-binding motif. Use of this protein to search the nonredundant protein databases showed that it was most similar (67% identity; E value = 10−51) to a predicted protein containing a LysM peptidoglycan-binding motif (accession no. YP_579785.1) from Psychrobacter cryohalolentis. Comparison of the outer membrane protein profiles of the wild-type strain O35E and the hfq deletion mutant (Fig. 7B) showed several bands that appeared to be slightly more abundant in the hfq mutant relative to the wild-type strain. These proteins, with apparent sizes of approximately 75, 25, and 18 kDa, were identified by N-terminal amino acid sequence analysis as the outer membrane proteins CopB (4), OMP G1b (1), and OMP J (24) (data not shown).
FIG. 7.
Effects of the hfq mutation on protein expression by M. catarrhalis. Proteins present in WCL (A) and outer membrane vesicles (OMV) (B) from the wild-type strain O35E and the hfq mutant O35EΔhfq were resolved by SDS-PAGE and stained with Coomassie blue. The positions of some of the bands that were more abundant in the hfq mutant are indicated by the black arrows. Proteins contained in the four selected bands were subjected to N-terminal amino acid sequence analysis. Protein molecular mass position markers (in kilodaltons) are indicated on the left side of each panel.
Effect of the hfq mutation on biofilm development in vitro.
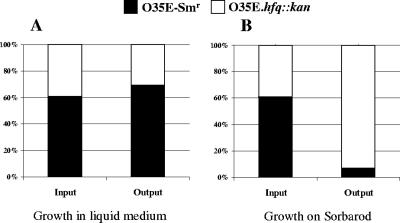
A competitive index approach (14, 29) was used to determine whether the hfq mutation affected the ability of M. catarrhalis to form a biofilm in vitro in a continuous flow system. Cells of the streptomycin-resistant mutant of the O35E parent strain (i.e., O35E-Smr) were mixed with an equivalent number of cells of the hfq mutant O35E.hfq::kan. When this mixture was used to inoculate broth and allowed to grow overnight, the proportions of the O35E-Smr strain and the O35E.hfq::kan mutant recovered after overnight growth were not different (P > 0.05) to those in the inoculum (Fig. 8A). These data are in agreement with the previous comparison of the growth curves of the wild-type strain and the hfq deletion mutant in broth, where the extent of growth of the hfq mutant in stationary phase was just slightly less than that of the wild-type strain (Fig. 4). In contrast, when a mixture of O35E-Smr and the O35E.hfq::kan mutant was used to inoculate a continuous-flow biofilm system, the hfq mutant clearly predominated (P < 0.05) in the population that was recovered from the biofilm after overnight growth (Fig. 8B).
FIG. 8.
The M. catarrhalis hfq mutant exhibits a growth advantage in a biofilm system. Cells of M. catarrhalis O35E-Smr (▪) and O35E.hfq::kan (□) were mixed together (input) and used to inoculate MH broth in a flask (A) and a Sorbarod filter in silicone tubing that was supplied with a continuous flow of MH broth (B). The cultures were allowed to grow overnight at 37°C and then harvested (output), serially diluted, and plated onto agar media containing the appropriate antibiotics to determine the relative percentages of each strain in the mixture. The data presented are the means of three independent experiments.
The M. catarrhalis Hfq protein can complement an E. coli hfq mutation.
The M. catarrhalis Hfq protein is considerably larger than the Hfq proteins of other bacteria, with the nonhomologous region in the C-terminal half being larger than the Hfq proteins of these other organisms. In order to investigate the possibility that these numerous additional amino acids in the C terminus of the M. catarrhalis Hfq protein might interfere with the ability of this protein to function in the same manner as other Hfq proteins, the wild-type M. catarrhalis hfq gene was cloned into the E. coli hfq mutant GS081 (67) on plasmid pAA105. As a negative control, this same E. coli hfq mutant was transformed with the same plasmid vector that had an irrelevant DNA insert (i.e., pAA105B).
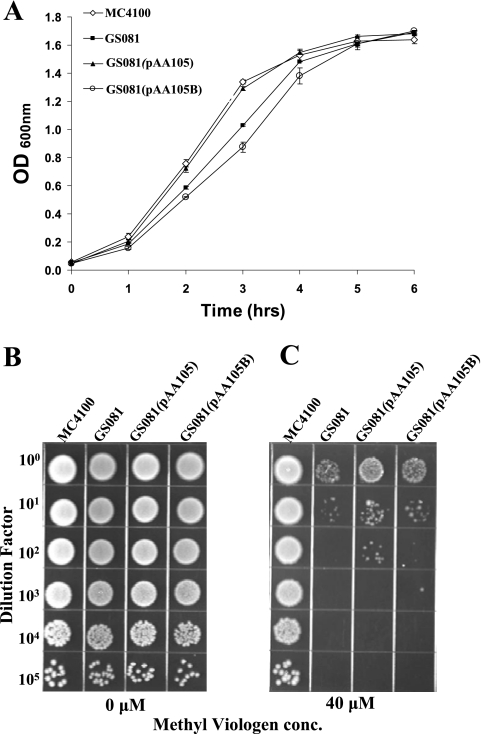
These two transformants, together with the E. coli parent strain MC4100 and the E. coli hfq mutant GS081, were each grown in LB broth to allow comparison of their growth rates (Fig. 9A). The E. coli hfq mutant GS081 (Fig. 9A, solid squares) showed a modest growth defect in LB broth relative to the parent strain (Fig. 9A, open diamonds). Expression of the M. catarrhalis Hfq protein in this mutant (Fig. 9A, solid triangles) reversed this growth defect, whereas the hfq mutant containing the control plasmid pAA105B (Fig. 9A, open circles) had a growth defect similar to that of the original GS081 hfq mutant. The E. coli hfq mutant GS081 showed increased susceptibility to 40 μM methyl viologen relative to the parent strain MC4100 (Fig. 9C). The expression of the M. catarrhalis Hfq protein in this E. coli hfq mutant partially reversed the decrease in the resistance of this mutant to methyl viologen (Fig. 9C), while the negative control strain E. coli GS081(pAA105B) was as sensitive to methyl viologen as GS081 (Fig. 9C). No obvious differences in the growth of these same four strains were apparent when they were spotted on plain BHI agar (Fig. 9B).
FIG. 9.
Complementation of an E. coli hfq mutant with the M. catarrhalis Hfq protein. (A) Growth curves of wild-type, mutant, and recombinant E. coli strains. The wild-type strain MC4100 (⋄), the hfq mutant GS081 (▪), the hfq mutant with the M. catarrhalis hfq gene provided in trans [GS081(pAA105)] (▴), and the hfq mutant containing the negative control plasmid [GS081(pAA105B)] (○) were suspended in LB broth to an OD600 of 1.0 and then diluted 1:20. These cultures were allowed to grow with aeration at 37°C, and the growth was monitored by measuring the absorbance at 600 nm every hour. (B and C) Cells of the wild-type, mutant, and recombinant E. coli strains were suspended to the same OD600 and then serially diluted and spotted onto BHI agar (B) and onto BHI agar containing 40 μM methyl viologen (C). The plates were dried and incubated overnight at 37°C.
The M. catarrhalis Hfq protein binds RNA.
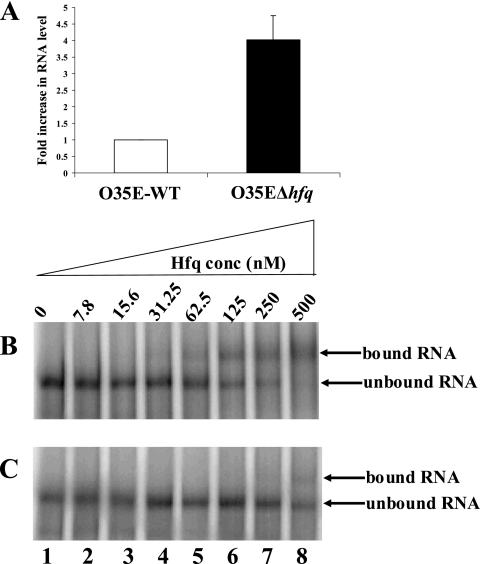
The hallmark of previously characterized Hfq proteins is their ability to bind RNA, acting as an RNA chaperone (20, 30, 38, 56). In order to test the ability of the M. catarrhalis Hfq protein to bind RNA, we chose a potential target that might be regulated through interaction with Hfq. One of the proteins that was highly expressed in the M. catarrhalis hfq mutant was the macromolecule that contained a LysM peptidoglycan-binding motif (Fig. 7A). This protein is encoded by a gene that was annotated as ORF 1068 in the M. catarrhalis ATCC 43617 genome (66). Real-time RT-PCR was performed to compare the levels of mRNA transcribed from this gene in the wild-type strain O35E and in the O35EΔhfq deletion mutant. The level of ORF 1068 transcript in the O35EΔhfq cells was at least four times greater than the amount of this same message in the wild-type strain (P = 0.027; Fig. 10A). It could be inferred from these data that Hfq is affecting the level of detectable mRNA derived from this particular gene.
FIG. 10.
The M. catarrhalis Hfq protein binds RNA. (A) Real-time RT-PCR analysis expression of the ORF 1068 gene that encodes the LysM motif-containing protein. Total RNA isolated from the wild-type O35E strain (□) and the O35EΔhfq mutant (▪) was used for real-time RT-PCR with oligonucleotide primers specific for the ORF 1068 gene and 16S RNA. The data are presented as the fold increase using the normalized level of the wild-type O35E ORF 1068 mRNA as the calibrator. These data represent the mean of two independent experiments (each performed with samples in triplicate), and the error bars represent the standard deviations. (B and C) EMSA. A radiolabeled RNA probe containing part of the ORF 1068 mRNA was incubated with increasing concentrations of the purified M. catarrhalis Hfq protein in the presence of 100 ng of E. coli tRNA/μl (B) or in the presence of 100 ng of E. coli tRNA/μl and 25 ng of unlabeled RNA probe (C). The binding reactions were then subjected to electrophoresis in a native 6% (wt/vol) polyacrylamide gel. The bands were visualized by exposing the gel to a storage phosphor intensifying screen that was then scanned by using a phosphorimager.
In order to determine whether this effect might involve an interaction between the Hfq protein and the ORF 1068 mRNA, in vitro transcription was used to transcribe a 317-nt RNA fragment containing 217 nt from the 5′ end of this ORF and 100 nt of the upstream region, which has a putative transcriptional start site. Equal amounts of this RNA probe were incubated with increasing concentrations of the purified, recombinant M. catarrhalis Hfq protein in an EMSA (Fig. 10B). The Hfq protein was shown to be able to bind this RNA fragment, as evidenced by the apparent retardation in the electrophoretic mobility of the radiolabeled RNA probe in the presence of concentrations of Hfq protein greater than 62.5 nM (Fig. 10B, lanes 5 to 8). The experiment was repeated in the presence of 25 ng of the same 317-nt RNA fragment that was not radiolabeled. The presence of the unlabeled RNA probe resulted in an increase in the Hfq protein concentration required to produce a change in the electrophoretic mobility of the radiolabeled probe, from 62.5 to 500 nM (Fig. 10C, lanes 5 to 8).
DISCUSSION
The Hfq protein has been characterized in several bacterial species in recent years (10, 13, 15, 16, 53, 55). It was first recognized as the E. coli host factor necessary for replication of RNA phage Qβ (17) and subsequently as a RNA chaperone and global effector molecular that orchestrates mRNA-sRNA interactions, frequently in response to environmental signals (for reviews, see references 11, 19, 32, and 61). Initial analysis of the M. catarrhalis hfq gene showed that this ORF is predicted to encode an Hfq protein that contained double the number of amino acids present in other Hfq homologues that have been previously studied (Fig. 1) and that the additional amino acids are located in the C-terminal half of the protein and form a highly hydrophilic sequence. Western plot analysis confirmed that M. catarrhalis expresses an Hfq protein which exhibits a mass in SDS-PAGE consistent with that predicted from the deduced amino acid sequence (Fig. 3). Searching the nonredundant protein databases for homologues of the M. catarrhalis Hfq protein showed that three Psychrobacter species have genes encoding an Hfq protein similar in size to that of M. catarrhalis, but the C-terminal region of these Psychrobacter proteins had only moderate identity with that of M. catarrhalis (data not shown). The redundancy and tandem repeat present in the amino acid sequence of the C-terminal portion of the M. catarrhalis Hfq protein suggest that this span of the hfq ORF arose from at least one duplication event. The exact role of this long, highly hydrophilic region in the C terminus of the M. catarrhalis Hfq protein is not clear at this time, but it apparently did not eliminate the ability of the M. catarrhalis Hfq protein to complement an E. coli hfq mutation (Fig. 9).
The ability of the M. catarrhalis Hfq protein to complement this E. coli hfq mutant suggested that this M. catarrhalis gene product could interact with RNA and led us to prove directly that this protein could bind at least one M. catarrhalis RNA species. Examination of the effect of an hfq mutation on the expression of proteins by M. catarrhalis revealed that several proteins detectable in either WCL or outer membranes were either upregulated or more abundant in the absence of the Hfq protein (Fig. 7). One of these genes whose mRNA was definitely more abundant in the hfq mutant, based on real-time RT-PCR assays (Fig. 10A), was ORF 1068, which encoded an 18-kDa macromolecule that contained a predicted LysM motif. LysM or lysine motif domains are thought to function as peptidoglycan-binding domains and are found in some enzymes that degrade this bacterial cell wall constituent (8, 9) but also in eukaryotic proteins (46).
Although sRNAs have not been described to date in M. catarrhalis, the observed effect of the hfq mutation on ORF 1068 mRNA levels raised the possibility that an sRNA might be involved in controlling expression of this gene. Identification of a possible sRNA that interacts with ORF 1068 mRNA was beyond the scope of the present study, but we took advantage of the fact that Hfq has been shown to bind to at least some mRNA molecules that are targets for sRNA (19, 36). When we used the 5′ end of the ORF 1068 transcript in an EMSA with purified, recombinant M. catarrhalis Hfq, this protein bound the RNA (Fig. 10B and C). It can be inferred from these data that the M. catarrhalis Hfq protein, either alone or in concert with sRNA components, affects the abundance of the ORF 1068 transcript.
Phenotypic characterization of the M. catarrhalis hfq mutant showed that it had similarities to hfq mutants of some other bacterial species, such as a slight growth deficiency in liquid medium (33, 54, 59) and increased sensitivity to oxidative and osmotic stresses (59, 63). The M. catarrhalis hfq mutant had an interesting and unexpected phenotype in a continuous-flow biofilm system where, in competitive index experiments, it became the predominant member of the biofilm after overnight growth in vitro (Fig. 8). To the best of the authors' knowledge, the effect of an hfq mutation on biofilm formation in vitro by other bacteria has not been reported to date, and a ready explanation for this apparent growth advantage of the M. catarrhalis hfq mutant is not apparent. However, the modest increase in expression or relative abundance of several outer membrane proteins, including CopB (4), OMP G1b (1), and OMP J (24), in the M. catarrhalis hfq mutant raises the possibility that overall outer membrane architecture may have been affected in this mutant. If so, then this change in the surface of the mutant may have had some effect on its ability to form a biofilm in this model system. Although hfq mutations frequently affect the virulence potential of some bacterial pathogens (13, 15, 47, 53, 54), the current lack of an animal model for M. catarrhalis disease (27, 62) precludes testing the effect of this mutation on the virulence of M. catarrhalis.
In conclusion, the data presented here indicated that the mucosal pathogen M. catarrhalis expresses an Hfq protein that is much larger than the Hfq proteins of other, well-studied bacteria. This Hfq protein can bind M. catarrhalis mRNA and likely interacts with sRNAs in a manner similar to that described for other Hfq proteins. Identification of sRNAs expressed by M. catarrhalis will be the subject of future research efforts.
Acknowledgments
This study was supported by U.S. Public Health Service grant no. AI36344 to E.J.H.
We thank Gisela Storz for helpful comments and for providing E. coli strains MC4100 and GS081 and both Kaiping Deng and Maria Labandeira for generous assistance and helpful discussions.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 24 March 2008.
REFERENCES
- 1.Adlowitz, D. G., T. Hiltke, A. J. Lesse, and T. F. Murphy. 2004. Identification and characterization of outer membrane proteins G1a and G1b of Moraxella catarrhalis. Vaccine 222533-2540. [DOI] [PubMed] [Google Scholar]
- 2.Adlowitz, D. G., C. Kirkham, S. Sethi, and T. F. Murphy. 2006. Human serum and mucosal antibody responses to outer membrane protein G1b of Moraxella catarrhalis in chronic obstructive pulmonary disease. FEMS Immunol. Med. Microbiol. 46139-146. [DOI] [PubMed] [Google Scholar]
- 3.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 654367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aebi, C., B. Stone, M. Beucher, L. D. Cope, I. Maciver, S. E. Thomas, G. H. McCracken, Jr., P. F. Sparling, and E. J. Hansen. 1996. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect. Immun. 642024-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arluison, V., P. Derreumaux, F. Allemand, M. Folichon, E. Hajnsdorf, and P. Regnier. 2002. Structural modelling of the Sm-like protein Hfq from Escherichia coli. J. Mol. Biol. 320705-712. [DOI] [PubMed] [Google Scholar]
- 6.Attia, A. S., and E. J. Hansen. 2006. A conserved tetranucleotide repeat is necessary for wild-type expression of the Moraxella catarrhalis UspA2 protein. J. Bacteriol. 1887840-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attia, A. S., E. R. Lafontaine, J. L. Latimer, C. Aebi, G. A. Syrogiannopoulos, and E. J. Hansen. 2005. The UspA2 protein of Moraxella catarrhalis is directly involved in the expression of serum resistance. Infect. Immun. 732400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from Escherichia coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 2991113-1119. [DOI] [PubMed] [Google Scholar]
- 9.Birkeland, N. K. 1994. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage phi LC3: a dual lysis system of modular design. Can. J. Microbiol. 40658-665. [DOI] [PubMed] [Google Scholar]
- 10.Bohn, C., C. Rigoulay, and P. Bouloc. 2007. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC. Microbiol. 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan, R. G., and T. M. Link. 2007. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10125-133. [DOI] [PubMed] [Google Scholar]
- 12.Caillet, J., and L. Droogmans. 1988. Molecular cloning of the Escherichia coli miaA gene involved in the formation of delta 2-isopentenyl adenosine in tRNA. J. Bacteriol. 1704147-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christiansen, J. K., M. H. Larsen, H. Ingmer, L. Sogaard-Andersen, and B. H. Kallipolitis. 2004. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 1863355-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 3251-62. [DOI] [PubMed] [Google Scholar]
- 15.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53345-354. [DOI] [PubMed] [Google Scholar]
- 16.Figueroa-Bossi, N., S. Lemire, D. Maloriol, R. Balbontin, J. Casadesus, and L. Bossi. 2006. Loss of Hfq activates the σE-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 62838-852. [DOI] [PubMed] [Google Scholar]
- 17.Franze de Fernandez, M. T., L. Eoyang, and J. T. August. 1968. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature 219588-590. [DOI] [PubMed] [Google Scholar]
- 18.Furano, K., and A. A. Campagnari. 2003. Inactivation of the Moraxella catarrhalis 7169 ferric uptake regulator increases susceptibility to the bactericidal activity of normal human sera. Infect. Immun. 711843-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58303-328. [DOI] [PubMed] [Google Scholar]
- 21.Gottesman, S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21399-404. [DOI] [PubMed] [Google Scholar]
- 22.Guillier, M., S. Gottesman, and G. Storz. 2006. Modulating the outer membrane with small RNAs. Genes Dev. 202338-2348. [DOI] [PubMed] [Google Scholar]
- 23.Hassan, H. M., and I. Fridovich. 1978. Superoxide radical and the oxygen enhancement of the toxicity of paraquat in Escherichia coli. J. Biol. Chem. 2538143-8148. [PubMed] [Google Scholar]
- 24.Hays, J. P., S. van Selm, T. Hoogenboezem, S. Estevao, K. Eadie, P. van Keelen, J. Tommassen, A. van Belkum, and P. W. Hermans. 2005. Identification and characterization of a novel outer membrane protein (OMP J) of Moraxella catarrhalis that exists in two major forms. J. Bacteriol. 1877977-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heiniger, N., R. Troller, P. S. Meier, and C. Aebi. 2005. Cold shock response of the UspA1 outer membrane adhesin of Moraxella catarrhalis. Infect. Immun. 738247-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8528-535. [PubMed] [Google Scholar]
- 27.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes. Infect. 2547-559. [DOI] [PubMed] [Google Scholar]
- 28.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 1831540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawlor, M. S., J. Hsu, P. D. Rick, and V. L. Miller. 2005. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol. Microbiol. 581054-1073. [DOI] [PubMed] [Google Scholar]
- 30.Majdalani, N., C. K. Vanderpool, and S. Gottesman. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 4093-113. [DOI] [PubMed] [Google Scholar]
- 31.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masse, E., H. Salvail, G. Desnoyers, and M. Arguin. 2007. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10140-145. [DOI] [PubMed] [Google Scholar]
- 33.McNealy, T. L., V. Forsbach-Birk, C. Shi, and R. Marre. 2005. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J. Bacteriol. 1871527-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier, P. S., S. Freiburghaus, A. Martin, N. Heiniger, R. Troller, and C. Aebi. 2003. Mucosal immune response to specific outer membrane proteins of Moraxella catarrhalis in young children. Pediatr. Infect. Dis. J. 22256-262. [DOI] [PubMed] [Google Scholar]
- 35.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 1755899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moll, I., D. Leitsch, T. Steinhauser, and U. Blasi. 2003. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 4284-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mollenkvist, A., T. Nordstrom, C. Hallden, J. J. Christensen, A. Forsgren, and K. Riesbeck. 2003. The Moraxella catarrhalis immunoglobulin D-binding protein MID has conserved sequences and is regulated by a mechanism corresponding to phase variation. J. Bacteriol. 1852285-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moller, T., T. Franch, P. Hojrup, D. R. Keene, H. P. Bachinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 923-30. [DOI] [PubMed] [Google Scholar]
- 39.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, T. F. 2000. Bacterial otitis media: pathogenetic considerations. Pediatr. Infect. Dis. J. 19S9-S15. [DOI] [PubMed] [Google Scholar]
- 41.Murphy, T. F., A. L. Brauer, C. Aebi, and S. Sethi. 2005. Antigenic specificity of the mucosal antibody response to Moraxella catarrhalis in chronic obstructive pulmonary disease. Infect. Immun. 738161-8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy, T. F., A. L. Brauer, C. Aebi, and S. Sethi. 2005. Identification of surface antigens of Moraxella catarrhalis as targets of human serum antibody responses in chronic obstructive pulmonary disease. Infect. Immun. 733471-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy, T. F., and M. R. Loeb. 1989. Isolation of the outer membrane of Branhamella catarrhalis. Microb. Pathog. 6159-174. [DOI] [PubMed] [Google Scholar]
- 45.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St.Geme, III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 704523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponting, C. P., L. Aravind, J. Schultz, P. Bork, and E. V. Koonin. 1999. Eukaryotic signalling domain homologues in archaea and bacteria: ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289729-745. [DOI] [PubMed] [Google Scholar]
- 47.Robertson, G. T., and R. M. Roop, Jr. 1999. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34690-700. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Sauter, C., J. Basquin, and D. Suck. 2003. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 314091-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schumacher, M. A., R. F. Pearson, T. Moller, P. Valentin-Hansen, and R. G. Brennan. 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 213546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seib, K. L., I. R. A. Peak, and M. P. Jennings. 2002. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol. Med. Microbiol. 32159-165. [DOI] [PubMed] [Google Scholar]
- 52.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347465-471. [DOI] [PubMed] [Google Scholar]
- 53.Sittka, A., V. Pfeiffer, K. Tedin, and J. Vogel. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63193-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonnleitner, E., S. Hagens, F. Rosenau, S. Wilhelm, A. Habel, K. E. Jager, and U. Blasi. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35217-228. [DOI] [PubMed] [Google Scholar]
- 55.Sonnleitner, E., M. Schuster, T. Sorger-Domenigg, E. P. Greenberg, and U. Blasi. 2006. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 591542-1558. [DOI] [PubMed] [Google Scholar]
- 56.Storz, G., S. Altuvia, and K. M. Wassarman. 2005. An abundance of RNA regulators. Annu. Rev. Biochem. 74199-217. [DOI] [PubMed] [Google Scholar]
- 57.Storz, G., and D. Haas. 2007. A guide to small RNAs in microorganisms. Curr. Opin. Microbiol. 1093-95. [Google Scholar]
- 58.Tjaden, B., S. S. Goodwin, J. A. Opdyke, M. Guillier, D. X. Fu, S. Gottesman, and G. Storz. 2006. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 342791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsui, H. C., H. C. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1335-49. [DOI] [PubMed] [Google Scholar]
- 60.Tzeng, Y. L., A. Datta, C. Strole, V. S. Kolli, M. R. Birck, W. P. Taylor, R. W. Carlson, R. W. Woodard, and D. S. Stephens. 2002. KpsF is the arabinose-5-phosphate isomerase required for 3-deoxy-d-manno-octulosonic acid biosynthesis and for both lipooligosaccharide assembly and capsular polysaccharide expression in Neisseria meningitidis. J. Biol. Chem. 27724103-24113. [DOI] [PubMed] [Google Scholar]
- 61.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 511525-1533. [DOI] [PubMed] [Google Scholar]
- 62.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. Van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wachi, M., A. Takada, and K. Nagai. 1999. Overproduction of the outer-membrane proteins FepA and FhuE responsible for iron transport in Escherichia coli hfq::cat mutant. Biochem. Biophys. Res. Commun. 264525-529. [DOI] [PubMed] [Google Scholar]
- 64.Wang, W., and E. J. Hansen. 2006. Plasmid pWW115, a cloning vector for use with Moraxella catarrhalis. Plasmid 56133-137. [DOI] [PubMed] [Google Scholar]
- 65.Wang, W., M. M. Pearson, A. S. Attia, R. J. Blick, and E. J. Hansen. 2007. A UspA2H-negative variant of Moraxella catarrhalis strain O46E has a deletion in a homopolymeric nucleotide repeat common to uspA2H genes. Infect. Immun. 752035-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, W., L. Reitzer, D. A. Rasko, M. M. Pearson, R. J. Blick, C. Laurence, and E. J. Hansen. 2007. Metabolic analysis of Moraxella catarrhalis and the effect of selected in vitro growth conditions on global gene expression. Infect. Immun. 754959-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 911-22. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, A., K. M. Wassarman, C. Rosenow, B. C. Tjaden, G. Storz, and S. Gottesman. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 501111-1124. [DOI] [PubMed] [Google Scholar]
- 69.Ziolkowska, K., P. Derreumaux, M. Folichon, O. Pellegrini, P. Regnier, I. V. Boni, and E. Hajnsdorf. 2006. Hfq variant with altered RNA binding functions. Nucleic Acids Res. 34709-720. [DOI] [PMC free article] [PubMed] [Google Scholar]