Abstract
The transcription factor p110 CUX1 was shown to stimulate cell proliferation by accelerating entry into S phase. As p110 CUX1 can function as a transcriptional repressor or activator depending on promoter context, we investigated its mechanism of transcriptional activation using the DNA polymerase α gene promoter as a model system. Linker-scanning analysis revealed that a low-affinity E2F binding site is required for transcriptional activation. Moreover, coexpression with a dominant-negative mutant of DP-1 suggested that endogenous E2F factors are indeed needed for p110-mediated activation. Tandem affinity purification, coimmunoprecipitation, chromatin immunoprecipitation, and reporter assays indicated that p110 CUX1 can engage in weak protein-protein interactions with E2F1 and E2F2, stimulate their recruitment to the DNA polymerase α gene promoter, and cooperate with these factors in transcriptional activation. On the other hand, in vitro assays suggested that the interaction between CUX1 and E2F1 either is not direct or is regulated by posttranslational modifications. Genome-wide location analysis revealed that targets common to p110 CUX1 and E2F1 included many genes involved in cell cycle, DNA replication, and DNA repair. Comparison of the degree of enrichment for various E2F factors suggested that binding of p110 CUX1 to a promoter will favor the specific recruitment of E2F1, and to a lesser extent E2F2, over E2F3 and E2F4. Reporter assays on a subset of common targets confirmed that p110 CUX1 and E2F1 cooperate in their transcriptional activation. Overall, our results show that p110 CUX1 and E2F1 cooperate in the regulation of many cell cycle genes.
CUX1 (cut homeobox) belongs to a family of transcription factors involved in the control of proliferation and differentiation (reviewed in reference 52). Several CUX1 isoforms can be expressed as a result of proteolytic processing or transcription initiation at alternative sites (21, 23, 45, 49, 70). Cux1 gene ablation in mice resulted in high perinatal lethality. Surviving mice exhibited a number of mutant phenotypes, including growth retardation, male infertility, curly whiskers, abnormal hair follicle morphogenesis, and a shortage of T and B cells (13, 43, 63, 73). Transgenic mice expressing p200 CUX1 exhibited multiorgan hyperplasia and organomegaly (36), whereas those expressing p75 or p110 CUX1 displayed enhanced susceptibility to malignancies in various tissues and cell types (4; C. Cadieux and A. Nepveu, unpublished data).
Initial studies of mammalian CUX1 revealed its role as a transcriptional repressor that is expressed in differentiating precursor cells and serves to down-regulate the expression of genes expressed only in terminally differentiated cells (41, 55, 64, 66, 67). CUX1 was also reported to regulate the expression of cell cycle-regulated genes such as those encoding p21WAF1 (7), histones H1, H2A, H2B, H3, and H4 (12, 27, 74, 76), and DNA polymerase (Pol) α (71). The full-length protein, p200 CUX1, interacts transiently with DNA and is expressed throughout the cell cycle (48). Dephosphorylation by the Cdc25A phosphatase and proteolytic cleavage by a nuclear isoform of cathepsin L at the end of the G1 phase yield an amino-terminally truncated isoform, p110 CUX1, which interacts stably with DNA (7, 20, 49). Constitutive expression of p110 CUX1 was shown to stimulate cell proliferation by accelerating entry into S phase (59). Genome-wide location analysis revealed that p110 CUX1 binds to the promoters of several genes that are involved in DNA replication and cell cycle progression (25). Multiple approaches including silencing RNA, transient or stable expression, and reporter assays demonstrated that most cell cycle targets are activated whereas a few are repressed or not affected by p110 CUX1 (25).
The E2F family of transcription factors is classically divided into activator E2Fs (E2F1, E2F2, and E2F3a), repressor E2Fs (E2F3b, E2F4, E2F5, E2F6, E2F7, and E2F8), and DP heterodimerization partners (DP1 and DP2) (reviewed in reference 42). Evidence from gene ablation studies has revealed some functional redundancy among E2F family members. While the loss of one E2F can be functionally compensated for by other E2Fs (15, 29, 30, 38), the combined loss of two E2Fs results in a more severe phenotype (6, 39). Loss of E2F1, E2F2, and E2F3 prevents mouse embryonic fibroblasts from reentering the cell cycle following quiescence (84). Indeed, E2Fs play a critical role in the control of cellular proliferation. In quiescence, pRB pocket protein family members bind E2Fs and repress transcription of target genes (58, 65). Following growth stimulation, cyclin-cyclin-dependent kinase (Cdk) complexes phosphorylate the pocket protein, which then dissociates from the promoter-bound E2F/DP heterodimer. This results in derepression and allows the transcriptional activation of numerous genes with roles in DNA replication and cell cycle progression. Importantly, however, while many target genes conform to this model, other genes clearly are regulated in a more complex manner (2, 31, 78, 80, 81).
Using classical approaches, such as transient reporter assays and overexpression systems, a number of E2F targets were identified, such as dihydrofolate reductase (DHFR), Cdc6, Orc1L, Cdc25A, B-myb, and cyclin A (37, 54). In more recent years, gene expression microarray profiling and chromatin immunoprecipitation-microarray (ChIP-chip) analysis have allowed the unbiased identification of target genes (3, 24, 51, 78, 80). Other studies have explored the promoter occupancy, at different stages of the cell cycle, of different E2F family members and pocket proteins (2, 57, 69). The molecular basis for the functional differences among E2F family members has been the subject of many recent studies (17, 18, 24, 62). Whereas no distinction in DNA binding specificity among E2Fs has been determined (86), specific protein-protein interactions were shown to contribute to promoter specificity. For example, cooperativity was documented between E2F3 and TFE3 (17) and both E2F2 and E2F3 and YY1 (62).
We began the present study by using the DNA Pol α gene promoter as a model system to investigate the mechanism of transcriptional activation by p110 CUX1. Linker-scanning analysis identified a region that is required for p110-mediated activation but is not bound by it. The homologous region in the mouse DNA Pol α gene promoter had been shown to be required for growth-dependent regulation and for E2F1-mediated stimulation (32). This finding as well as results of reporter assays in the presence of a dominant-negative mutant of DP1 suggested that the binding of an E2F factor was necessary for CUX1-mediated activation. ChIP confirmed the recruitment of E2F factors to the DNA Pol α reporter plasmid and showed that coexpression with p110 CUX1 enhances the recruitment of E2F1 and E2F2 but not of E2F3 and E2F4. Genome-wide location analysis identified a number of targets that are common to p110 CUX1 and E2F1. Gene ontology analysis revealed that functions related to cell cycle progression, DNA replication and DNA repair were overrepresented in the list of common targets. Reporter assays confirmed that p110 CUX1 and E2F1 cooperate in the regulation of their common targets. Overall, our results suggest that weak protein-protein interactions contribute to the recruitment of distinct E2F factors to specific promoters.
MATERIALS AND METHODS
Plasmid and lentivirus construction.
CUX1 and luciferase reporter constructs were described in our previous studies (25, 26, 60, 71). The lentivirus vector was constructed by inserting the CUX1 short-hairpin RNA (shRNA) sequence, AAGAATCTTCTCGTTTGAAACTT, into pLVTHM (82). Lentiviruses were produced by transient transfection of 293FT cells by using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen).
Cell culture, transfection, and synchronization.
Hs 578T cells were grown in Dulbecco's modified Eagle medium supplemented with 5% fetal bovine serum (FBS) (Gibco). Transient transfections were performed with GeneJuice (Novagen) according to the manufacturer's instructions. For synchronization by thymidine block, Hs 578T cells were cultured overnight in Dulbecco's modified Eagle medium plus 5% fetal bovine serum supplemented with 2 mM thymidine and harvested.
Luciferase assay.
Luciferase assays were performed as previously described (49). Because the internal control plasmid is itself often repressed by CUX1, as a control for transfection efficiency the purified β-galactosidase protein (Sigma) was included in the transfection mix, as previously described (28). The luciferase activity was then normalized based on β-galactosidase activity.
ChIP.
Thymidine-blocked Hs 578T cells (2 ×108) were used for each ChIP. Immunoprecipitation was performed with affinity-purified CUX1 antibodies 861 and 1300 (49) or an E2F1 antibody (05-379; Upstate). The nuclei were lysed as described in reference 79, lysed in radioimmunoprecipitation assay M buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5 mM EGTA, 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate [DOC], 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride, protease inhibitors), and sonicated on ice to obtain 250- to 800-bp DNA fragments. After preclearing for 1 h and incubation with antibodies overnight, immunocomplexes were washed three times each in wash buffer I (20 mM Tris-HCl [pH 8], 2 mM EDTA, 2 mM EGTA, 150 mM NaCl, 1% NP-40, 0.5% DOC, 0.2% SDS), wash buffer II (20 mM Tris-HCl [pH 9], 2 mM EDTA, 2 mM EGTA, 500 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS), and wash buffer III (50 mM Tris-HCl [pH 7.5], 2 mM EDTA, 1 mM EGTA, 0.5 M LiCl, 1% NP-40, 0.7% DOC) and then washed once in Tris-EDTA. Cross-linked DNA was eluted with 1% SDS, 10 mM Tris-HCl (pH 8), 10 mM EDTA at 65°C for 30 min. After reversal of formaldehyde cross-linking, chromatin-immunoprecipitated DNAs were treated with RNase A and proteinase K.
Enrichment calculation.
Enrichment levels of genes were determined by real-time PCR using G6PDH as an internal control and chromatin-immunoprecipitated DNAs obtained by immunoprecipitation with either CUX1 antibodies or no antibody. Specific enrichment of a promoter was calculated as follows: (CUX1 immunoprecipitation target/no-antibody immunoprecipitation target) × (no-antibody G6PDH/CUX1 immunoprecipitation G6PDH).
Probe generation and microarray hybridization.
The generation of labeled DNAs from individual chromatin affinity purification (ChAP) samples was performed following the protocol of linker-mediated PCR as detailed previously (57). Briefly, chromatin-immunoprecipitated and chromatin affinity-purified DNAs and input DNA were blunted, ligated to a unidirectional linker and amplified by PCR for 24 cycles to generate a sufficient amount of DNA. ChIP or ChAP DNAs and input DNAs were fluorescently labeled with Cy5 fluorophore and Cy3 fluorophore, respectively, by using a BioPrime array CGH genomic labeling kit following the manufacturer's instructions (Invitrogen). Prior to hybridization, microarray slides were incubated in a blocking solution, 1.6% succinic anhydride in 1-methyl-2-pyrrolidinone, for 20 min at room temperature. After washing, labeled DNAs were added to the hybridization buffer (25% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS, 0.2% bovine serum albumin, 0.4 μg/μl of human Cot-1 DNA, 0.8 μg/μl of yeast tRNA) and hybridized at 55°C for 20 h. The slides were washed once with 2× SSC, 0.1% SDS for 15 min, twice for 2 min with 0.1× SSC, 0.1% SDS, and twice for 1 min with 0.1× SSC and then spin dried. Hybridized slides were scanned with an Axon 4000b scanner, and the acquired images were analyzed with the software GenePix Pro, version 4.1. Each set of hybridizations was performed three times with independent ChIP or ChAP materials.
Microarray design.
A microarray containing 19,000 human promoters was generated as reported elsewhere (34). In brief, the regions ranging from 800 bp upstream and 200 bp downstream of the transcription start sites from 18,660 human genes were amplified by PCR, quality-control tested, and applied to poly-l-lysine glass slides.
Microarray data analysis.
The ChIP-chip or ChAP-chip results were analyzed as previously described (53). Promoters were considered bound when the binding P value in the error model was <0.005. Functional categories were established using programs from Expression Analysis Systematic Explorer (EASE) (http://david.abcc.ncifcrf.gov/). The list of genes from the 19,000-gene microarray was used as the background.
Tandem affinity purification (TAP) and Western blot analysis.
Hs 578T cells stably expressing a recombinant p110-Tag2 protein or vector control were transfected with the expression plasmid pCMV/HA-E2F1, pCMV/HA-E2F2, pCMV/HA-E2F3a, or pCMV/HA-E2F5. A total of 2 × 108 to 4 × 108 cells were used for purification by the Taptag purification method (56). Western blots were performed using the antibodies 861 and 1300 (data not shown) or HA-11 (Covance).
In vivo DNA binding to transfected reporter plasmids.
Hs 578T cells were transfected with pGL3-Pol α (−65/+47) or pCADluc, either pXJ42 or pXJ42/Myc-CUX1 878-1336, and either pcDNA3, pCMV/E2F1, or pCMV/E2F2. DNA was extracted approximately 24 h posttransfection and was processed as described in reference 71.
Electrophoretic mobility shift assay.
Electrophoretic mobility shift assay was performed with various quantities of purified protein. Samples were incubated at room temperature for 20 min in a final volume of 30 μl of 25 mM NaCl, 10 mM Tris (pH 7.5), 1 mM MgCl2, 5 mM EDTA (pH 8.0), 5% glycerol, 1 mM DTT, 1 mg BSA with 0.2 pmol of radiolabeled oligonucleotide probes. Where added, samples were incubated with antibody for 10 min prior to addition of probe. Samples were loaded on a 5% polyacrylamide (29:1), 0.5× Tris-borate-EDTA gel and separated by electrophoresis at 8 V/cm in 1× Tris-borate-EDTA. Gels were dried and visualized by autoradiography. The DNA Pol α (−40/−14) probe was GGGCCGCTGATTGGCTTTCAGGCTGGCGCCTCGA, and the E2F probe was TCGAGAAAAGAAGCTTTTCGCGCCCGTCTCGA. The CUX1 binding site is indicated by bold type; the E2F binding site is underlined.
Immunoprecipitation.
Lysates from Hs 578T cells (10 mM Tris [pH 7.5], 5 mM EDTA, 100 mM NaCl, 1% Brij 97, protease inhibitor cocktail [Roche]) were treated with ethidium bromide at a final concentration of 100 μg/ml for 1 h, precleared with protein G-agarose (Sigma), and incubated overnight with the appropriate antibody. Immunoprecipitates were washed three times with lysis buffer, and Western blot analysis was performed.
RESULTS
The dominant-negative DP1Δ103-126 mutant prevents transcriptional activation by CUX1.
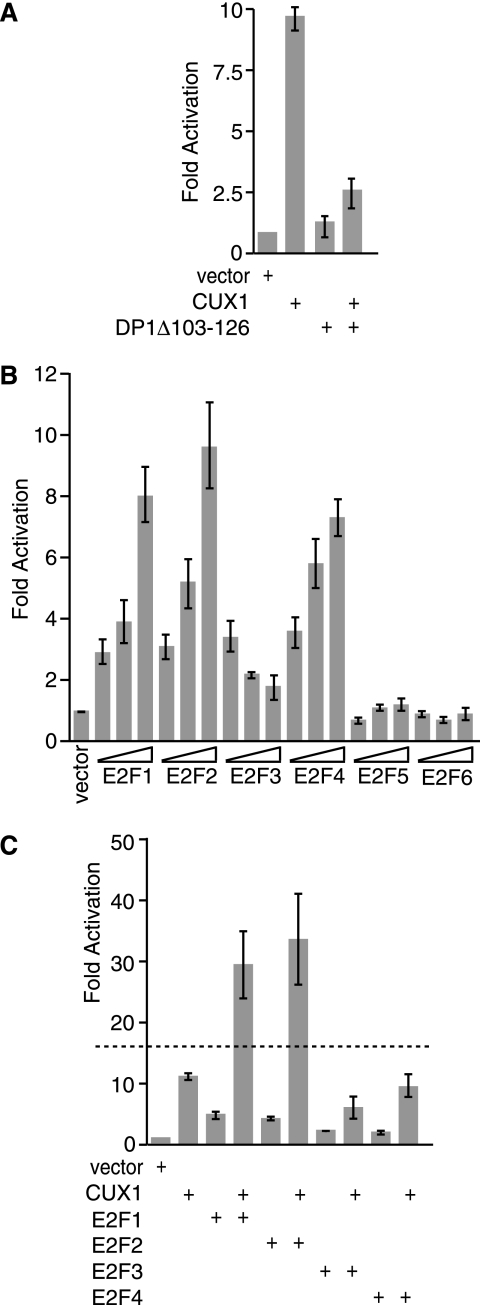
Previous experiments using linker-scanning mutations identified distinct regions of the DNA Pol α gene promoter that were necessary for transcriptional activation by p110 CUX1 (71). The −35/−26 and the −25/−16 regions, respectively, contain binding sites for p110 CUX1 and E2F (32, 71) (see the introduction). The requirement for an E2F binding site raised the possibility that an endogenous E2F factor might participate in the transcriptional activation mediated by p110 CUX1. As a preliminary approach to test this hypothesis, we measured the activity of the DNA Pol α gene reporter in the presence of CUX1 and a dominant-negative mutant of DP1, DP1Δ103-126. This mutant was previously shown to interact with E2Fs but to be unable to bind DNA, thereby keeping its E2F partners away from DNA (83). Transcriptional activation was reduced from 10-fold down to 2.5-fold in the presence of DP1Δ103-126 (Fig. 1A). These results suggested that functional endogenous E2F factors are necessary for CUX1 to transactivate the DNA Pol α gene promoter.
FIG. 1.
p110 CUX1 cooperates with E2F1 and E2F2 in the stimulation of the DNA Pol α reporter. Hs 578T cells were transfected with the DNA Pol α (−65/+47)/luciferase reporter construct and the indicated vectors expressing CUX1, DP1Δ103-126 (A), or E2F1, E2F2, E2F3, E2F4, E2F5, or E2F6 (B and C). Cytoplasmic extracts were prepared and processed to measure luciferase activity. The means of three or more transfections are shown. The dashed line (C) represents the border between additive and synergistic cooperation between CUX1 and E2F1. Synergistic activation is defined as activation greater than the sum of activation by each factor on its own.
CUX1 cooperates with E2F1 and E2F2 in the stimulation of the DNA Pol α gene promoter.
We next investigated which E2F factors, if any, were able to transactivate the DNA Pol α gene promoter. Hs 578T cells were transfected with the reporter construct and increasing amounts of expression plasmids for E2F1 to E2F6. Dose-dependent stimulation was observed for E2F1, E2F2, and E2F4 (Fig. 1B).
We then asked if any E2Fs could cooperate with p110 CUX1 (Fig. 1C). Hs 578T cells were cotransfected with suboptimal amounts of various combinations of effector plasmids. Under these experimental conditions, p110 CUX1 and E2F1 on their own mediated 11- and 5-fold activation, respectively (Fig. 1C). When the factors were expressed in combination, synergy was observed between CUX1 and either E2F1 or E2F2 but not E2F4 (Fig. 1C). Therefore, directly or indirectly, E2F1 and E2F2 cooperate with p110 CUX1 in the activation of the DNA Pol α gene promoter.
Cooperation between p110 CUX1 and E2F1 requires binding sites for both factors.
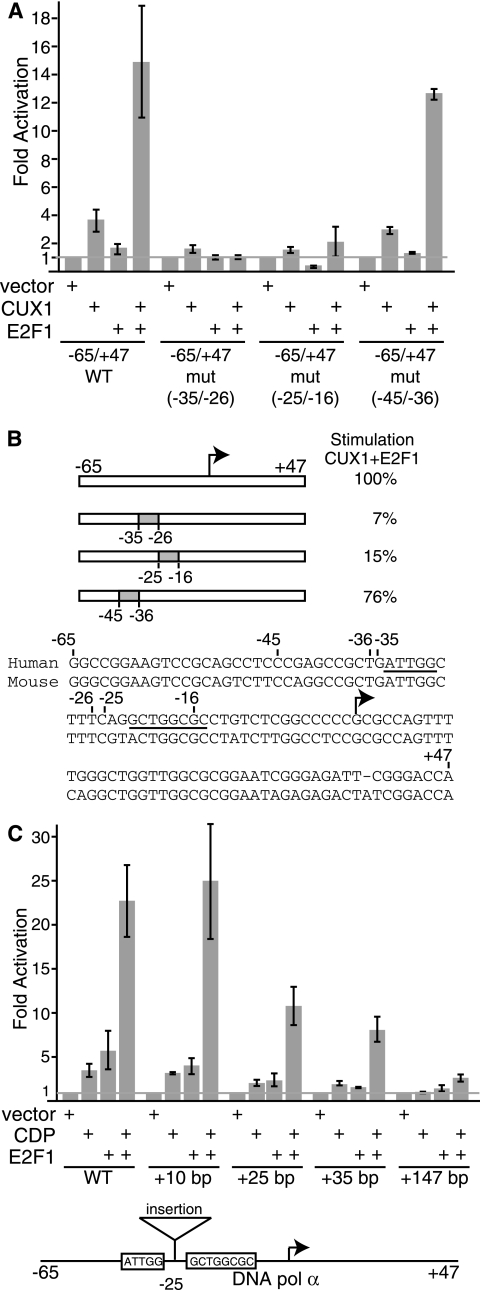
The above results indicated that some E2F factors were able to potentiate the transcriptional activation mediated by p110 CUX1. The effect of E2F could be indirect or could involve a direct interaction with the DNA Pol α promoter. To begin to investigate the mechanism by which E2F and p110 CUX1 cooperate, we repeated the reporter assay using linker-scanning mutants in which the CUX1 or the E2F binding site, or other sequences, was replaced. The same replacement sequence used in the −35/−26 and −25/−16 mutants was used to replace the −45/−36 promoter sequence, which was near, but did not contain, CUX1 and E2F binding sites. We noted changes in the basal activity of the different constructs, which are likely due to regulation by endogenous factors. In particular, replacement of the −45/−36 region resulted in a 2.3-fold reduction in promoter activity, which likely prevented the binding of an endogenous activator (Fig. 2). Interestingly, when CUX1 and E2F1 were coexpressed, transcriptional activation was reduced 14.2- and 6.6-fold relative to the wild-type promoter, upon replacement of the CUX1 or E2F binding site, respectively (Fig. 2A). In contrast, replacement of the −45/−36 region resulted in only a 1.3-fold reduction in stimulation by p110 CUX1 and E2F1 (Fig. 2A). We conclude that the cooperation between p110 CUX1 and E2F1 requires that the DNA Pol α gene promoter contain binding sites for both transcription factors. Interestingly, the cooperation was not affected following the insertion of 10 bp between the two binding sites (Fig. 2C). However, insertions of 25 and 35 bp weakened the cooperativity, while an insertion of 147 bp annihilated it (Fig. 2C).
FIG. 2.
p110 CUX1 cooperates with E2F1 in the activation of the DNA Pol α gene promoter in a binding site-dependent manner. (A and C) Hs 578T cells were transfected with wild-type or mutant DNA Pol α (−65/+47)/luciferase reporter constructs and the indicated vectors expressing CUX1 or E2F1. Cytoplasmic extracts were prepared and processed to measure luciferase activity. Results were expressed as relative light units and were normalized to β-galactosidase activity from an internal control. The means of three transfections are shown. Results are representative of three separate experiments. (B) The diagram and DNA sequence show the position of the linker scanner and insertion mutations within the DNA Pol α gene promoter. The sequences of CUX1 and E2F sites are underlined. Stimulation is expressed relative to the wild-type reporter construct. The transcription start site is indicated with an arrow.
p110 CUX1 interacts with E2F1 and E2F2 in vivo.
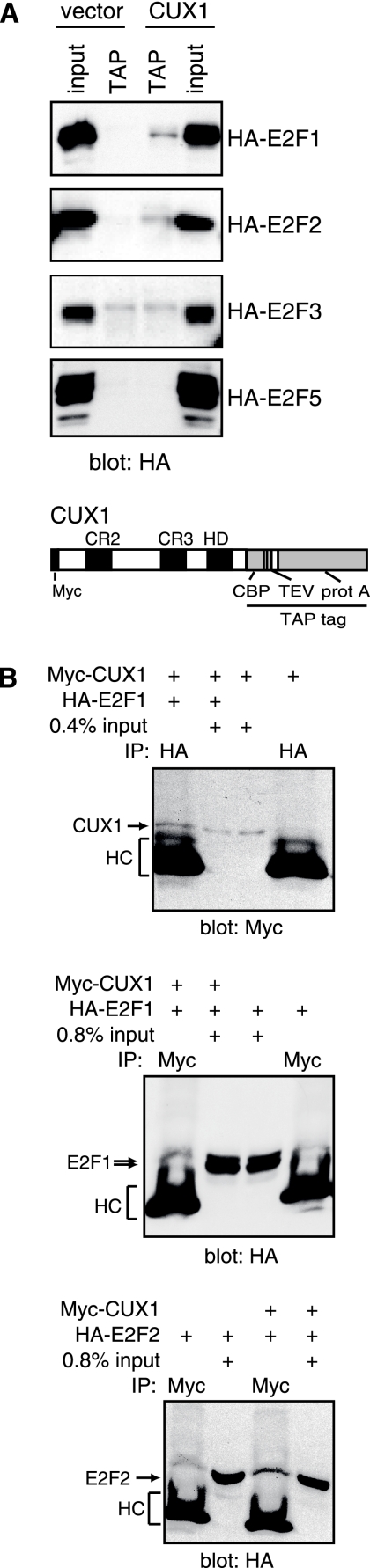
E2F and p110 CUX1 may each bind to the promoter independently. Alternatively, cooperation may involve physical interaction between the two factors. To investigate this possibility, TAP was performed with Hs 578T cells stably carrying a retroviral vector expressing CUX1 831-1336/tag2 and transiently transfected with vectors for hemagglutinin (HA)-tagged E2F proteins. As controls, transient transfections and TAP were performed in parallel using Hs 578T cells carrying an empty retroviral vector. Western blot analysis revealed weak protein-protein interactions between CUX1 and E2F1 and between CUX1 and E2F2 (Fig. 3). In contrast, no band was observed for E2F5 (Fig. 3). A band for E2F3a was also observed in the purified fraction; however, since this band was also present in the empty-vector control, we dismissed this result as being evidence for interaction. Thus, in affinity chromatography CUX1 specifically interacted with E2F1 and E2F2. The interactions between CUX1 and E2F1 or E2F2 were confirmed by coimmunoprecipitation assays in the presence of ethidium bromide (Fig. 3B). However, in pull-down assays with purified recombinant proteins, we did not observe a specific interaction between CUX1 and E2F1, whether in the presence or absence of DP-1 (data not shown). We conclude that an interaction between CUX1 and E2F1 or E2F2 may occur only in vivo and necessitates either the presence of additional proteins or the posttranslational modification of one partner or the other.
FIG. 3.
CUX1 interacts with E2F1 and E2F2 in vivo. (A) Hs 578T cells stably carrying an empty vector or a vector expressing CUX1 831-1336/tag2 were transfected with the indicated HA-E2F constructs. Cellular extracts were subjected to TAP, followed by Western blot analysis with an HA antibody. Input (0.2%) was loaded as a protein expression control. A schematic of CUX1 831-1336/tag2 is shown at the bottom. (B) Hs 578T cells were transfected with Myc-CUX1 878-1336 and HA-E2F1 or HA-E2F2. Whole-cell lysates were immunoprecipitated with the indicated antibodies in the presence of 100 μg/ml ethidium bromide, followed by Western blot analysis.
p110 recruits E2F1 and E2F2 to the DNA Pol α gene promoter.
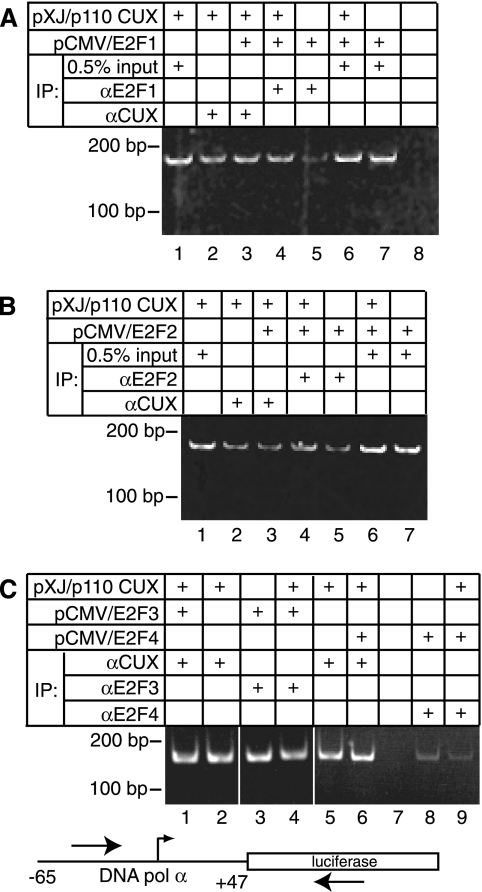
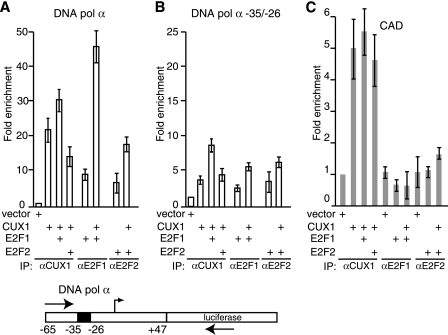
Given the proximity of the CUX1 and E2F binding sites and the observed protein-protein interactions, we next asked if coexpression of p110 and E2F would strengthen their interaction with the DNA Pol α gene promoter. ChIP assays were performed to measure the interaction in vivo between E2Fs or CUX1 and the DNA Pol α gene reporter plasmid. Immunoprecipitation using E2F1 or E2F2 antibodies indicated that each factor was able to bind to the reporter plasmid in vivo (Fig. 4A and B, lanes 5). Interestingly, coexpression with p110 CUX1 increased the interaction of either E2F1 or E2F2 with the reporter plasmid (Fig. 4A, compare lane 5 with 4; Fig. 4B, compare lane 4 with 5). Quantitative real-time PCR revealed 5.4-fold and 2.8-fold increases in promoter binding for E2F1 and E2F2, respectively, upon coexpression of CUX1 (Fig. 5A). However, the recruitment of E2F1 and E2F2 by CUX1 was significantly reduced by a linker-scanning mutation that overlapped the CUX1 binding site (Fig. 5A). Importantly, coexpression of p110 did not increase the steady-state level of E2F1 and E2F2 (data not shown). These results demonstrate that overexpression of p110 CUX1 can help recruit E2F1 and E2F2 to the DNA Pol α reporter in vivo. This effect appears to be specific to E2F1 and E2F2, since CUX1 did not enhance the recruitment of E2F3 or E2F4 to the same reporter (Fig. 4C, compare lane 3 with lane 4 and lane 5 with lane 6).
FIG. 4.
CUX1 enhances the recruitment of E2F1 and E2F2 to the DNA Pol α gene promoter. Hs 578T cells were cotransfected with the DNA Pol α reporter. The following day, ChIP assays were performed using the indicated antibodies. PCRs were performed in parallel using the immunoprecipitated chromatin and 0.5% total chromatin, as indicated.
FIG. 5.
CUX1 enhances the recruitment of E2F1 and E2F2 to specific promoters. Hs 578T cells were cotransfected with the wild-type DNA Pol α reporter (A), the same reporter with the −35/−26 linker scanning mutation (B), or the CAD reporter (C) and the indicated expression vectors. The following day, ChIP assays were performed using the indicated antibodies. Quantitative PCR was performed using primers recognizing the CAD or DNA Pol α gene promoters and luciferase cDNA, and results were normalized to a sample transfected with reporter and empty vector DNA. The values are means of three measurements, and error bars represent standard deviations.
To ensure that the recruitment of E2F1 and E2F2 by p110 is specific for promoters activated by E2Fs, we repeated the ChIP with a CAD reporter plasmid which had previously been shown not to be regulated by E2Fs (40). Immunoprecipitation with CUX1 antibodies gave fivefold enrichment in binding; however, no binding by E2F1 or E2F2 was observed, whether they were transfected alone or with p110 CUX1 (Fig. 5B). These results indicate that p110 CUX1 does not recruit E2F1 and E2F2 to every promoter to which p110 CUX1 binds.
Cell cycle genes are overrepresented among ChIP-chip targets common to p110 CUX1 and E2F1.
Genome-wide location analysis was recently performed to identify transcriptional targets of p110 CUX1 (26). The same 19,000-gene promoter microarray was employed in location analysis with an E2F1 antibody. Two hundred sixty-two E2F1 targets were identified with a P value below 0.005 (see Table S1 in the supplemental material). Comparison of p110 CUX1 and E2F1 targets indicated that 86 targets were common to E2F1 and p110 (see Table S2 in the supplemental material). To this list, we added 22 genes that were identified as E2F targets in previous studies (5, 8, 18, 46, 57, 81, 85, 87) (see Table S2 in the supplemental material). Of the common targets, 20% were genes involved in cell cycle progression, DNA replication, or DNA repair (see Table S3 in the supplemental material). Gene ontology analysis using EASE revealed that these functions were clearly overrepresented in the list of targets common p110 and E2F1 (Table 1). To validate these results, ChIP was performed with CUX1 and E2F1 antibodies, and the enrichment of 14 promoters was measured by was quantitative PCR. Enrichment was observed for all tested targets (Table 2). ChIP with antibodies specific for E2F2, E2F3, and E2F4 indicated that each of these factors is recruited to a subset of targets (Table 2). These results are in agreement with previous studies showing that a given promoter could be bound by several E2F proteins (75). Interestingly, comparison of the enrichment for each E2F factor suggests that the interaction of p110 CUX1 with a given promoter favors the specific recruitment of E2F1, and to a lesser extent E2F2, over that of other E2F factors (Table 2).
TABLE 1.
CUX1 and E2F1 targets include an overrepresentation of cell cycle genesa
| Category | P value | No. of genes |
|---|---|---|
| Cell cycle | 3.59E−9 | 22 |
| DNA replication | 8.96E−7 | 9 |
| DNA repair | 2.45E−4 | 7 |
| Nuclear organization | 6.60E−4 | 7 |
Targets common to CUX1 and E2F1 from ChIP-chip analysis were analyzed for distribution among functional categories using EASE. The P values, calculated with the Fisher exact test, represent the probability that the number of targets common to CUX1 and E2F1 would be randomly found. Shown are categories from biological level 5 with at least five genes and P values below 0.001.
TABLE 2.
CUX1 and E2F1 bind many common cell cycle gene targetsa
| Function | Gene symbol |
P value (location array)
|
Referenceb | Enrichment (ChIP-PCR)c
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| CUX1 | E2F1 | CUX1 | E2F1 | E2F2 | E2F3 | E2F4 | |||
| Cell cycle, S | CCNA2 | 6.0E−5 | 7.3E−3 | 5, 18, 56, 85 | 5.1 | 9.3 | 9.0 | 2.3 | 1.7 |
| CDC25A | 2.6E−4 | 9.0E−4 | 8, 56 | 8.4 | 64.4 | 49.3 | 10.3 | 2.8 | |
| Cell cycle, G2/M | MAD2L1 | 2.1E−7 | 2.0E−3 | 56 | 3.8 | 16.1 | 13.9 | 9.9 | 1.9 |
| DNA replication | CDC7 | 3.0E−4 | 8.8E−4 | 5.4 | 14.2 | 5.5 | 2.0 | 2.0 | |
| MCM3 | 1.1E−9 | 6.7E−6 | 56 | 13.0 | 9.9 | 1.7 | 4.2 | 5.7 | |
| MCM7 | 1.9E−3 | 2.0E−2 | 36, 53 | 6.2 | 13.4 | 12.5 | 4.6 | 1.4 | |
| ORC1L | 5.5E−14 | 3.9E−3 | 56 | 7.2 | 2.3 | 2.3 | 0.7 | 1.2 | |
| POLA | 3.2E−3 | 4.4E−1 | 45, 56 | 2.5 | 7.8 | 3.4 | 8.9 | 1.7 | |
| POLD3 | 4.7E−3 | 1.4E−3 | 3.8 | 3.8 | 1.0 | 1.3 | 0.8 | ||
| RPA3 | 6.2E−2 | 6.2E−2 | 56 | 3.0 | 29.6 | 14.0 | 11.4 | 9.6 | |
| Repair, checkpoint | MLH1 | 2.1E−4 | 1.4E−3 | 56 | 5.2 | 5.2 | 2.4 | 1.4 | 2.0 |
| PMS1 | 3.5E−3 | 3.2E−4 | 4.8 | 22.3 | 4.8 | 4.6 | 2.0 | ||
| RAD51 | 2.5E−3 | 2.2E−3 | 56, 80 | 5.5 | 4.4 | 3.3 | 1.6 | 2.6 | |
| TP53 | 4.4E−4 | 7.2E−2 | 56 | 3.0 | 1.9 | 1.8 | 0.8 | 0.7 | |
A subset of gene targets from ChIP-chip analysis and their P values are shown.
References for E2F1 targets previously found by chromatin immunoprecipitation.
ChIP-quantitative PCR validation was performed with thymidine-blocked Hs 578T cells. Results are expressed as enrichment relative to a no-antibody ChIP control, normalized for enrichment of G6PDH.
Inhibition of CUX1 expression reduces the recruitment of E2F1 and E2F2 to their endogenous targets.
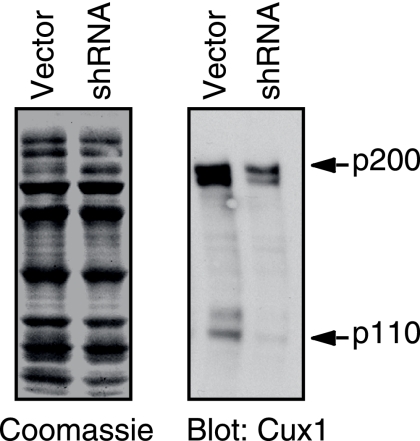
To verify whether CUX1 is required for the efficient recruitment of E2F1 and E2F2 to some of these endogenous targets, we sought to inhibit CUX1 expression by infecting cells with a lentiviral vector expressing CUX1 shRNA. T98G cells were preferred in this experiment because we achieve a better inhibition of CUX1 expression in these cells than in the Hs 578T cells. Immunoblotting analysis showed that expression of CUX1, and in particular its p110 isoform, was reduced after a week in culture (Fig. 6). At this time, chromatin was immunoprecipitated with E2F1 or E2F2 antibodies and quantitative PCR was performed to measure the recruitment of each factor to five target genes: the CCNA2, MCM7, ORC1L, POLA, and PMS1 genes. The results revealed a two- to threefold reduction in the recruitment of E2F1 and E2F2 (Table 3). We conclude that the optimal recruitment of these factors to some of their targets requires the presence of CUX1.
FIG. 6.
shRNA-mediated inhibition of CUX1. T98G cells were infected with pLVTHM lentivirus expressing CUX1 shRNA or with the empty vector. EGFP-expressing cells were subjected to fluorescence-activated cell sorting 3 days postinfection and cultured for 5 days. Nuclear extracts were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Coomassie blue staining and immunoblotting with CUX1 antibodies.
TABLE 3.
Inhibition of CUX1 expression reduces the recruitment of E2F1 to endogenous gene targetsa
| Gene | Enrichment
|
|||
|---|---|---|---|---|
| E2F1
|
E2F2
|
|||
| Vector | Cux1 shRNA | Vector | Cux1 shRNA | |
| CCNA2 | 9.2 | 2.9 | 8.7 | 2.8 |
| MCM7 | 16.5 | 9.0 | 20.2 | 8.5 |
| ORC1L | 8.1 | 3.4 | 9.2 | 3.6 |
| POLA | 11.5 | 5.6 | 4.3 | 2.5 |
| PMS1 | 7.7 | 2.5 | 3.8 | 2.1 |
T98G cells were infected with pLVTHM lentivirus expressing CUX1 shRNA or with the empty vector. EGFP-expressing cells were sorted by fluorescence-activated cell sorting 3 days postinfection, cultured for 5 days, and then processed for ChIP assay with E2F1 antibodies. Quantitative PCR results are expressed as enrichment relative to no antibody ChIP control and normalized for enrichment of G6PDH. Expression of CUX1 is shown in Fig. 6.
p110 CUX1 and E2F cooperate in the stimulation of cell cycle-regulated gene promoters.
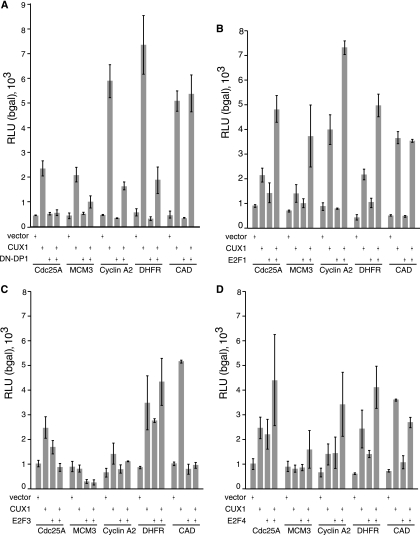
We recently showed, using transient reporter assays, that p110 CUX1 can stimulate expression from a number of cell cycle-regulated gene promoters that were identified in ChIP-chip analysis (25). We asked whether E2F activity was necessary for the stimulation of cell cycle-regulated genes by p110 CUX1. Cotransfection of DP1Δ103-126 with p110 caused a significant decrease in the stimulation of Cdc25A, MCM3, cyclin A2, and DHFR (Fig. 7A). However no change was observed with the CAD reporter, which has previously been shown not to be an E2F target (Fig. 7A) (40). This suggested that E2F cooperates with p110 CUX1 in the stimulation of many CUX1-regulated gene promoters. Reporter assays were repeated with p110 CUX1 and E2F1. Coexpression of E2F1 resulted in greater-than-additive effects on the stimulation of Cdc25A, MCM3, cyclin A2, and DHFR (Fig. 7B). No increase in CAD promoter activity was observed, confirming that the effect of E2F1 is specific to E2F-regulated gene promoters. No cooperation was observed between p110 CUX1 and E2F3 (Fig. 7C); however, we observed an additive effect when p110 CUX1 was coexpressed with E2F4 (Fig. 7D). Together, these results suggest that p110 CUX1 specifically cooperates with E2F1 to stimulate the expression of many cell cycle-regulated gene promoters.
FIG. 7.
CUX1 and E2F1 cooperate in the activation of their common target genes. Hs 578T cells were transfected with the reporter plasmids for Cdc25A, MCM3, cyclin A2, DHFR, and CAD and vectors expressing p110 CUX1 and DP1Δ103-126 (A), E2F1 (B), E2F3 (C), or E2F4 (D). Cytoplasmic extracts were prepared and processed to measure luciferase activity. Results are expressed as relative light units (RLU) normalized to β-galactosidase (bgal) activity from an internal control. The means from three transfections are shown, and the values are representative of three separate experiments.
DISCUSSION
CUX1 was originally characterized as a transcriptional repressor, but recent results indicated that its shorter isoforms could also function in transcriptional activation (22, 49, 61, 71, 72). In particular, p110 CUX1 was shown to stimulate expression from the DNA Pol α gene promoter, whether in reporter assays or following the infection of cells with a high-titer retroviral vector (71). Direct involvement of CUX1 in activation was demonstrated from the correlation between the stimulation of gene expression and the binding of p110 CUX1 to the DNA Pol α gene promoter, both in vitro and in vivo (71). A similar correlation has now been established using a number of promoters from other genes, including those encoding cyclin A2 (59), MCM3, Cdc25a, and Orc1l (25).
The mechanism by which short isoforms of CUX1 function in transcriptional activation was not immediately apparent, since in the Gal4 DNA binding domain fusion assay two active repression domains were identified downstream of the Cut homeodomain, but no region of CUX1 was found to function as an activation domain (44). One clue, however, was suggested from the finding that CUX1 is the DNA binding subunit of the HiNF-D protein complex that regulates transcription of cell cycle-regulated histone genes (1, 27, 35, 74-77). These results suggested that CUX1 could be part of larger nucleoprotein complexes that regulate transcription. This line of reasoning led us to investigate a replacement mutation, at position −25/−16 of the DNA Pol α gene promoter, that prevented transcriptional activation by p110 CUX1 without affecting its DNA binding site (71). In the present study, we present evidence that E2F is the factor that binds to this region and cooperates with p110 CUX1 to transactivate the DNA Pol α gene. Briefly, in reporter assays we observed an increase in transcriptional activation upon coexpression of p110 CUX1 and E2F1 or E2F2 (Fig. 1C). Replacement mutations of the CUX1 or E2F binding sites reduced stimulation by each factor individually (Fig. 6B and C in reference 71) (Fig. 2) and also reduced cooperative stimulation when both factors were coexpressed (Fig. 2A and B). Importantly, coexpression of a dominant negative DP1 significantly reduced the transcriptional activation mediated by p110 CUX1, implying that the activity of endogenous E2F factors was necessary for the stimulatory effect of p110 CUX1 (Fig. 1A). Results from TAP, coimmunoprecipitation, and ChIP suggested a potential mechanism for the cooperation between p110 CUX1 and E2F factors. Firstly, E2F1 and E2F2 were found to interact with a tagged version of p110 CUX1 (Fig. 3A), and these interactions were confirmed by coimmunoprecipitation (Fig. 3B). Secondly, ChIP assays had previously shown that both p110 and E2F1 could bind to the DNA Pol α gene promoter (18, 46, 71) (data not shown). In the present study, we demonstrated that coexpression with p110 CUX1 leads to an increase in the recruitment of E2F1 and E2F2 to this promoter (Fig. 4). In contrast, we did not observe cooperation in the recruitment to, nor the activation of, the CAD promoter, which in previous studies was shown not to be a target of E2F (Fig. 4C and 7). Altogether, the accumulated data suggest a scenario in which the DNA Pol α gene promoter contains suboptimal binding sites for p110 CUX1 and E2F. Consequently, each factor exhibits a low affinity for its binding site and, at physiological concentration, would not be expected to bind to the promoter on its own. However, the proximity of the two binding sites makes it possible for the two proteins to interact with each other or with another partner as they bind to their respective sites on DNA. Thus, when present together, E2F and p110 CUX1 would bind to the promoter with an affinity that is equal to the sum of their protein-protein and protein-DNA interactions. Since purified E2F1, DP1, and CUX1 did not bind to each other in pull-down assays (data not shown), we speculate that their interaction in vivo requires the presence of one or more additional proteins or the posttranslational modification of one partner or the other.
Interestingly, the cooperation between E2F and p110 CUX1 in the regulation of cell cycle genes was independently brought to light by using a genomic approach: the location array. Gene ontology analysis of the common targets between E2F1 and p110 CUX1 showed a striking overrepresentation of genes that play a role in cell cycle progression, DNA replication, and DNA repair. Indeed, a role for both E2F and p110 CUX1 in cell cycle regulation was previously established by using cell-based assays and transgenic models (4, 59; reviewed in reference 9). Importantly, using CUX1 shRNA we demonstrated that CUX1 expression was needed for the efficient recruitment of E2F1 and E2F2 to some of these targets (Fig. 6; Table 3). We note that genes involved in apoptosis were not overrepresented among the targets common to p110 CUX1 and E2F1. This result is also in accordance with the known cellular functions of E2F1 and p110 CUX1. While overexpression of E2F1 was shown to induce quiescent cells to enter S phase and then to undergo apoptosis, p110 CUX1 was unable to stimulate quiescent cells to reenter the cell cycle. In the presence of growth factors, however, cells overexpressing p110 CUX1 were able to enter S phase more rapidly and proliferated faster than control cells, with no evidence of apoptosis. Future experiments should verify whether the induction of apoptosis by E2F1 could be circumvented by overexpressing p110 CUX1.
Results from location array analyses confirmed that E2F1 and p110 CUX1 do not cooperate in the induction of apoptotic genes but do cooperate in the regulation of cell cycle genes. These findings confirm that the location array analysis is an unbiased method that can effectively reveal the biological functions of a transcription factor. Moreover, comparative analysis of data obtained with several transcription factors can point out the cellular activities in which two or more transcription factors cooperate. As transcriptional regulation is a combinatorial process involving the concerted action of several factors and cofactors, a better understanding of how transcriptional programs are established will require the completion of a repository of all overlapping sets of targets for various transcription factors. The location array will be essential in the accomplishment of this task.
We obtained evidence that p110 CUX1 cooperates with E2F1 and E2F2, but we did not observe cooperation with E2F3. The interaction with p110 CUX1, therefore, appears to be specific to some E2F factors, but the significance of this specificity is not immediately apparent. Recent results using RNAi-mediated knockdown in mouse embryo fibroblasts suggested that E2F3 is the primary E2F factor responsible for the expression of genes involved in cell proliferation (33). These findings, however, do not exclude the possibility that E2F1 and E2F2 play an essential role in promoting proliferation in distinct cell types or in specific situations. Moreover, the activator E2Fs are likely to have partially redundant functions, as revealed from the various knockout mouse models (15, 29, 30, 38). One particular situation where the stimulation of cell proliferation could be induced by any of the activator E2Fs is cancer. While deregulation of the cyclin D/pRb pathway was most often reported, amplification and/or overexpression of E2F1 and E2F3 has been observed in erythroleukemia cell lines, primary human acute lymphoid or myeloid leukemias, gastric and colorectal carcinomas, non-small-cell lung carcinomas, esophageal squamous cell carcinomas, and bladder and prostate cancers (11, 14, 16, 19, 68). On the other hand, from mRNA and immunohistochemical analyses, CUX1 was found to be overexpressed in breast tumors and in malignant plasma cells, and studies addressing the specific isoforms of CUX1 established that p110 and p75 were overexpressed in some uterine leiomyomas and breast tumor cell lines, respectively (10, 23, 47, 50). Moreover, in transgenic mice, both p110 and p75 CUX1 exhibited oncogenic potential (4; C. Cadieux et al., unpublished data). Therefore, we envision that the combined expression of both CUX1 and E2F factors in cancer cells may contribute to the aberrant stimulation of cell proliferation at the expense of differentiation.
Targeting of transcription factors to specific regulatory sites does not rely exclusively, or even primarily, on their interactions with high-affinity binding sites. Indeed, location array analysis has revealed that a sizeable fraction of targets do not include high-affinity binding sites. In these cases, targeting can be accomplished by the formation of a larger nucleoprotein complex that is stabilized by the accumulation of weak protein/DNA and protein/protein interactions. The results presented here support a model whereby p110 CUX1 recruits E2F1 to a subset of cell cycle-regulated promoters in order to stimulate gene expression.
Supplementary Material
Acknowledgments
We gratefully acknowledge the gift of the pLVTHM lentivirus vector from Didier Trono.
A.N. was the recipient of a scholarship from the Fonds de la Recherche en Santé du Québec. M.T. was the recipient of a studentship from the Terry Fox Foundation through the National Cancer Institute of Canada during the initial stages of this study and a studentship from the Fonds de Recherches en Santé Québec during the latter part of the study. This research was supported by grant MT-11590 from the Canadian Institute of Health Research of Canada to A.N.
Footnotes
Published ahead of print on 17 March 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aziz, F., A. J. Vanwijnen, P. S. Vaughan, S. J. Wu, A. R. Shakoori, J. B. Lian, K. J. Soprano, J. L. Stein, and G. S. Stein. 1998. The integrated activities of Irf-2 (Hinf-M), Cdp/Cut (Hinf-D) and H4tf-2 (Hinf-P) regulate transcription of a cell cycle controlled human histone H4 gene—mechanistic differences between distinct H4 genes. Mol. Biol. Rep. 251-12. [DOI] [PubMed] [Google Scholar]
- 2.Balciunaite, E., A. Spektor, N. H. Lents, H. Cam, H. Te Riele, A. Scime, M. A. Rudnicki, R. Young, and B. D. Dynlacht. 2005. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol. 258166-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, E. P., T. Hallstrom, H. K. Dressman, M. West, and J. R. Nevins. 2005. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc. Natl. Acad. Sci. USA 10215948-15953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadieux, C., S. Fournier, A. C. Peterson, C. Bédard, B. J. Bedell, and A. Nepveu. 2006. Transgenic mice expressing the p75 CCAAT-displacement protein/Cut homeobox isoform develop a myeloproliferative disease-like myeloid leukemia. Cancer Res. 669492-9501. [DOI] [PubMed] [Google Scholar]
- 5.Chang, W. Y., D. M. Bryce, S. J. D'Souza, and L. Dagnino. 2004. The DP-1 transcription factor is required for keratinocyte growth and epidermal stratification. J. Biol. Chem. 27951343-51353. [DOI] [PubMed] [Google Scholar]
- 6.Cloud, J. E., C. Rogers, T. L. Reza, U. Ziebold, J. R. Stone, M. H. Picard, A. M. Caron, R. T. Bronson, and J. A. Lees. 2002. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol. Cell. Biol. 222663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coqueret, O., G. Berube, and A. Nepveu. 1998. The mammalian Cut homeodomain protein functions as a cell-cycle dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 174680-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dasgupta, P., V. Betts, S. Rastogi, B. Joshi, M. Morris, B. Brennan, D. Ordonez-Ercan, and S. Chellappan. 2004. Direct binding of apoptosis signal-regulating kinase 1 to retinoblastoma protein: novel links between apoptotic signaling and cell cycle machinery. J. Biol. Chem. 27938762-38769. [DOI] [PubMed] [Google Scholar]
- 9.DeGregori, J., and D. G. Johnson. 2006. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 6739-748. [DOI] [PubMed] [Google Scholar]
- 10.De Vos, J., T. Thykjaer, K. Tarte, M. Ensslen, P. Raynaud, G. Requirand, F. Pellet, V. Pantesco, T. Reme, M. Jourdan, J. F. Rossi, T. Orntoft, and B. Klein. 2002. Comparison of gene expression profiling between malignant and normal plasma cells with oligonucleotide arrays. Oncogene 216848-6857. [DOI] [PubMed] [Google Scholar]
- 11.Ebihara, Y., M. Miyamoto, T. Shichinohe, Y. Kawarada, Y. Cho, A. Fukunaga, S. Murakami, H. Uehara, H. Kaneko, H. Hashimoto, Y. Murakami, T. Itoh, S. Okushiba, S. Kondo, and H. Katoh. 2004. Over-expression of E2F-1 in esophageal squamous cell carcinoma correlates with tumor progression. Dis. Esophagus 17150-154. [DOI] [PubMed] [Google Scholar]
- 12.el-Hodiri, H. M., and M. Perry. 1995. Interaction of the CCAAT displacement protein with shared regulatory elements required for transcription of paired histone genes. Mol. Cell. Biol. 153587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis, T., L. Gambardella, M. Horcher, S. Tschanz, J. Capol, P. Bertram, W. Jochum, Y. Barrandon, and M. Busslinger. 2001. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 152307-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feber, A., J. Clark, G. Goodwin, A. R. Dodson, P. H. Smith, A. Fletcher, S. Edwards, P. Flohr, A. Falconer, T. Roe, G. Kovacs, N. Dennis, C. Fisher, R. Wooster, R. Huddart, C. S. Foster, and C. S. Cooper. 2004. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene 231627-1630. [DOI] [PubMed] [Google Scholar]
- 15.Field, S. J., F.-Y. Tsai, F. Kuo, A. M. Zubiaga, J. W. G. Kaelin, D. M. Livingston, S. H. Orkin, and M. E. Greenberg. 1996. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85549-561. [DOI] [PubMed] [Google Scholar]
- 16.Foster, C. S., A. Falconer, A. R. Dodson, A. R. Norman, N. Dennis, A. Fletcher, C. Southgate, A. Dowe, D. Dearnaley, S. Jhavar, R. Eeles, A. Feber, and C. S. Cooper. 2004. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene 235871-5879. [DOI] [PubMed] [Google Scholar]
- 17.Giangrande, P. H., T. C. Hallstrom, C. Tunyaplin, K. Calame, and J. R. Nevins. 2003. Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol. Cell. Biol. 233707-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giangrande, P. H., W. Zhu, R. E. Rempel, N. Laakso, and J. R. Nevins. 2004. Combinatorial gene control involving E2F and E box family members. EMBO J. 231336-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorgoulis, V. G., P. Zacharatos, G. Mariatos, A. Kotsinas, M. Bouda, D. Kletsas, P. J. Asimacopoulos, N. Agnantis, C. Kittas, and A. G. Papavassiliou. 2002. Transcription factor E2F-1 acts as a growth-promoting factor and is associated with adverse prognosis in non-small cell lung carcinomas. J. Pathol. 198142-156. [DOI] [PubMed] [Google Scholar]
- 20.Goulet, B., A. Baruch, N. S. Moon, M. Poirier, L. L. Sansregret, A. Erickson, M. Bogyo, and A. Nepveu. 2004. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell 14207-219. [DOI] [PubMed] [Google Scholar]
- 21.Goulet, B., and A. Nepveu. 2004. Complete and limited proteolysis in cell cycle progression. Cell Cycle 3986-989. [PubMed] [Google Scholar]
- 22.Goulet, B., M. Truscott, and A. Nepveu. 2006. A novel proteolytically processed CDP/Cux isoform of 90 kDa is generated by cathepsin L. Biol. Chem. 3871285-1293. [DOI] [PubMed] [Google Scholar]
- 23.Goulet, B., P. Watson, M. Poirier, L. Leduy, G. Berube, S. Meterissian, P. Jolicoeur, and A. Nepveu. 2002. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Research 626625-6633. [PubMed] [Google Scholar]
- 24.Hallstrom, T. C., and J. R. Nevins. 2003. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc. Natl. Acad. Sci. USA 10010848-10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada, R., C. Vadnais, L. Sansregret, L. Leduy, G. Berube, F. Robert, and A. Nepveu. 2008. Genome-wide location analysis and expression studies reveal a role for p110 CUX1 in the activation of DNA replication genes. Nucleic Acids Res. 36189-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harada, R., C. Vadnais, L. L. Sansregret, L. Leduy, G. Bérubé, F. Robert, and A. Nepveu. 2007. Genome-wide location analysis and expression studies reveal a role for p110 CDP/Cux in the activation of DNA replication genes. Nucleic Acids Res. doi: 10.1093/nar/gkm970. [DOI] [PMC free article] [PubMed]
- 27.Holthuis, J., T. A. Owen, A. J. van Wijnen, K. L. Wright, A. Ramsey-Ewing, M. B. Kennedy, R. Carter, S. C. Cosenza, K. J. Soprano, J. B. Lian, J. L. Stein, and G. S. Stein. 1990. Tumor cells exhibit deregulation of the cell cycle histone gene promoter factor HiNF-D. Science 2471454-1457. [DOI] [PubMed] [Google Scholar]
- 28.Howcroft, T. K., S. L. Kirshner, and D. S. Singer. 1997. Measure of transient transfection efficiency using beta-galactosidase protein. Anal. Biochem. 24422-27. [DOI] [PubMed] [Google Scholar]
- 29.Humbert, P. O., C. Rogers, S. Ganiatsas, R. L. Landsberg, J. M. Trimarchi, S. Dandapani, C. Brugnara, S. Erdman, M. Schrenzel, R. T. Bronson, and J. A. Lees. 2000. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol. Cell 6281-291. [DOI] [PubMed] [Google Scholar]
- 30.Humbert, P. O., R. Verona, J. M. Trimarchi, C. Rogers, S. Dandapani, and J. A. Lees. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14690-703. [PMC free article] [PubMed] [Google Scholar]
- 31.Iwanaga, R., H. Komori, S. Ishida, N. Okamura, K. Nakayama, K. I. Nakayama, and K. Ohtani. 2006. Identification of novel E2F1 target genes regulated in cell cycle-dependent and independent manners. Oncogene 251786-1798. [DOI] [PubMed] [Google Scholar]
- 32.Izumi, M., M. Yokoi, N. S. Nishikawa, H. Miyazawa, A. Sugino, M. Yamagishi, M. Yamaguchi, A. Matsukage, F. Yatagai, and F. Hanaoka. 2000. Transcription of the catalytic 180-kDa subunit gene of mouse DNA polymerase alpha is controlled by E2F, an Ets-related transcription factor, and Sp1. Biochim. Biophys. Acta 1492341-352. [DOI] [PubMed] [Google Scholar]
- 33.Kong, L. J., J. T. Chang, A. H. Bild, and J. R. Nevins. 2007. Compensation and specificity of function within the E2F family. Oncogene 26321-327. [DOI] [PubMed] [Google Scholar]
- 34.Laganiere, J., G. Deblois, C. Lefebvre, A. R. Bataille, F. Robert, and V. Giguere. 2005. Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. USA 10211651-11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Last, T. J., M. Birnbaum, A. J. van Wijnen, G. S. Stein, and J. L. Stein. 1998. Repressor elements in the coding region of the human histone H4 gene interact with the transcription factor CDP/cut. Gene 221267-277. [DOI] [PubMed] [Google Scholar]
- 36.Ledford, A. W., J. G. Brantley, G. Kemeny, T. L. Foreman, S. E. Quaggin, P. Igarashi, S. M. Oberhaus, M. Rodova, J. P. Calvet, and G. B. Vanden Heuvel. 2002. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev. Biol. 245157-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leone, G., J. DeGregori, Z. Yan, L. Jakoi, S. Ishida, R. S. Williams, and J. R. Nevins. 1998. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 122120-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leone, G., R. Sears, E. Huang, R. Rempel, F. Nuckolls, C. H. Park, P. Giangrande, L. Wu, H. I. Saavedra, S. J. Field, M. A. Thompson, H. Yang, Y. Fujiwara, M. E. Greenberg, S. Orkin, C. Smith, and J. R. Nevins. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8105-113. [DOI] [PubMed] [Google Scholar]
- 39.Li, F. X., J. W. Zhu, C. J. Hogan, and J. DeGregori. 2003. Defective gene expression, S phase progression, and maturation during hematopoiesis in E2F1/E2F2 mutant mice. Mol. Cell. Biol. 233607-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, Y., J. E. Slansky, D. J. Myers, N. R. Drinkwater, W. G. Kaelin, and P. J. Farnham. 1994. Cloning, chromosomal location, and characterization of mouse E2F1. Mol. Cell. Biol. 141861-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lievens, P. M. J., J. J. Donady, C. Tufarelli, and E. J. Neufeld. 1995. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J. Biol. Chem. 27012745-12750. [DOI] [PubMed] [Google Scholar]
- 42.Logan, N., A. Graham, X. Zhao, R. Fisher, B. Maiti, G. Leone, and N. B. La Thangue. 2005. E2F-8: an E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene 245000-5004. [DOI] [PubMed] [Google Scholar]
- 43.Luong, M. X., C. M. van der Meijden, D. Xing, R. Hesselton, E. S. Monuki, S. N. Jones, J. B. Lian, J. L. Stein, G. S. Stein, E. J. Neufeld, and A. J. van Wijnen. 2002. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol. Cell. Biol. 221424-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mailly, F., G. Berube, R. Harada, P. L. Mao, S. Phillips, and A. Nepveu. 1996. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol. Cell. Biol. 165346-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maitra, U., J. Seo, M. M. Lozano, and J. P. Dudley. 2006. Differentiation-induced cleavage of Cutl1/CDP generates a novel dominant-negative isoform that regulates mammary gene expression. Mol. Cell. Biol. 267466-7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marlowe, J. L., E. S. Knudsen, S. Schwemberger, and A. Puga. 2004. The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J. Biol. Chem. 27929013-29022. [DOI] [PubMed] [Google Scholar]
- 47.Michl, P., A. R. Ramjaun, O. E. Pardo, P. H. Warne, M. Wagner, R. Poulsom, C. D'Arrigo, K. Ryder, A. Menke, T. Gress, and J. Downward. 2005. CUTL1 is a target of TGFβ signaling that enhances cancer cell motility and invasiveness. Cancer Cell 7521-532. [DOI] [PubMed] [Google Scholar]
- 48.Moon, N. S., G. Berube, and A. Nepveu. 2000. CCAAT displacement activity involves Cut repeats 1 and 2, not the Cut homeodomain. J. Biol. Chem. 27531325-31334. [DOI] [PubMed] [Google Scholar]
- 49.Moon, N. S., P. Premdas, M. Truscott, L. Leduy, G. Berube, and A. Nepveu. 2001. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol. Cell. Biol. 216332-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moon, N. S., W. Rong Zeng, P. Premdas, M. Santaguida, G. Berube, and A. Nepveu. 2002. Expression of N-terminally truncated isoforms of CDP/CUX is increased in human uterine leiomyomas. Int. J. Cancer 100429-432. [DOI] [PubMed] [Google Scholar]
- 51.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 2701-15. [DOI] [PubMed] [Google Scholar]
- 53.Nishio, H., and M. J. Walsh. 2004. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl. Acad. Sci. USA 10111257-11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohtani, K., R. Iwanaga, M. Nakamura, M. Ikeda, N. Yabuta, H. Tsuruga, and H. Nojima. 1999. Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F. Oncogene 182299-2309. [DOI] [PubMed] [Google Scholar]
- 55.Pattison, S., D. G. Skalnik, and A. Roman. 1997. Ccaat displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5′ end of the human papillomavirus type 6 long control region. J. Virol. 712013-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24218-229. [DOI] [PubMed] [Google Scholar]
- 57.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ross, J. F., A. Naar, H. Cam, R. Gregory, and B. D. Dynlacht. 2001. Active repression and E2F inhibition by pRB are biochemically distinguishable. Genes Dev. 15392-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sansregret, L., B. Goulet, R. Harada, B. Wilson, L. Leduy, J. Bertoglio, and A. Nepveu. 2006. The p110 isoform of the CDP/Cux transcription factor accelerates entry into S phase. Mol. Cell. Biol. 262441-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santaguida, M., Q. Ding, G. Berube, M. Truscott, P. Whyte, and A. Nepveu. 2001. Phosphorylation of the CCAAT displacement protein (CDP)/Cux transcription factor by cyclin A-Cdk1 modulates its DNA binding activity in G2. J. Biol. Chem. 27645780-45790. [DOI] [PubMed] [Google Scholar]
- 61.Santaguida, M., and A. Nepveu. 2005. Differential regulation of CDP/Cux p110 by cyclin A/Cdk2 and cyclin A/Cdk1. J. Biol. Chem. 28032712-32721. [DOI] [PubMed] [Google Scholar]
- 62.Schlisio, S., T. Halperin, M. Vidal, and J. R. Nevins. 2002. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 215775-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinclair, A. M., J. A. Lee, A. Goldstein, D. Xing, S. Liu, R. Ju, P. W. Tucker, E. J. Neufeld, and R. H. Scheuermann. 2001. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood 983658-3667. [DOI] [PubMed] [Google Scholar]
- 64.Skalnik, D. G., E. C. Strauss, and S. H. Orkin. 1991. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J. Biol. Chem. 26616736-16744. [PubMed] [Google Scholar]
- 65.Smith, E. J., G. Leone, J. DeGregori, L. Jakoi, and J. R. Nevins. 1996. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol. Cell. Biol. 166965-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stunkel, W., Z. Huang, S. H. Tan, M. J. O'Connor, and H. U. Bernard. 2000. Nuclear matrix attachment regions of human papillomavirus type 16 repress or activate the E6 promoter, depending on the physical state of the viral DNA. J. Virol. 742489-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Superti-Furga, G., A. Barberis, E. Schreiber, and M. Busslinger. 1989. The protein CDP, but not CP1, footprints on the CCAAT region of the γ-globulin gene in unfractionated B-cell extracts. Biochim. Biophys. Acta 1007237-242. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki, T., W. Yasui, H. Yokozaki, K. Naka, T. Ishikawa, and E. Tahara. 1999. Expression of the E2F family in human gastrointestinal carcinomas. Int. J. Cancer 81535-538. [DOI] [PubMed] [Google Scholar]
- 69.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14804-816. [PMC free article] [PubMed] [Google Scholar]
- 70.Truscott, M., J. B. Denault, B. Goulet, L. Leduy, G. S. Salvesen, and A. Nepveu. 2007. Carboxy-terminal proteolytic processing of CUX1 by a caspase enables transcriptional activation in proliferating cells. J. Biol. Chem. 28230216-30226. [DOI] [PubMed] [Google Scholar]
- 71.Truscott, M., L. Raynal, P. Premdas, B. Goulet, L. Leduy, G. Berube, and A. Nepveu. 2003. CDP/Cux stimulates transcription from the DNA polymerase alpha gene promoter. Mol. Cell. Biol. 233013-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Truscott, M., L. Raynal, Y. Wang, G. Berube, L. Leduy, and A. Nepveu. 2004. The N-terminal region of the CCAAT displacement protein (CDP)/Cux transcription factor functions as an autoinhibitory domain that modulates DNA binding. J. Biol. Chem. 27949787-49794. [DOI] [PubMed] [Google Scholar]
- 73.Tufarelli, C., Y. Fujiwara, D. C. Zappulla, and E. J. Neufeld. 1998. Hair defects and pup loss in mice with targeted deletion of the first cut repeat domain of the Cux/CDP homeoprotein gene. Dev. Biol. 20069-81. [DOI] [PubMed] [Google Scholar]
- 74.van den Ent, F. M., A. J. van Wijnen, J. B. Lian, J. L. Stein, and G. S. Stein. 1994. Cell cycle controlled histone H1, H3, and H4 genes share unusual arrangements of recognition motifs for HiNF-D supporting a coordinate promoter binding mechanism. J. Cell Physiol. 159515-530. [DOI] [PubMed] [Google Scholar]
- 75.van Wijnen, A. J., T. A. Owen, J. Holthuis, J. B. Lian, J. L. Stein, and G. S. Stein. 1991. Coordination of protein-DNA interactions in the promoters of human H4, H3, and H1 histone genes during the cell cycle, tumorigenesis, and development. J. Cell Physiol. 148174-189. [DOI] [PubMed] [Google Scholar]
- 76.van Wijnen, A. J., M. F. van Gurp, M. C. de Ridder, C. Tufarelli, T. J. Last, M. Birnbaum, P. S. Vaughan, A. Giordano, W. Krek, E. J. Neufeld, J. L. Stein, and G. S. Stein. 1996. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc. Natl. Acad. Sci. USA 9311516-11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Wijnen, A. J., K. L. Wright, J. B. Lian, J. L. Stein, and G. S. Stein. 1989. Human H4 histone gene transcription requires the proliferation-specific nuclear factor HiNF-D. Auxiliary roles for HiNF-C (Sp1-like) and HiNF-A (high mobility group-like). J. Biol. Chem. 26415034-15042. [PubMed] [Google Scholar]
- 78.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 216820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weinmann, A. S., and P. J. Farnham. 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 2637-47. [DOI] [PubMed] [Google Scholar]
- 80.Wells, J., C. R. Graveel, S. M. Bartley, S. J. Madore, and P. J. Farnham. 2002. The identification of E2F1-specific target genes. Proc. Natl. Acad. Sci. USA 993890-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wells, J., P. S. Yan, M. Cechvala, T. Huang, and P. J. Farnham. 2003. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sites during S phase. Oncogene 221445-1460. [DOI] [PubMed] [Google Scholar]
- 82.Wiznerowicz, M., and D. Trono. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 778957-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu, C. L., M. Classon, N. Dyson, and E. Harlow. 1996. Expression of dominant-negative mutant DP-1 blocks cell cycle progression in G1. Mol. Cell. Biol. 163698-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414457-462. [DOI] [PubMed] [Google Scholar]
- 85.Yoshida, K., and I. Inoue. 2004. Expression of MCM10 and TopBP1 is regulated by cell proliferation and UV irradiation via the E2F transcription factor. Oncogene 236250-6260. [DOI] [PubMed] [Google Scholar]
- 86.Zheng, T. S., S. Hunot, K. Kuida, and R. A. Flavell. 1999. Caspase knockouts: matters of life and death. Cell Death Differ. 61043-1053. [DOI] [PubMed] [Google Scholar]
- 87.Zhu, W., P. H. Giangrande, and J. R. Nevins. 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 234615-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.