SUMMARY
The LTR-retrotransposon Tf1 preserves the coding capacity of its host Schizosaccharomyces pombe by integrating upstream of open reading frames (ORFs). To determine which features of the target sites were recognized by the transposon, we introduced plasmids containing candidate insertion sites into S. pombe and mapped the positions of integration. We found that Tf1 was targeted specifically to the promoters of pol II transcribed genes. A detailed analysis of integration in plasmids that contained either ade6 or fbp1 revealed insertions occurred in the promoters at positions where transcription factors bound. Further experiments revealed that the activator Atf1p and its binding site were required for directing integration to the promoter of fbp1. An interaction between Tf1 integrase and Atf1p was observed indicating that integration at fbp1 was mediated by the activator bound to its promoter. Surprisingly we found Tf1 contained sequences that activated transcription and these substituted for elements of the ade6 promoter disrupted by integration.
Keywords: Integrase, pol II promoters, fbp1, ade6, Atf1p, retrotransposon, Tf1
The relationship between transposable elements and their hosts depends on integration that does not impair the fitness of the host. Many transposable elements protect the coding capacity of their host by directing integration to nonessential regions of the genome. The long terminal repeat (LTR) retrotransposons of Saccharomyces cerevisiae possess efficient mechanisms for targeting integration. Ty3 integrates precisely one or two nucleotides upstream of the transcription start sites of tRNAs (Chalker and Sandmeyer, 1990). The integration of Ty1 also shows strong preference for regions upstream of pol III transcribed genes (Bachman et al., 2004; Bachman et al., 2005; Devine and Boeke, 1996; Ji et al., 1993). Ty5 integrates specifically into heterochromatin (Zou et al., 1996). In comparison to the transposons of S. cerevisiae, the LTR-retrotransposon Tf1 of Schizosaccharomyces pombe exhibits a different preference for integration sites. Studies of Tf1 demonstrated that the region 500 bp upstream of ORFs is strongly preferred for integration (Behrens et al., 2000; Bowen et al., 2003; Singleton and Levin, 2002).
To identify what factors determine the positions of integration we developed a plasmid-based targeting assay in cells induced for Tf1 transposition. Five genes were tested and in each case, the insertions clustered in the promoter regions. Detailed studies of ade6 and fbp1 revealed that integration occurred in their promoter elements. In the case of fbp1, the transcription activator Atf1p and its binding site were required for insertion. In addition, we detected an interaction between Tf1 IN and Atf1p, indicating that integration was mediated by the tethering of IN to the promoter by Atf1p. Although disruption of promoters by Tf1 was expected to reduce transcription, we found Tf1 introduced its own promoter elements that compensated for the damage it caused to the ade6 promoter.
RESULTS
A plasmid based targeting assay for Tf1 transposition identified preferred sites of integration upstream of ORFs
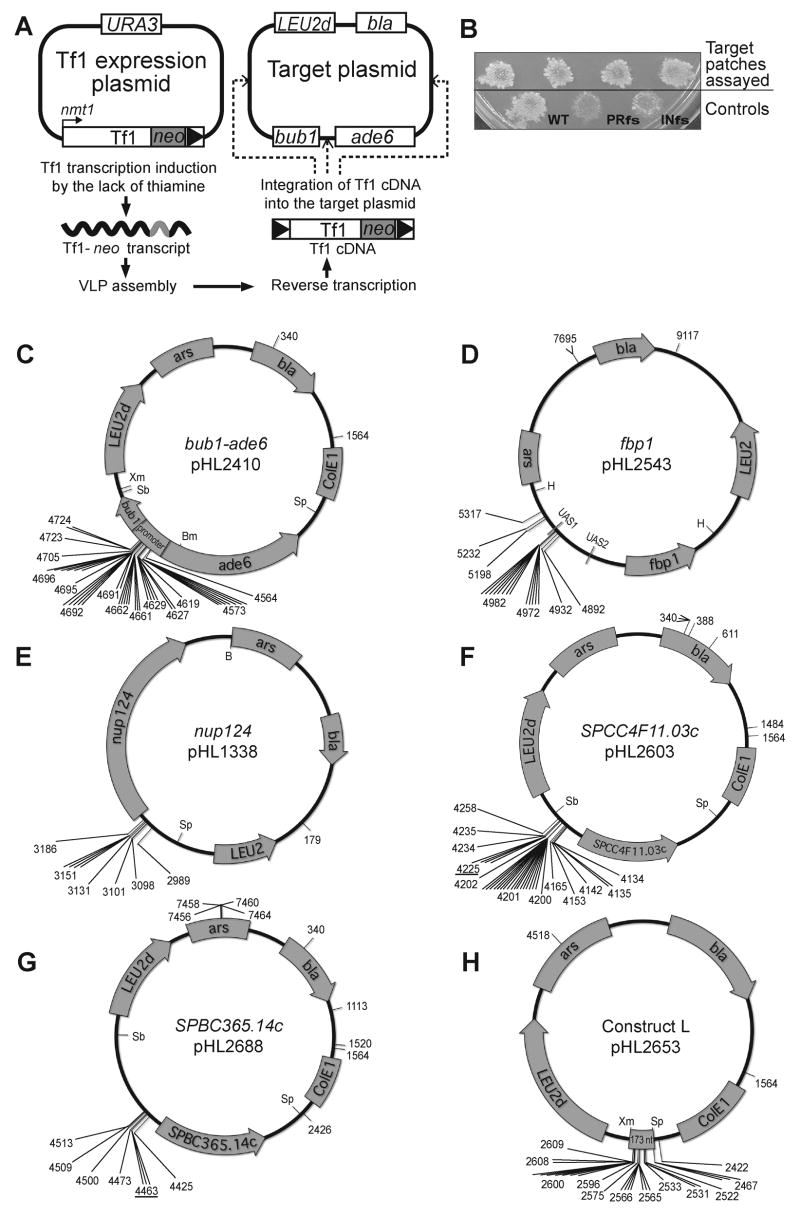
To identify what factors and sequence elements determine the insertion sites of Tf1 we developed a plasmid based targeting assay that measured the integration activity of specific DNAs (Fig. 1A). A strain of S. pombe was generated that contained a plasmid for the expression of Tf1 and a plasmid for target sequences. Tf1 was marked with neo so that integration would result in resistance to G418. After transcription of Tf1-neo was induced, the expression plasmid was removed from cells and insertion events were selected on medium containing G418 (Fig. 1B). Tf1-neo with mutations that block the expression of IN (INfs) or RT and IN (PRfs) produced significantly less integration (Lin and Levin, 1997). Target plasmids extracted from individual patches were introduced into bacteria by selecting for resistance to ampicillin. Due to the presence of neo in Tf1, target plasmids with insertions resulted in colonies that were also resistant to kanamycin. These plasmids were readily identified and the position of their insertions sequenced.
Figure 1.
A plasmid based assay for integration targets. A. Transposition was induced by activating transcription of Tf1 from an expression plasmid. The Tf1-neo mRNA was packaged into virus like particles, reverse transcription occurred, and IN inserted the Tf1-neo cDNA. A target plasmid present in the same cells had the potential to be the target of Tf1 integration. The target plasmid shown contained bub1-ade6 sequence as a potential region for integration. LEU2d was used to select for cells with high copy numbers of the target plasmid. B. Cells with inserts of Tf1-neo were identified on medium containing G418. Frameshift mutations that block expression of PR and RT (PRfs) or IN (INfs) inhibit transposition. The position of insertions in target plasmids are shown with the plasmid coordinates. The plasmids contained bub1-ade6 (C), fbp1 (D), nup124 (E), SPCC4F11.03c (F), SPBC365.14c (G), and the 173 bp insertion window of bub1-ade6 (H). Restriction sites for Xma I (Xm), Sbf I (Sb), Bmg BI (Bm), Spe I (Sp), Hind III (H), and Bam HI (B) are shown.
To test the integration preferences of Tf1 for a selected fragment of DNA, a target plasmid was created that contained the ORF of ade6, its upstream intergenic region, and a portion of the adjacent ORF, bub1 (Fig. 1C). This particular segment of the pombe genome was selected because its chromatin structure, as measured by digestion with micrococcal nuclease, is maintained when placed on a plasmid (Bernardi et al., 1991; Mizuno et al., 1997). The only other portion of the target plasmid that contained a promoter was LEU2d, a selection gene from S. cerevisiae that has a damaged promoter. The bub1-ade6 target plasmid was introduced into a strain of S. pombe and individual patches of cells were induced for the expression of Tf1-neo. No selection was placed on the function of ade6. Target plasmids were isolated by preparing DNA from each G418R patch and electroporating it into bacteria. Of 43 independent insertions of Tf1 isolated in the plasmid, 41 (95%) were in the intergenic region between bub1 and ade6 (Fig. 1C). These results demonstrated that the preference of Tf1 for integration upstream of ORFs was reconstituted within the context of a plasmid. Interestingly, all 41 insertions in the intergenic region occurred within a 160 bp window. The insertions clustered at specific nucleotides where no preferences for orientation were observed.
To test whether integration would cluster upstream of other ORFs we created four additional target plasmids that contained different genes. Each plasmid exhibited a clustering of insertions at different positions upstream of the ORFs (Fig. 1D–G). Two of the plasmids contained genes not previously known to be targets of Tf1 (Fig. 1D and E) and two contained sequences that were known to be targets in their chromosomal context (Fig. 1F and G). As chromosomal genes SPCC4F11.03c and SPBC365.14c had single base pair positions upstream of their ORFs that were repeatedly selected as positions for integration (Singleton and Levin, 2002). The same base pairs were among insertions isolated in the target plasmids (#4225 of Fig. 1F and #4463 of Fig. 1G).
The 160 bp window was the only sequence of bub1-ade6 required for efficient integration
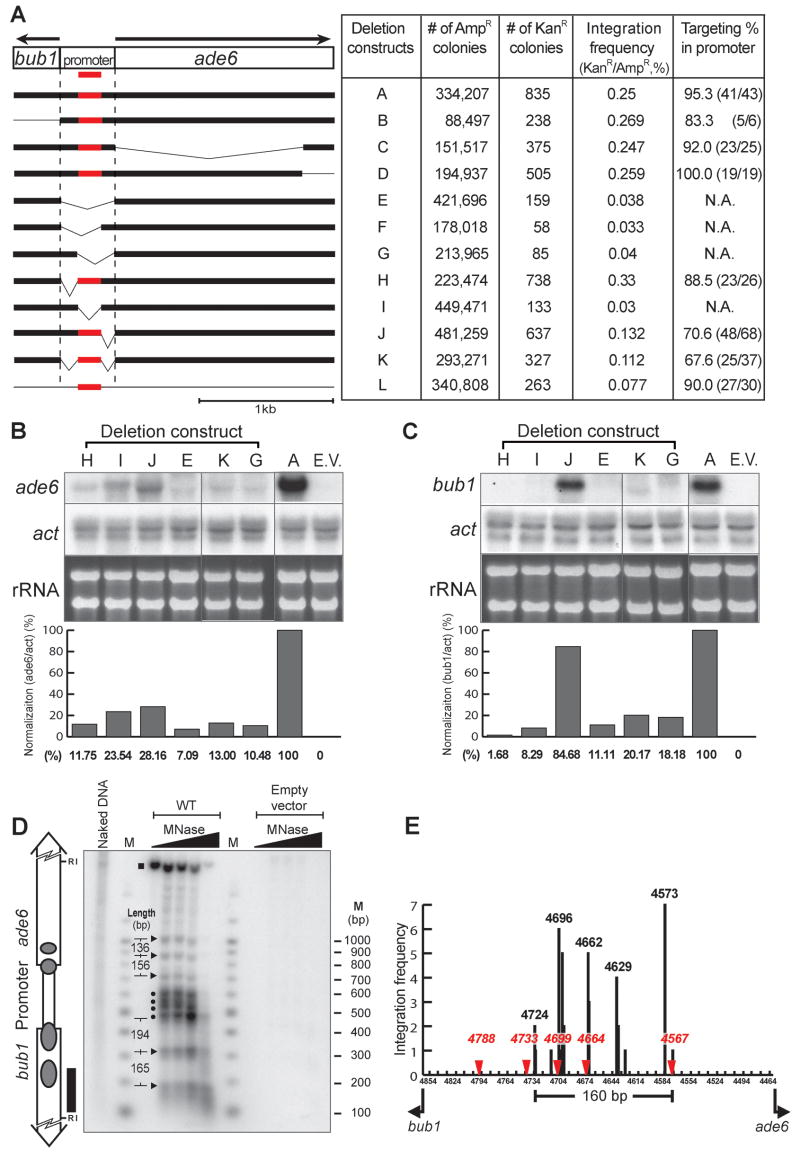
To identify what type of sequence elements were recognized by Tf1 we made a series of deletions throughout the bub1-ade6 fragment in the target plasmid. The integration frequency of the plasmid with the intact target, defined as the fraction of the target plasmids with insertions (ampR colonies that were also kanR ), was 0.25% (Fig. 2A, constr. A). Deletions that removed the ORFs of either ade6 or bub1 did not reduce the integration frequency (Fig. 2A, constr. B, C, and D). However, removal of the entire intergenic sequence resulted in a significant reduction in integration frequency (Fig. 2A, constr. E). Additional deletions within the intergenic region revealed that sequences upstream and downstream of the 160 bp window of insertion were not important for integration (Fig. 2A, constr. H and J). Whereas plasmids that lacked the 160 bp window exhibited substantial reductions in integration (Fig. 2A, constr. F, G, and I). These data revealed that the 160 bp target window was the only portion of bub1-ade6 that was important for efficient integration.
Figure 2.
A 160 bp window was the only sequence of bub1-ade6 required for efficient integration. A. Deletion constructs that lacked portions of the bub1-ade6 sequence were tested with the plasmid assay for defects in integration frequency and targeting. The number of target plasmids screened for insertions is indicated by the number of AmpR colonies. The proportion of target plasmids that have an insertion is the number of KanR colonies divided by the AmpR colonies (integration frequency). The percent of the insertions that mapped to the intergenic sequence is in the column labeled targeting % in promoter. The strains used for this data were YHL1101 containing pHL414-2 (constr. A) or pHL2541, and the deleted versions of pHL2410 (Table S2). B. Blots of RNA from cells containing the deletion constructs were probed for mRNA produced by ade6 and actin. The ribosomal RNA stained with ethidium bromide is also shown. The RNA levels were quantified by phosphoimaging and the amount of ade6 mRNA is shown normalized to actin mRNA. C. RNA from the same strains was probed for bub1 mRNA and, as in B, the amounts were normalized to actin mRNA. D. Proteins bound to bub1-ade6 sequence were mapped by digesting chromatin fractions with increasing amounts of MNase. The positions of MNase cleavages were detected on DNA blots probed by indirect end label (black rectangle). The samples containing plasmids with the intact bub1-ade6 target (constr. A) are labeled WT (YHL8964) and the samples containing plamids with no target sequence are labeled empty vector (YHL9085). Naked DNA indicates the purified DNA that was treated with increasing amounts of MNase and pooled. M indicates the molecular weight markers and the triangles are the nucleosome specific cleavages. The supersensitive cleavages are marked with black circles. The distances between the MNase cleavages are shown in bp. The grey ovals on the diagram of the bub1-ade6 sequence represent nucleosomes. E. The insertions in the plasmid with bub1-ade6 (Fig. 1C) are shown in a histogram. The positions of the MNase sensitive cleavages are indicated in red with triangles.
In addition to their effects on integration frequency, the deletions were also tested for their impact on targeting the promoter region. As reported above, 95% of the insertions in the plasmid with bub1-ade6 occurred within the 405 bp promoter region between the bub1 and ade6 ORFs (Fig. 2A, constr. A). This high percentage of insertions targeted to the intergenic sequence was not significantly reduced by the deletions that removed sequences on either side of the 160 bp window (Fig. 2A, constr. H and J). Thus, the 160 bp window of integration was the only region of bub1-ade6 required for efficient targeting.
To test whether the insertion window was sufficient to be an integration target, a plasmid was created that contained just the 160 bp window plus 13 bp of flanking sequence (Fig. 2A constr. L). Of 30 insertions isolated, 27 (90%) occurred specifically within the target window (Fig. 1H and Fig. 2A constr. L). These data show that just 173 bp of the promoter sequence functioned as a highly specific target for integration. However, the integration frequency of this plasmid was 3.5 fold lower than the intact bub1-ade6, suggesting that the 173 bp window lacked an element important for integration efficiency.
The efficiency and specificity of Tf1 integration was unrelated to the transcription levels of ade6 and bub1
The targeting of Tf1 to the region of the ade6 and bub1 promoters suggests that the transcription of these two genes may play a critical role in integration. This possibility was tested by comparing the amounts of mRNA produced by the deletion plasmids to their efficiency of targeted integration. Each of the deletions in the intergenic region significantly reduced the levels of ade6 mRNA (Fig. 2B). In addition, most of the deletions greatly reduced the levels of mRNA produced by the bub1 promoter (Fig. 2C). Interestingly, deletions H and K exhibited sharp reductions in the transcription of both ade6 and bub1, and yet both plasmids supported efficient targeting. In particular, deletion plasmid H produced 12% of the ade6 mRNA and 2% of the bub1 mRNA that the intact plasmid produced. Nevertheless, plasmid H supported wild-type levels of integration frequency and targeting (Fig. 2A, constr. H). This comparison indicates that the transcription activity of the promoters per se did not play an important role in targeted integration.
The preferred sites of integration in the intergenic region of bub1-ade6 correspond to specific chromatin structures
The result that transcription was not important for efficient integration into the promoters of bub1-ade6 suggests that static features such as DNA binding proteins may be recognized by Tf1. To determine whether nucleosomes or DNA-bound transcription factors were positioned at the integration sites of bub1-ade6 we digested chromatin with micrococcal nuclease, cut it with Eco RI, and blotted it. Hybridization with an end specific probe revealed a number of nuclease sensitive sites spaced 136 bp to 165 bp apart (Fig. 2D, triangles). These sites are the result of positioned nucleosomes along the open reading frames at positions previously reported (Bernardi et al., 1991; Mizuno et al., 1997). Interestingly, four supersensitive sites were clustered together within the intergenic sequence (Fig. 2D, circles). The intensity of these bands and their close spacing within the promoter region suggest they were created by transcription factors bound to regulatory sequences. The bands produced by nucleosomes and the supersensitive bands were generated by chromatin on the target plasmid since these signals were absent from the isogenic strain that had a plasmid lacking bub1-ade6 (Fig. 2D, empty vector).
Precise measurements with molecular weight standards were used to map the positions of the micrococcal cleavage sites (Supplemental Fig. S1) and these were plotted against the position of the insertion sites (Fig. 2E). Of the five major sites of insertion, three corresponded closely with positions of micrococcal supersensitivity (4664, 4699, and 4733) and one matched the site of a nucleosome within the intergenic sequence (4567). The distances between the four major insertion sites and the micrococcal cleavage sites were each less than 10 nt. This close association suggests the promoter binding proteins responsible for the micrococcal sites played a role in recruiting IN to the sites of integration. Since the transcription factors that bind the promoters of bub1-ade6 are unknown we chose to study the integration activity of another promoter.
The promoter of fbp1 was a target of Tf1 IN
The promoter of fbp1 is highly regulated and contains an upstream activating sequence (UAS1) that consists of an eight bp binding site for the transcription activator Atf1p (Neely and Hoffman, 2000). UAS2 is a short binding sequence for the transcription activator Rst2p as well as other factors (Higuchi et al., 2002; Neely and Hoffman, 2000).
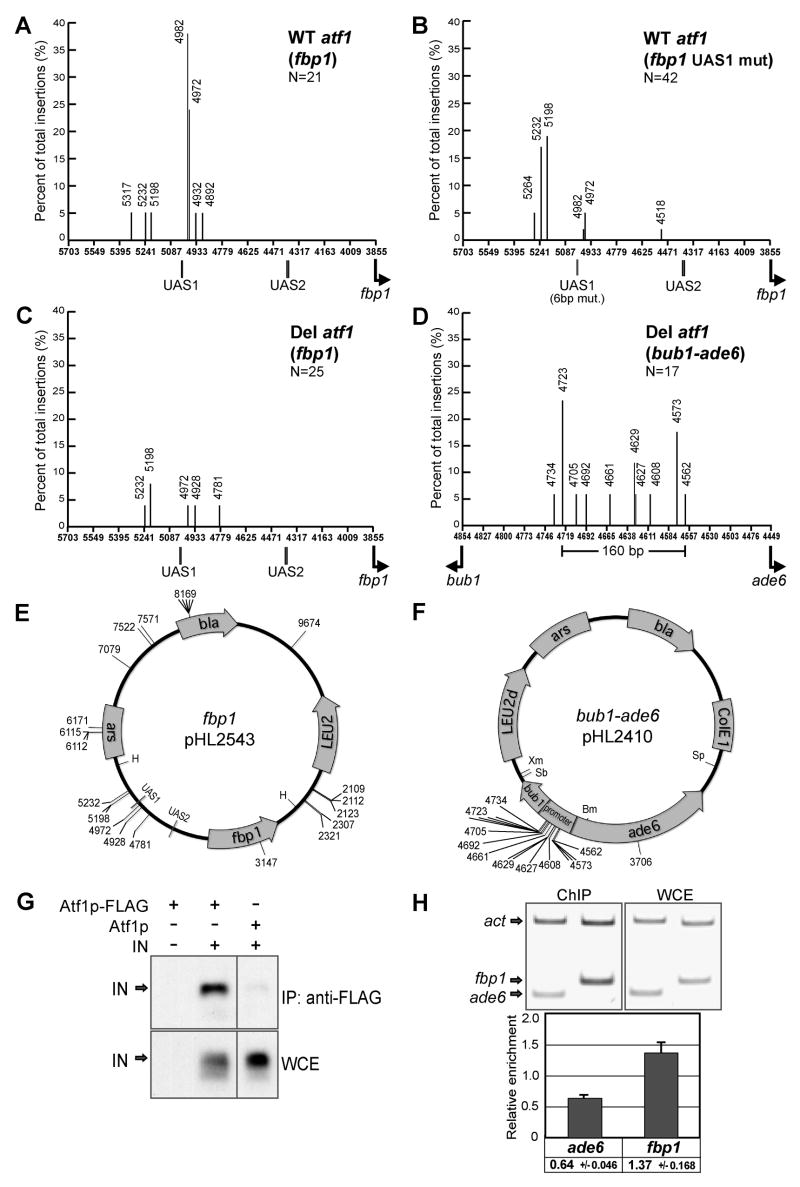
Eighty-six percent of the insertions in the plasmid with fbp1 occurred in sequence upstream of the open reading frame (Fig. 1D). The role of transcription factors in integration was supported by the concentration of integrations that occurred adjacent to the binding site of Atf1p, UAS1. Of the 18 inserts upstream of the ORF, 15 clustered within a 90 bp window. The most prominent of the insertions sites were coordinates 4972 and 4982, two positions that were just 40 bp and 30 bp downstream of UAS1, respectively. In a separate target plasmid that contained the entire fbp1 sequence in the opposite orientation, 19 of 23 insertions (83%) occurred in the promoter region and the same two positions were the dominant sites (data not shown).
To test whether UAS1 was sufficient for directing targeted integration, a target plasmid was constructed with just a 274 bp sequence that contained UAS1. Of 10 inserts isolated, four (40%) occurred at the dominant positions adjacent to UAS1 (data not shown). This 40% efficiency of directing integration to UAS1 is not significantly different from 62%, the proportion of the inserts in the full sized fbp1 plasmid that occurred at UAS1.
Targeted integration at UAS1 of fbp1 required a functional binding site for Atf1p
The integration activity of fbp1 allowed us to ask whether the clustering of inserts in the promoter region was the result of UAS1 function. A single nucleotide mutation in the eight bp binding sequence of UAS1 resulted in a significant reduction in fbp1 mRNA that correlated with a drop of Atf1p binding as measured with gel shift assays (Neely and Hoffman, 2000). We introduced this point mutation (CTACGTCA to CTACCTCA) into the fbp1 plasmid and used the target assay to ask whether the pattern of integration was changed. In the plasmid with the wild-type promoter, 13 ( 62%) of the 21 insertions isolated occurred at the two dominant positions adjacent to UAS1, 4972 and 4982 (plasmid coordinates) (Fig. 3A). In the plasmid with the single point mutation in UAS1, just nine of 36 insertions (25%) occurred at positions 4972 and 4982, a 2.5-fold reduction in integration at these two dominant positions. We also tested whether changing six of the eight nucleotides in UAS1 (CTACGTCA to GATGGAGA) would cause a more severe defect in integration at 4972 and 4982. Of 42 insertions isolated in this plasmid, just three occurred at positions 4972 and 4982 (7%). As a result, the six nucleotide mutation in UAS1 caused a nine-fold reduction in integration at these key positions (Fig. 3B).
Figure 3.
Targeted integration in the promoter of fbp1 requires UAS1 and Atf1p. A. The positions of Tf1 integration in the plasmid that contained fbp1 (Fig. 1D) were represented on a histogram of the promoter. The height of each line is the percent of all the insertions in the plasmid that occurred at the specified position. B. The target plasmid assay was used to map Tf1 integration in fbp1 that contained a six bp mutation in UAS1. C. The target plasmid assay was used to map integration in fbp1 in a strain that lacked atf1 (YHL9526/pHL2541+pHL2543). The positions of insertion were represented on a histogram of the promoter. D. Insertion sites were mapped in bub1-ade6 in the strain that lacked atf1 (YHL9526/pHL2541+pHL2410). E. The insertions in fbp1 in the strain lacking atf1 are shown on a map of the plasmid. F. The insertions in bub1-ade6 in the strain lacking atf1 are shown on a map of the plasmid. G. Immunoprecipitation was performed on extracts from cells expressing Atf1p-FLAG. The whole cell extract (WCE) and the protein precipitated with anti-FLAG antibody were immunoblotted and probed with an anti-IN antibody. H. The binding of Atf1p to the UAS1 of fbp1 and to the target window of ade6 was measured by ChIP. The amount of fbp1 and ade6 precipitated was measured relative to the amount of the actin gene and was normalized using the ratio of fbp1 or ade6 from the whole cell extract (WCE) divided by actin. The quantified results from three separate experiments were averaged and are shown with error bars indicating one standard deviation.
The defect in the ability of UAS1 to be recognized by Tf1 allowed the secondary targets 5198 and 5232 to become major sites of integration. This result demonstrated that a functional binding site for the transcription factor Aft1p plays an important role in the targeting of Tf1 integration to the two major insertion sites in the fbp1 promoter.
Atf1p interacts with IN and directs integration to the promoter of fbp1
The importance of UAS1 in the integration at the two key positions in the promoter of fbp1 suggested that Atf1p, the factor that binds UAS1, may mediate integration. To test this possibility we deleted the gene encoding Atf1p and assayed the strain for integration in the plasmid containing fbp1. Strikingly, the absence of Atf1p caused integration at UAS1 to be reduced 15.5-fold (Fig. 3C). The lack of Atf1p caused the insertions to be redistributed to other regions in the vector (Fig. 1D vs. Fig. 3E). These results indicated that Atf1p is required to position integration at the promoter of fbp1. To determine whether Atf1p played a similar role in directing integration to the promoter of ade6, we isolated insertions in the ade6 target plasmid using the strain lacking Atf1p. Surprisingly, we found 94% (16/17) of these inserts were targeted to the promoter region (Figs. 3D and F). This highly clustered pattern was very similar to the insertions isolated from the strain that expressed Atf1p (Figs. 1C vs. 3F), indicating that for ade6, Atf1p was not required for targeting integration.
One model for the specific role of Atf1p in positioning integration at the promoter of fbp1 is that Atf1p bound to UAS1 mediates integration there by forming a complex with IN. A second prediction is that integration at the promoter of ade6 was independent of Atf1p because a different transcription factor recruited IN to this promoter. We first tested this model by determining whether IN formed a complex with Atf1p. We used a strain of S. pombe that expressed a FLAG tagged Atf1p from its native promoter and introduced a plasmid that expressed a TAP tagged IN. Extracts from this strain were immunoprecipitated with anti-FLAG antibody and the precipitate was immunoblotted to test for the presence of TAP-IN (Fig. 3G). A substantial level of IN was precipitated by FLAG-Atf1p. No TAP-IN was precipitated when the same experiment was conducted with a strain that lacked the FLAG tag on Atf1p, indicating that the TAP-IN precipitated by FLAG-Atf1p was due to a specific interaction.
The model that Atf1p directly mediated integration at fbp1 and not at ade6 was tested by measuring binding of Atf1p at the promoters of fbp1 and ade6. We performed chromatin immunoprecipitation (ChIP) and measured binding of FLAG-Atf1p to the chromosomal copies of the fbp1 and ade6 promoters and normalized the signals to binding at the actin gene (Fig. 3H). The average of three independent experiments revealed a modest but consistent enrichment of Atf1p at the fbp1 promoter that was 2.1 greater than at the ade6 promoter. This is consistent with the model that Atf1p mediates integration at the promoter of fbp1 and another transcription factor mediates integration at ade6.
Targeted integration into the promoter of ade6 did not reduce transcription
Integration is a form of DNA damage that might be expected to disrupt promoter function. For example, deletion H in the promoter of ade6 revealed key promoter elements were upstream of all the major sites of integration (Fig. 2B). We tested whether the integration of Tf1-neo at a dominant site of insertion, nucleotide 4573, disrupted the function of the ade6 promoter. Target plasmids with Tf1-neo inserted in left (L) and right (R) orientations were introduced into S. pombe and the levels of ade6 mRNA from four independent transformants were measured on RNA blots (Fig. 4A and 4B, constr. A-L and A-R). The ade6 mRNA from each strain was quantified relative to the amounts of actin mRNA. The copy number of the plasmids in each transformant was determined on DNA blots (Fig. S2) and these values were used to adjust the levels of ade6 mRNA (Fig. 4B, only one transformant shown). For each plasmid, the adjusted levels of mRNA from the four transformants were averaged. Surprisingly, insertion of Tf1-neo actually stimulated ade6 transcription. Insertion in the L orientation increased ade6 mRNA by 2.1-fold while insertion in the R orientation increased ade6 expression by 3.6-fold. The absence of defects in the expression of ade6 together with the increases observed suggests the intriguing possibility that Tf1 carries promoter elements that are capable of repairing the promoters disrupted by the transposon.
Figure 4.
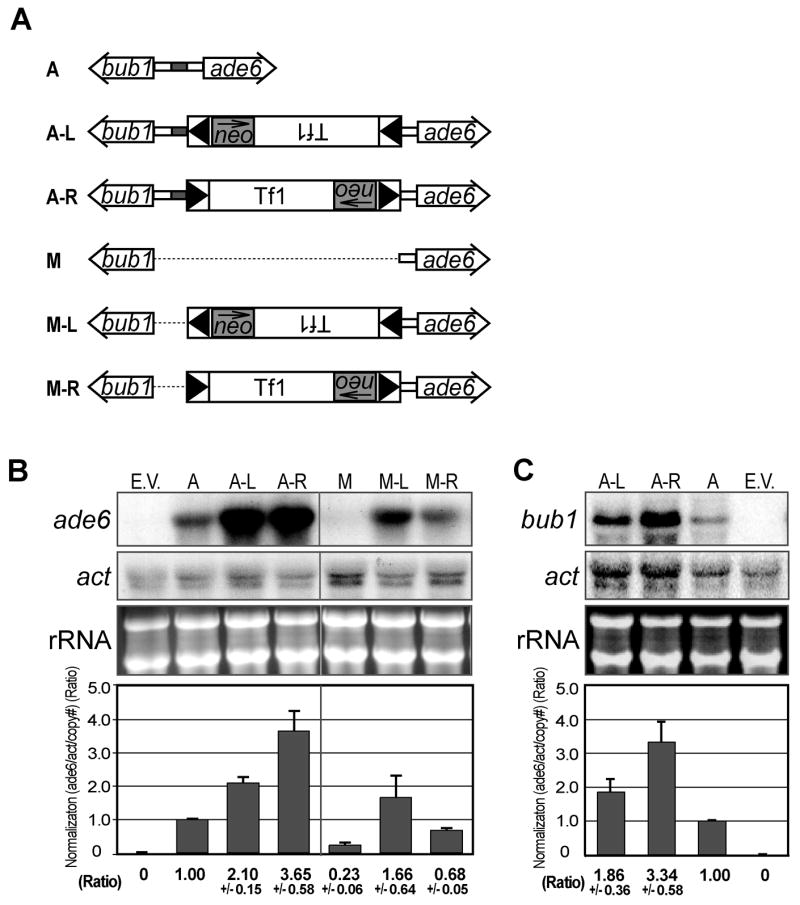
Integration of Tf1-neo activates the transcription of ade6 and bub1. A. Plasmids derived from construct A (A) include: construct A with an insertion of Tf1-neo in the left orientation at position 4573 (A-L), construct A with an insertion of Tf1-neo in the right orientation at position 4573 (A-R), construct A with a deletion from position 4573 up to the ATG of bub1 (M), construct M with an insertion of Tf1-neo in the left orientation at position 4573 (M-L), and construct M with an insertion of Tf1-neo in the right orientation at position 4573 (M-R). Neo is indicated by a grey rectangle and the direction of its transcription is indicated by an arrow. B. The levels of ade6, actin, and ribosomal RNA produced by strains (YHL9435-YHL9441) with these construct plasmids was measured on an RNA blot. The RNA levels from four independent yeast transformants were quantified by phosphoimaging and averaged. The amount of ade6 mRNA was normalized to actin mRNA and adjusted for differences in plasmid copy number (Fig. S2 and S3). The variation in mRNA levels between transformants is depicted with error bars indicating one standard deviation. C. The levels of bub1 and actin mRNA were measured and normalized as described for ade6.
Tf1 integration restored transcription to a damaged promoter
We tested directly whether Tf1 was capable of activating a damaged version of the ade6 promoter that lacked key promoter elements. Deletion construct M lacks the sequences between the target site 4573 and the ATG of bub1. This plasmid produced 4.3-fold less ade6 mRNA than the plasmid with the intact bub1-ade6 sequence (Fig. 4B). Deletion construct M-L was the same as construct M except it contained Tf1-neo inserted in the L orientation at nucleotide 4573. Construct M-R contained the Tf1-neo in the R orientation. As described above, the mRNA levels from RNA blots were normalized to actin expression and adjusted for variations in plasmid copy number (Fig. S3). Significantly, the ade6 expression from constructs M-L and M-R was 7.2 and 2.9 times greater than that from construct M, respectively (Fig. 4B). These data demonstrate that Tf1-neo introduced promoter elements that substituted for the transcription elements located upstream of nucleotide 4573. The levels of rescue provided by Tf1-neo in the M-L and M-R plasmids was sufficient to compensate for the damaged promoter since the mRNA produced was 1.7 and 0.7 times that generated by the intact bub1-ade6 promoter, respectively.
A separate experiment was designed to test whether Tf1-neo was capable of restoring expression to an ade6 promoter lacking the 114 bp between the ade6 ATG and the 160 bp target window (Fig. 2A, constr. J). This deletion included the sequence 42 and 43 bp upstream of the ATG, the sites where ade6 transcription initiates (N. Kon and W.P. Wahls, personal communication). The insertion of Tf1-neo in both orientations at nucleotide 4619 provided substantial increases in ade6 transcription compared to the levels produced by construct J (Fig. S4). These data indicate that Tf1 was capable of repairing the damage to the ade6 promoter that resulted from integration.
To test whether Tf1 integration would activate a different promoter we measured the transcription of bub1, the gene in divergent orientation from ade6 in the bub1-ade6 target plasmid. We found that the insertion at nt. 4573 resulted in a 1.7-fold or 3.3-fold increase in bub1 mRNA depending on the orientation of Tf1 (Fig. 4C, constr. A compared to constrs. A-L and AR). We also examined whether insertion of Tf1 increased the levels of transcription of fbp1, nup124, and SPCC4F11.03c. In these cases, integration of Tf1 did not cause any significant increase in transcription (data not shown).
DISCUSSION
The integration patterns of the five genes tested were similar in a variety of features. In each case the majority of insertions clustered within a small window in the region of the promoters (Fig. 1). Each target window contained dominant sites of insertion. This clustering of integration in the promoter regions and the dominant positions of insertion suggested integration was mediated by transcription factors bound to their promoter elements. Our finding that the major insertion sites of fbp1 depended on a functional UAS1 provided strong evidence that factors bound to this promoter element were responsible for targeting integration. This integration likely results from a tethering of IN to UAS1 by Aft1p. Support for this model is that deletion of atf1 resulted in a disruption in targeted insertion and that IN formed a complex with Atf1p.
For the promoters of bub1-ade6 a similar role for transcription factors was observed. Deletions in bub1-ade6 mapped promoter elements to the same 160 bp window that functioned as the target site for integration. The strong correlation between sites of MNase sensitivity and the dominant positions of integration supported the role of DNA bound transcription factors in the targeting of integration. Taken together, our data is strong evidence that it is pol II promoters that are recognized by Tf1. This is significant since previous work only demonstrated that integration occurred upstream of ORFs (Behrens et al., 2000; Singleton and Levin, 2002).
Although we do not known which other promoters are targeted for integration by Atf1p, it is reasonable to conclude that the other genes regulated by Atf1p are also targets of integration. Atf1p is responsible for regulating transcription of 224 stress response genes (Chen et al., 2003) and 40% of 33 insertion targets (Singleton and Levin, 2002) analyzed by microarray occurred at promoters that are regulated by stress (H. Levin, unpublished).
Our data clearly show that Atf1p is not required for directing integration to the target window of bub1-ade6. We therefore proposed that a different transcription factor served this function. This is supported by our ChIP experiment that showed Atf1p preferentially bound to the insertion sites of fbp1 but not to the target sites of bub1-ade6. The possibility that different transcription factors are capable of mediating integration is an intriguing departure from the tethering mechanism of Ty5 which relies on a specific interaction between IN and the Sir4p factor of heterochromatin (Dai et al., 2007; Zhu et al., 2003).
The mechanisms that direct the integration of LTR-retrotransposons likely evolved to protect the integrity of host genes (Sandmeyer, 2003). Since pol III promoters are internal, integration of Ty3 and Ty1 upstream of pol III genes prevents damage to regulatory elements (Bolton and Boeke, 2003). Why then does Tf1 integrate into the promoter elements of genes? This mechanism does avoid the disruption of ORFs. However, the insertion of 6 kb of transposon DNA damages promoters by separating UAS sequences from downstream elements such as the TBP binding sites and the start sites of transcription. Nevertheless, our analyses of ade6 and bub1 mRNA indicated that Tf1 itself introduced activation sequences that functioned in place of the UASs displaced by the insertions. Recent studies of a closely related transposon of S. pombe, Tf2, showed clearly that solo LTRs have potent promoter activity that can transcribe neighboring genes (Sehgal et al., 2007). If the Tf1 LTRs transcribe downstream genes, their ability to induce targeted genes would be limited by the presence of intervening out of frame ATGs that would trigger nonsense mediated mRNA decay (NMD) (Chang et al., 2007; Isken and Maquat, 2007). Out of frame ATGs exist upstream of fbp1, nup124, and SPCC4F11.03c that could explain why Tf1 did not induce these genes. Whereas, ade6, the gene with the strongest induction, had no intervening ATGs.
The process of gene activation at insertion sites is typical of retroviruses and these events have been studied extensively for their ability to induce malignancies (Rosenberg and Jolicoeur, 1997). As LTR-retrotransposons are close relatives of retroviruses it is not surprising that they too induce genes at insertion sites. What is unique about Tf1 is its specific targeting of pol II promoters which in turn provides each insertion the opportunity to alter the expression of the target gene.
The targeting of pol II promoters by Tf1 provides an important model for the integration preferences of retroviruses. The insertion of MuLV and HIV-1 into pol II transcription units is likely the result of host proteins that tether IN to the target sites. The transcriptional coactivator LEDGF is one factor that binds HIV-1 IN (Cherepanov et al., 2004; Cherepanov et al., 2003; Llano et al., 2004). Recent results indicate that LEDGF contributes to the integration preference of HIV-1 IN for pol II transcription units (Ciuffi et al., 2005; Shun et al., 2007). Our demonstration that binding of Atf1p to UAS1 was required to target integration and the role of LEDGF in HIV-1integration suggest that transcription factors may play a direct and general role in recruiting preintegration complexes to sites of insertion.
EXPERIMENTAL PROCEDURES
Detailed methods are presented in the supplement. The following is a summary of key techniques.
Target plasmid assay
S.pombe containing a target plasmid and the Tf1-neo expression plasmid were grown on medium lacking vitamin B1 to induce Tf1 transcription. Patches of cells were then grown in medium containing 5-fluoroorotic acid (FOA) to select against the Tf1 plasmid. Cells with transposition events were selected on medium containing G418. DNA purified from individual patches of these cells were introduced into bacteria and colonies resistant to kanamycin contained target plasmids with insertions.
Micrococcal nuclease mapping
Strains of S. pombe with and without bub1-ade6 in the target plasmid were grown without expression of Tf1. Chromatin extracts were treated with micrococcal nuclease, digested with Eco RI, and blotted. Hybridization with an end specific probe revealed a number of nuclease sensitive sites.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH from the National Institute of Child Health and Human Development. H. E. was supported by a fellowship from the Japan Society for the Promotion of Science. We thank Dr. S. Jia for providing the FLAG tagged allele of atf1. We are grateful to Dr. W.P. Wahls for communicating unpublished results on the transcription start site of ade6.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bachman N, Eby Y, Boeke JD. Local definition of Tyl target preference by long terminal repeats and clustered tRNA genes. Genome Research. 2004;14:1232–1247. doi: 10.1101/gr.2052904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman N, Gelbart ME, Tsukiyama T, Boeke JD. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 2005;19:955. doi: 10.1101/gad.1299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens R, Hayles J, Nurse P. Fission yeast retrotransposon Tf1 integration is targeted to 5 ′ ends of open reading frames. Nucleic Acids Research. 2000;28:4709–4716. doi: 10.1093/nar/28.23.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi F, Koller T, Thoma F. The Ade6-Gene of the Fission Yeast Schizosaccharomyces-Pombe Has the Same Chromatin Structure in the Chromosome and in Plasmids. Yeast. 1991;7:547–558. doi: 10.1002/yea.320070603. [DOI] [PubMed] [Google Scholar]
- Bolton EC, Boeke JD. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: A genomic point of view. Genome Research. 2003;13:254–263. doi: 10.1101/gr.612203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, Jordan I, Epstein J, Wood V, Levin HL. Retrotransposons and their Recognition of pol II Promoters: A Comprehensive Survey of the Transposable Elements derived from the Complete Genome sequence of Schizosaccharomyces pombe. Genome Research. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Sandmeyer SB. Transfer RNA genes are genomic targets for de novo transposition of Ty3. Genetics. 1990;126:837–850. doi: 10.1093/genetics/126.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. 2007 doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Chen DR, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bahler J. Global transcriptional responses of fission yeast to environmental stress. Molecular Biology of the Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. Journal of Biological Chemistry. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. Journal of Biological Chemistry. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. 1227. doi: 10.1038/nm1329. Epub 2005 Nov. [DOI] [PubMed] [Google Scholar]
- Dai J, Xie W, Brady TL, Gao J, Voytas DF. Phosphorylation Regulates Integration of the Yeast Ty5 Retrotransposon into Heterochromatin. Molecular Cell. 2007;27:289–299. doi: 10.1016/j.molcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- Higuchi T, Watanabe Y, Yamamoto M. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Molecular and Cellular Biology. 2002;22:1–11. doi: 10.1128/MCB.22.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Ji H, Moore DP, Blomberg MA, Braiterman LT, Voytas DF, Natsoulis G, Boeke JD. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- Lin JH, Levin HL. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes Dev. 1997;11:270–285. doi: 10.1101/gad.11.2.270. [DOI] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. Journal of Virology. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes & Development. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- Neely LA, Hoffman CS. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Molecular and Cellular Biology. 2000;20:6426–6434. doi: 10.1128/mcb.20.17.6426-6434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N, Jolicoeur P. In: Retroviral Pathogenesis. Retroviruses JM, Coffin SH, Hughes, Varmus HE, editors. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 475–587. [PubMed] [Google Scholar]
- Sandmeyer S. Integration by design. Proc Natl Acad Sci Usa. 2003;100:5586–5588. doi: 10.1073/pnas.1031802100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Lee CY, Espenshade PJ. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 2007;3:e131. doi: 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes & Development. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton TL, Levin HL. A Long Terminal Repeat Retrotransposon of Fission Yeast Has Strong Preferences for Specific Sites of Insertion. Eukaryotic Cell. 2002;1:44–55. doi: 10.1128/EC.01.1.44-55.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Dai J, Fuerst PG, Voytas DF. From the Cover: Controlling integration specificity of a yeast retrotransposon. Proc Natl Acad Sci U S A. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Ke N, Kim JM, Voytas DF. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.