Abstract
In mammals, small multigene families generate spliceosomal U snRNAs that are nearly as abundant as rRNA. Using the tandemly repeated human U2 genes as a model, we show by footprinting with DNase I and permanganate that nearly all sequences between the enhancer-like distal sequence element and the initiation site are protected during interphase whereas the upstream half of the U2 snRNA coding region is exposed. We also show by chromatin immunoprecipitation that the SNAPc complex, which binds the TATA-like proximal sequence element, is removed at metaphase but remains bound under conditions that induce locus-specific metaphase fragility of the U2 genes, such as loss of CSB, BRCA1, or BRCA2 function, treatment with actinomycin D, or overexpression of the tetrameric p53 C terminus. We propose that the U2 snRNA promoter establishes a persistently open state to facilitate rapid reinitiation and perhaps also to bypass TFIIH-dependent promoter melting; this open state would then be disassembled to allow metaphase chromatin condensation.
U2 small nuclear RNA (snRNA) is the catalytic RNA component of the U2 small nuclear ribonucleoprotein particle (snRNP). Along with the U1, U4/U6, and U5 snRNPs, the U2 snRNP assembles onto eukaryotic mRNA precursors to form a spliceosome, the large multisubunit molecular machine responsible for mRNA splicing (81). The genes encoding these U snRNAs are single copy in budding yeast, where introns are rare and U snRNPs are scarce, but in mammals, where almost all mRNAs have multiple introns, the major spliceosomal U snRNAs are encoded by multigene families and the U snRNPs are nearly as abundant as rRNA. U snRNA genes have been characterized for many species, and the major transcriptional signals and trans-acting factors are well defined, especially in mammals (35, 41, 42). U1 to U5 are transcribed by RNA polymerase II (Pol II). The 20-bp proximal sequence element (PSE) centered at position −50 upstream from the transcription initiation site serves many of the functions of a TATA element and binds the U snRNA-specific transcription factor SNAPc (snRNA-activating protein complex; also known as PSE-binding transcription factor [PTF] and PSE-binding protein). The bipartite distal sequence element (DSE) is centered at position −235 upstream of the initiation site, serves as an enhancer, and typically binds the Sp1 and Oct-1 (or Staf/SBF) factors (90). The distance between the DSE and PSE is almost exactly one nucleosome in length and is highly conserved in metazoa; indeed, the evidence for a positioned nucleosome between the DSE and PSE is strong for U2, U6, and 7SK, suggesting that this nucleosome juxtaposes factors bound to the DSE and PSE (7, 112).
In mammals, transcription apparently continues for approximately 800 bp past the U snRNA coding region (19, 73). The 3′ end of the U snRNA precursor is generated by an endonucleolytic cleavage—presumably by the Int11 component of the Integrator complex (5, 66)—just upstream from the 3′-end formation signal (also known as the 3′ box) located 10 to 20 bp beyond the mature U snRNA coding region. Intriguingly, recognition of the 3′ end formation signal requires a U snRNA promoter (20, 36); mRNA promoters cannot substitute, suggesting that U snRNA promoters either prevent loading of mRNA-specific factors, such as the CPSF component of the polyadenylation apparatus, or facilitate loading of snRNA-specific factors, such as Integrator, both of which bind to the Pol II CTD (15, 24). Curiously, although U1 to U5 are transcribed by Pol II, the presence of a TATA box just downstream of the U6 PSE switches the promoter specificity from Pol II to Pol III; accordingly, U6 genes have a Pol III oligo(dT) termination signal instead of a Pol II 3′-end formation signal (35, 41, 42).
In the primate lineage, where the abundance of U1 and U2 snRNA approaches that of the rRNAs, the genes encoding U1 and U2 are tandemly repeated. The human RNU1 locus spans 1.35 Mbp and contains about 30 tandemly repeated U1 snRNA genes; the individual repeat units are >45 kb in size and contain a single U1 snRNA gene interspersed with numerous tRNA genes (6, 101). The RNU2 locus spans 30 to 150 kb and contains 5 to 25 tandemly repeated U2 snRNA genes; the individual repeat units are 6.1 kb and contain a single U2 snRNA gene but encode no other stable RNA species (70, 86, 100). The 45-kb U1 repeats are slightly heterogeneous, but the 6.1-kb U2 repeat units are homogeneous except for a hypervariable CT dinucleotide repeat region (Fig. 1A) which may play a role in the stability (4) and/or concerted evolution of the array (54, 57). Although U2 arrays differ in gene copy number from individual to individual, the arrays are stably inherited (55) and subject to dosage compensation (3, 68).
FIG. 1.
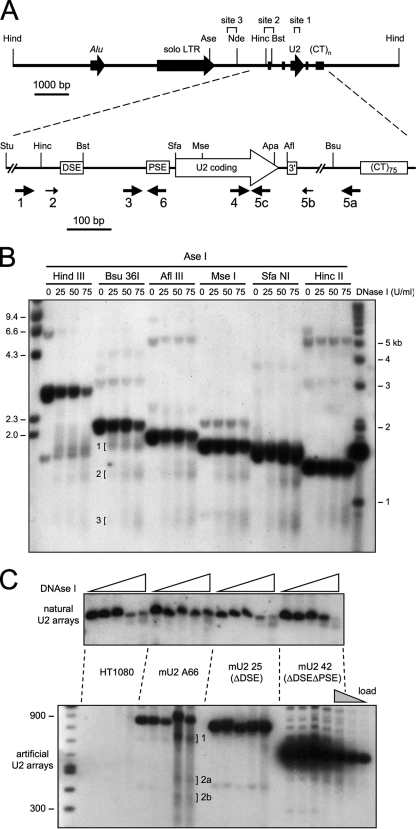
DNase I-hypersensitive sites in the U2 snRNA genes mapped by genomic blotting. (A) Upper, restriction map of the 6.1-kb U2 tandem repeat unit. The three DNase I-hypersensitive sites 1, 2, and 3 identified in panel B are shown. Restriction sites, from left to right, are HindIII, AseI, NdeI, HincII, and BstBI. Lower, enlarged view of the U2 snRNA coding region showing LM-PCR oligonucleotide sets. Key features are the DSE and PSE and the 3′-end formation signal (“3′” box). Restriction sites, from left to right, are StuI, HincII, BstBI, SfaNI, MseI, ApaLI, AflIII, and Bsu36I. Large arrows indicate LM-PCR oligonucleotide sets, each consisting of a primer extension, PCR, and labeling oligonucleotide. Smaller arrows indicate additional labeling primers; primer 2 was used with oligonucleotide set 1, primer 5b with set 5a. (B) Identification of DNase I-hypersensitive sites in the U2 tandem repeat unit by indirect end labeling. HT1080 cells were treated with DNase I in vivo. Genomic DNA was digested with AseI, redigested with the indicated restriction enzymes, and resolved by native agarose gel electrophoresis, and blots were probed with the AseI/NdeI fragment. The secondary restriction enzymes also generate unique, apparently single-copy bands which are unaffected by DNase I digestion; these may be orphan U2 repeat units or previously characterized junction fragments where the U2 tandem repeat meets flanking DNA (85). (C) Deletion of the DSE or PSE abolishes DNase I hypersensitivity of U2 genes. HT1080 cells and derivatives containing artificial tandem arrays of U2 minigenes (3) were treated with DNase I as in panel B. Genomic DNA was digested with AflIII and resolved by electrophoresis through 0.8% agarose (natural gene assay) or 1.5% agarose (minigene assay), and blots were probed with the StuI/HincII fragment. Site 2 resolves into two bands on the higher-percentage gel. The three rightmost lanes represent the 1, 0.25, and 0.125 standard sample loads (gray triangle). Cell line mU2 42 has 10-fold as many minigenes as natural U2 genes (3). The faint unmarked bands seen for mU2 25 and mU2 42 were disregarded since these do not increase significantly with the DNase I concentration.
U2 genes not only are tandemly repeated but are transcribed at an unusually high rate. For comparison, the genes encoding the 35S precursor of the large ribosomal RNAs (18S, 5.8S, and 28S rRNAs) are also tandemly repeated in mammals, present in 1,000 copies per diploid genome, and transcribed by the dedicated RNA Pol I (33, 88, 91). Only a few hundred of these genes appear to be active in most cell types, yet the genes produce approximately 5 × 106 rRNA molecules per generation. Similarly, the small 5S rRNA, which is stoichiometric with the 18, 5.8, and 28S rRNAs, is encoded by approximately 400 tandemly repeated genes per diploid human genome and transcribed by the specialized RNA Pol III, which also transcribes many other small RNAs, including tRNAs, 7SL and 7SK RNA, and U6 snRNA (71). Although the U1 and U2 snRNA components of the spliceosome are nearly as abundant as the rRNAs (0.5 × 106 to 1 × 106 molecules per cell) and are also encoded by tandemly repeated genes in humans, these genes are present in only 20 (U2) or 60 (U1) copies per diploid genome (64, 100, 104). Moreover, the heritable haploid U2 gene copy number varies from 5 to 30 in humans (55), suggesting that no more than half the genes are likely to be active. Assuming a cell generation time of 12 h and that half of the genes are active and continuously transcribed throughout the cell cycle, the calculated transcription initiation rates would be 0.2/s for the large rRNAs transcribed by Pol I, 0.6/s for 5S rRNA transcribed by Pol III, and 0.8/s for U1 or 1.2/s for U2 snRNA transcribed by the same Pol II that transcribes mRNAs. How do the dedicated U snRNA promoters enable Pol II to initiate more frequently on U snRNA genes than the dedicated Pol I does on rRNA genes?
We previously found that the four sites of adenovirus 12-induced metaphase chromosome fragility as originally described by zur Hausen (113) in fact coincide with the human U1 genes (RNU1), U2 genes (RNU2), U1 pseudogenes (PSU1), and 5S genes (RN5S) (59). Remarkably, each of these four loci contains a tandem array of short, powerful transcription units encoding a small, highly structured RNA. We therefore proposed that adenovirus infection causes locus-specific metaphase chromosome fragility by interfering locally with metaphase shutdown of U1, U2, and 5S transcription. While pursuing this hypothesis, we found that U1 and U2 transcription are required for metaphase fragility (3, 53); that the Ad12 55K protein alone can induce fragility, apparently by interacting with p53 (53, 58); and that activation of p53 (110) or defects in the Cockayne syndrome group B (CSB) protein also induce fragility, presumably because direct binding of p53 to CSB phenocopies loss of the CSB protein (111). We concluded that modulation or inactivation of CSB function is the mechanism by which adenovirus 12 infection induces locus-specific fragility of the RNU1, RNU2, and RN5S loci.
The CSB protein is a 1,493-residue DNA-dependent ATPase of the SWI2/SNF2 family with demonstrated chromatin remodeling activity both in vitro (17) and in vivo (79). CSB also plays a role in global and transcription-coupled nucleotide excision repair, as well as global base excision repair of oxidative damage (105); however, the relationship between the chromatin remodeling and repair activities of CSB is not yet clear. Cockayne syndrome (CS), a devastating progeroid disease leading to early death, is most frequently caused by mutations in CSB; the remaining cases of CS are caused by CSA mutations or by rare alleles of the xeroderma pigmentosum genes XPB, XPD, and XPG (89).
To explore the functions of the CSB protein in metaphase condensation of U1 and U2 snRNA genes transcribed by Pol II, we examined the chromatin structure of U2 genes. We used genomic blotting and ligation-mediated PCR (LM-PCR) (37, 77) to map DNase I-sensitive sites and permanganate reactivity (1) in permeabilized cells at nucleotide resolution and chromatin immunoprecipitation (ChIP) to identify proteins bound to U2 genes during interphase and metaphase. We found that the U2 snRNA promoter and the downstream half of the U2 coding region are protected but the upstream half of the U2 coding region is relatively exposed and partially single stranded during interphase. We also found by ChIP that the SNAPc complex which binds the TATA-like PSE is removed at metaphase but remains bound under conditions that induce locus-specific metaphase fragility of the U2 genes, such as a loss of CSB, BRCA1, or BRCA2 function, treatment with actinomycin D, or overexpression of the tetrameric C terminus of p53. We propose that the U2 snRNA promoter establishes a persistently open state in which TFIIH is required for capping (72) but not for the potentially slow promoter melting step (34, 35, 41, 42, 48); this open transcriptional state would facilitate rapid initiation during interphase but require disassembly during metaphase to allow proper chromatin condensation.
(The experiment described in Fig. 1B was first presented by C. P. Elco as part of a senior thesis for the Molecular Biophysics and Biochemistry Department at Yale University in 1998 [25].)
MATERIALS AND METHODS
Cell culture.
HT1080 cells and the mU2 25 (ΔDSE) and mU2 42 (ΔDSE ΔPSE) lines derived from them (3) were grown in minimal essential medium alpha (MEMα) with 10% fetal bovine serum (FBS). Primary cells (GM00739B, GM01629, GM10903, GM10905, and WI38) and telomerase-immortalized primary cells (GM00739Bhtert, GM01629htert, GM10903htert, GM10905htert, and WI38htert) were grown in MEMα with 10% FBS, 2× vitamins and amino acids, and 25 mM HEPES, pH 8.0. GM00739B and GM01629 are primary fibroblasts derived from patients with severe CS; GM00739B is derived from patient CS1AN and is also the source of the well-studied, simian virus 40-transformed CS1ANSV line (97). GM10903 and GM10905 are primary fibroblasts from patients with CSB mutations causing DeSanctis-Cacchione syndrome, an XP variant displaying substantial clinical overlap with CSB-CS. PG-13/htert immortalized cells were selected with 1 mg/ml G418. CS1ANSV was grown in Dulbecco's modified Eagle medium with 10% FBS. CAPAN 1 and HCC1937 cells were grown in RPMI with 15 and 10% FBS, respectively. MCF7 cells were grown in MEMα with 10% FBS.
DNase I and permanganate treatment.
HT1080 cells were grown to 50 to 80% confluence on 150-mm plates, washed with cold phosphate-buffered saline (PBS), and overlaid at room temperature with 4 ml of DNase I digestion buffer (15 mM Tris-HCl [pH 7.5], 15 mM NaCl, 5 mM MgCl2, 60 mM KCl, 300 mM sucrose, 0.5 mM EGTA, 0.1% NP-40) (50) containing 0 to 200 U/ml of DNase I or 0 to 20 mM KMnO4 in PBS (37). The plates were gently agitated for 5 min and the reactions stopped by addition of 2 ml 3× DNase I stop/lysis solution (3% sodium dodecyl sulfate, 300 mM sodium EDTA, pH 8.0) preheated to 65°C or permanganate stop solution. DNA was purified by phenol-chloroform extraction followed by ethanol precipitation. DNase I-digested DNA was analyzed by genomic blotting using the AseI/NdeI restriction fragment as an indirect end-labeled probe (86). Naked duplex DNA controls were as described previously (37); single-stranded DNA controls were heated to 95°C for 5 min in PBS, quick-chilled on ice, and then brought to room temperature before addition of permanganate.
LM-PCR.
The LM-PCR protocol was generally carried out as described previously (29). The sequence, position, Tm, and use of all oligonucleotide primers are available upon request. To ensure specificity, all primers were first tested in a standard PCR using genomic DNA as the template and then in mock LM-PCRs using genomic DNA digested with a restriction enzyme that cuts within the interval under examination (Fig. 2A, labels 3 and 4, lanes M and A). Naked DNA controls were selected from a series of reactions in which the time and/or reagent concentration was varied. The permanganate concentration in all double- and single-stranded controls was 0.2 mM.
FIG. 2.
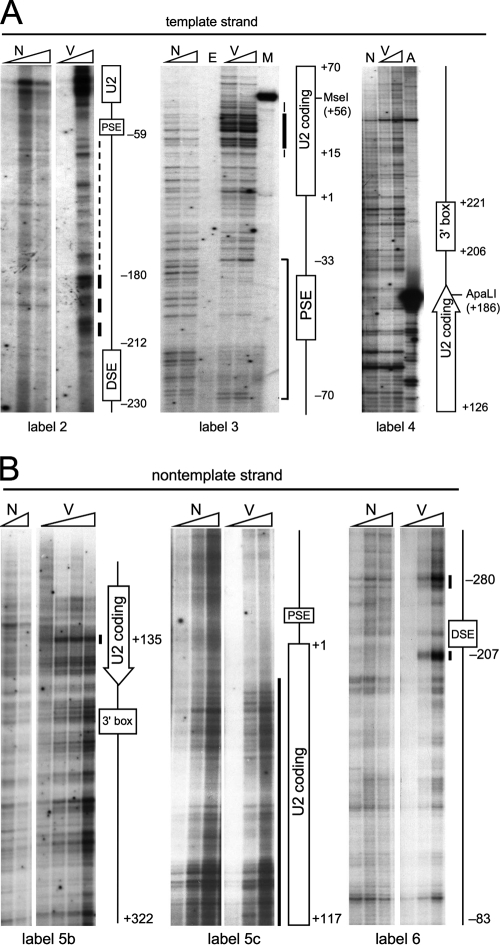
DNase I sensitivity of U2 snRNA genes in HT1080 cells assayed by LM-PCR. (A) Analysis of the U2 template strand. DNase I-hypersensitive site 2 (Fig. 1A) can be seen downstream from the DSE (label 2) and hypersensitive site 1 within the U2 coding region (label 3). The upstream boundary of the large PSE footprint (bracketed) is not visible here because primer set 3 lies just upstream from the PSE. (B) Analysis of the U2 nontemplate strand. The labeling primer used in the final two rounds of PCR is indicated below each panel; label 4 used a previously published primer set (19). DNA sequence features and positions are numbered relative to the U2 transcription start site at +1. Heavy and light vertical bars indicate the degree of DNase I sensitivity. V, genomic DNA treated in vivo with 50 to 100 U/ml DNase I, deproteinized, and digested with EcoRI, which does not cut within the U2 repeat unit (70, 100)]; N, naked genomic DNA treated in vitro with increasing DNase I; E, M, and A, untreated naked genomic DNA cut with EcoRI, MseI, or ApaLI. MseI and ApaI cut within the U2 coding region (Fig. 1A).
Mitotic cells.
HT1080 cells were synchronized by a double thymidine block. A nearly confluent plate was split 1:40 and grown for 48 h. Cells were incubated overnight in 0.3 μg/ml thymidine, released for 8 h in fresh medium, incubated again overnight in thymidine, and then released for 3 h in fresh medium. After addition of 100 ng/ml Colcemid, cells were incubated for 3 h before harvest. See below for drug treatments.
Drug treatments.
Mitotic cells for ChIP were synchronized by double thymidine block (0.3 μg/ml) and collected after incubation with 100 ng/ml Colcemid for 3 h (HT1080 and HeLa) or 16 h (GM00739Bhtert). For asynchronous cultures treated with 50 ng/ml actinomycin D, cells were harvested after 5 h; for mitotic cultures, cells were harvested after treatment for 2 h with actinomycin D alone, followed by 3 h with actinomycin D and Colcemid. Cells were oxidatively stressed by treatment for 2 h with menadione sodium bisulfite (MSB), 15 min with KBrO4, or 30 min with H2O2 and then incubated with Colcemid for an additional 3 h.
Chromosome fragility assays.
Metaphase chromosome fragility assays were performed with the U1 and U2 probes as described previously (3, 110, 111). The 5S probe was an 875-bp NaeI-to-PvuI fragment from the 2.23-kb 5S repeat unit (61).
Altered U2 genes.
All U2 constructs were derived from a U2 minigene spanning 824 bp from the StuI site upstream of the U2 coding region to the HinfI site downstream; HT1080 cell lines containing artificial tandem arrays of these altered U2 genes were then generated as described previously (3). The MR construct deletes U2 coding sequences from the MseI site to the RsaI site (nt +57 to +167). The FslR construct replaces U2 coding sequences from MseI to RsaI (nt +55 to +167) with the artificial stem-loop sequence (5′-AACGCTTGCGTTAACTGATAAGAAGCTTATAAATGGATTTTTTTTGGCCATGGTGACCTTCGGGTCA-3′), and MslR replaces the same U2 coding sequence with 5′-AACGCTTGCGTTAACAAGCTTATAAATGGATTTTTTTTGGCCATG GTGACCTTCGGGTCA-3′. Construct U2Ad3′ was described previously (27, 36).
TATp53 transduction.
A DNA fragment encoding C-terminal residues 303 to 393 of p53 was cloned into the pTAT·HA expression vector (78), and the hexahistidine-tagged TATp53(303-393) peptide was purified from BL21(DE3)pLysS cells by affinity chromatography on a Talon cobalt column (Clontech). TATp53(303-393) was transduced into HT1080 and GM10903htert cells by addition of the protein directly to medium. For mitotic cells, Colcemid was added 2 h after TATp53(303-393) transduction and cells collected 3 h later. For unsynchronized cells and ChIP chromatin, cells were harvested 5 h after transduction with TATp53(303-393).
ChIP.
Chromatin was prepared as described previously (9, 102) with some modifications. Adherent cells were grown to 70 to 80% confluence and cross-linked with 1% formaldehyde on plates. Mitotic cells were collected and cross-linked with 1% formaldehyde in complete medium at a concentration of 2 × 106 cells/ml. Buffers contained 10 mM pyrophosphate (Na2H2P2O7, pH 7.4), 1 mM phenylmethylsulfonyl fluoride, and a cocktail of protease inhibitors (Roche catalog no. 1873580). ChIP was performed as described previously (82, 83) using a 50% (vol/vol) slurry of protein A-Sepharose/protein G-agarose beads. After washes the pelleted beads were then resuspended in 100 μl of Tris-EDTA (TE), pH 7.5.
Immunoprecipitated chromatin was digested with 50 μg/ml DNase-free RNase (Roche) for 30 min at 37°C and then with 0.8 mg/ml proteinase K for 1.5 h at 37°C by addition of 25 μl 5× PK buffer (50 mM Tris-HCl, 25 mM EDTA, 1.25% sodium dodecyl sulfate, pH 7.5) and 5 μl of 20-mg/ml proteinase K. The beads were pelleted, and washed again with 175 μl of TE buffer; the two supernatants were pooled and the NaCl concentration brought to 0.3 M. For the chromatin control (“input”), which contained >500 ng DNA compared to only a few pg of DNA in the immunoprecipitated experimental lanes, 500 μl of chromatin was digested with 100 μg/ml DNase-free RNase for 1.5 h at 37°C and then for >3 h at 37°C with 0.8 mg/ml proteinase K following addition of 120 μl 5× PK buffer and 30 μl of proteinase K. NaCl was added to 0.3 M.
To reverse the cross-links, chromatin samples and controls were incubated for 5 h at 67°C and then extracted twice with phenol-chloroform and once with chloroform alone. After addition of 10 μg glycogen (1 μl of a 10-mg/ml stock) and 2.5 volumes of ethanol, DNA was precipitated overnight at −20°C, pelleted at 13,000 rpm, washed once with 70% ethanol, air dried, and resuspended in 60 μl TE buffer. For the chromatin control, 2 volumes ethanol was added but no glycogen. After precipitation and air drying, the DNA pellet was resuspended in 300 μl of TE buffer and the DNA concentration determined by the optical density at 260 nm (OD260). PCR analysis of chromatin was performed as described previously (23) using 2.5 μCi of [α-32P]dCTP per reaction. The PCR primers, annealing temperatures, cycle numbers, and magnesium concentrations are available upon request. For each PCR, 2 out of 60 μl of immunoprecipitated DNA or 50 ng of control chromatin DNA was amplified using Taq polymerase (New England Biolabs). Repetitive genes were amplified for 25 cycles and single-copy genes for 28 cycles. Aliquots of the PCRs (one-fifth for repetitive genes and one-third for single-copy genes) were analyzed by nondenaturing 6% polyacrylamide gel electrophoresis. The gel was then dried and band intensities quantified by using a Storm PhosphorImager (Molecular Dynamics).
Genomic oxidative damage assay.
We used formamidopyrimidine-DNA glycosylase (FPG) (New England Biolabs) to cleave chromosomal DNA at sites of oxidative damage (63, 94). The cell lines HT1080, GM00739Bhtert, and WI38htert were treated with the oxidizing agent MSB (30), KBrO4 (76), or H2O2 and allowed to recover for 2 h in fresh medium. Chromosomal DNA was then isolated using the DNeasy tissue kit (Qiagen). All subsequent buffers contained 50 μM α-phenyl-N-tert-butyl nitrone (Aldrich) to inhibit oxidation in vitro. To assay endogenous oxidative damage in specific genes or gene regions, the FPG-treated DNA was amplified by PCR as described above for the ChIP assay but stopped at two cycle intervals and the products analyzed by using 6% native polyacrylamide gels followed by phosphorimaging. To analyze DNA from cells treated with oxidizing agents, 10 ng of genomic DNA from each condition was analyzed by continuous fluorescent PCR (DNA Engine Opticon System; MJ Research) using the Sybr Green PCR master mix (Applied Biosystems).
RESULTS
Identification of DNase I-hypersensitive sites in U2 snRNA genes.
We investigated the chromatin structure of the tandemly repeated U2 snRNA genes in HT1080, a human pseudodiploid fibrosarcoma line with well-characterized RNU2 alleles (56, 57, 85, 86). HT1080 has two and only two U2 tandem arrays, one spanning 9 and the other 13 6.1-kb repeat units. Although the U2 repeats in HT1080 are undermethylated compared to those in primary cells (43), the U2 tandem arrays are stable over thousands of cell generations (86), thus reducing background signals from the unstable, fragmented, and possibly inactive U2 arrays seen in HeLa cells (3). DNase I digestion experiments examining the entire U2 repeat unit identified three hypersensitive regions by genomic blotting (25). These sites were then mapped at higher resolution by genomic blotting with an indirect end label as a probe. Adherent, subconfluent HT1080 cells were permeabilized with NP-40 detergent and treated with a range of DNase I concentrations. Genomic DNA was then purified, digested with AseI and a second restriction enzyme, resolved by agarose gel electrophoresis, blotted, and probed with the labeled AseI-to-NdeI fragment as described previously (86).
Three DNase I-hypersensitive sites in the U2 genes are seen in permeabilized HT1080 cells (Fig. 1). Site 1 is located between the MseI site at +56 bp downstream from the 5′ end of the U2 coding region and the AflIII site at +202 just upstream of the 3′ end formation signal (also known as the “3′ box”) (Fig. 1A, top, and B). Site 2 spans the DSE from the HincII site to the BstBI site (also see Fig. 2B, below), and site 3 maps about 800 bp downstream from the solo long terminal repeat. The hypersensitive sites represent double-strand breaks or tightly clustered nicks because the DNA was not denatured before gel electrophoresis. Site 1 appears at the lowest DNase I concentration; sites 2 and 3 are of equal intensity, although site 3 is more diffuse. The relative intensity of the AseI/Bsu36I band at 0 and 75 U/ml of DNase I indicates that roughly two-thirds of the U2 snRNA genes are cleaved, and the relative intensities of the intact AseI/MseI and AseI/AflIII bands at 75 U/ml DNase I suggest that most genes are cleaved at site 1 (Fig. 1C).
Surprisingly, all sites seemed to be insensitive to the Pol II transcription inhibitor α-amanitin (data not shown), which abolishes U2 snRNA transcription (40); however, this may simply reflect the ability of α-amanitin to freeze Pol II in place by blocking elongation as well as initiation (14). To examine whether U2 transcription is required for DNase I hypersensitivity, we used HT1080 cell lines with artificial tandem arrays of U2 minigenes lacking either the DSE or both the DSE and PSE (3). Consistent with previous evidence that the U2 DSE strongly activates the basal PSE but is not absolutely essential for U2 transcription (2, 67), we found that the ΔDSE deletion reduced the intensity of site 1 whereas the ΔDSE ΔPSE deletion abolished site 1 completely (Fig. 1C). As expected, the ΔDSE deletion also abolishes site 2, which maps near or within the DSE (Fig. 1A and C). We conclude that U2 transcriptional control elements are required for site 1 hypersensitivity.
LM-PCR analysis of DNase I-hypersensitive sites.
DNase I-hypersensitive sites are regions of DNA where chromatin structure (or the lack of it) allows nuclease access. We did not pursue hypersensitive sites 3 and 2 because site 3 is located considerably upstream of the essential U2 snRNA promoter (2, 41, 42) and site 2 lies within the promoter, which is known to be highly structured (8).
Instead, we chose to focus on the novel hypersensitive site 1 because the signal was strong and located directly over the U2 coding region. We first examined the pattern of DNase I sensitivity at the nucleotide level on both strands of the U2 transcription unit by LM-PCR. HT1080 cells were treated with DNase I in vivo as for Fig. 1B; the viscosity of the purified genomic DNA was reduced by digestion with EcoRI, which does not cut within the U2 tandem repeat unit (70, 100), and the DNA was subjected to LM-PCR as described previously (37, 77) using multiple primer sets (Fig. 1A) for the template (Fig. 2A) and nontemplate (Fig. 2B) strands. The resulting data set is nearly complete except downstream of the 3′ end formation signal, where numerous primers located between the signal and the downstream CT dinucleotide repeat failed to give specific products in the genomic U2 PCR assay.
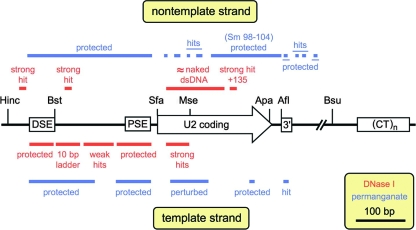
As summarized in Fig. 3, the DSE and PSE are protected on the template strand, presumably by Sp1 and Oct1 bound at the DSE and SNAPc at the PSE (Fig. 2A, label 3), with a 10-bp ladder and weak DNase sensitivity extending from the DSE to the PSE (Fig. 2A, label 2); this pattern has been interpreted as a positioned nucleosome (8), although DNA wrapping by other snRNA- or U2-specific factors cannot be strictly excluded. (Note that the “nontranscribed strand” of Boyd et al. [8] is in fact the transcribed or U2 template strand.) Consistent with the DSE footprint, two strong DNase I-hypersensitive sites flank the DSE on the nontemplate strand (Fig. 2B, right panel, label 6). In addition, a strong, broad region of DNase I hypersensitivity is seen within the U2 coding region extending from +15 to +56 on the template strand (Fig. 2A, labels 2 and 3). The hypersensitive site at +15, which was seen with micrococcal nuclease but apparently not with DNase I (8), presumably corresponds to the 5′ boundary of DNase I sensitivity within the U2 coding region. Taken together, these data suggest that DNase I-hypersensitive site 1, visualized without DNA denaturation (Fig. 1B), reflects nicking on both strands between +15 and the DNase I-hypersensitive site at +135 (Fig. 2B, label 5b). No hypersensitivity or DNase I protection is seen over the 3′-end formation signal, consistent with current evidence that it functions as an RNA processing site (5, 19, 40, 66, 99).
FIG. 3.
Summary of DNase I (Fig. 2) and permanganate sensitivity data (Fig. 4).
Intriguingly, despite dramatic DNase I hypersensitivity of the U2 template strand from +15 to +56, the DNase I sensitivity of nontemplate U2 sequences is very similar in vivo and in the naked duplex DNA controls (Fig. 2B, left and middle panels), suggesting that this DNA retains significant duplex character even in active U2 genes. Although this could in principle reflect two populations of U2 snRNA genes, one active and one inactive, most genes appear to be cleaved at site 1 (Fig. 1C), leading us to agree with Boyd et al. (8), who argue for different reasons that most if not all U2 snRNA genes are likely to be active; and even if there were two such distinct populations, it would still be difficult to understand how one strand could be hypersensitive while the other appeared indistinguishable from duplex. Careful examination of the DNase I-hypersensitive sites in the U2 coding region, however, reveals that all of these sites are also cut in the duplex DNA controls, albeit at very low levels (Fig. 2A, middle panel). Thus, this DNA may adopt a conformation in vivo which is sufficiently strained to induce hypersensitive sites on the template strand, while the nontemplate strand remains nearly duplex; the existence of both permanganate-sensitive and protected sites within this region of the nontemplate strand also provides evidence that it is not a true duplex (Fig. 3 and 4).
FIG. 4.
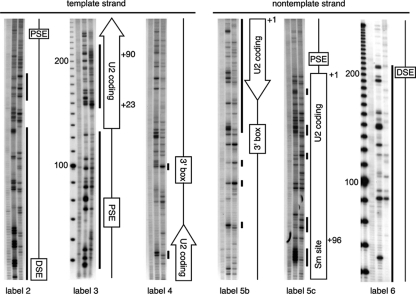
Permanganate sensitivity of U2 snRNA genes in HT1080 cells assayed by LM-PCR. In each panel, the three lanes from left to right are duplex genomic DNA treated with permanganate in vitro as a control for DNA breathing, single-stranded genomic DNA treated with permanganate in vitro as a control for context-dependent differences in reactivity (39) and to provide a T sequence ladder, and genomic DNA treated in vivo. An additional leftmost lane in two panels contains a 10-bp ladder. The labeling primer used in the final two rounds of PCR is indicated below each panel. Vertical bars indicate regions of DNA protection, sensitivity, or perturbation.
Since hypersensitive sites 1, 2, and 3 are all insensitive to α-amanitin, we conclude that hypersensitivity from +15 to +56 is likely to be caused not by transcription itself but rather by the ability of the dedicated U2 snRNA promoter to maintain downstream DNA in a persistently open state. We speculate below, based on this and other experiments, that the open state may facilitate polymerase engagement, initiation, and elongation by circumventing the TFIIH-dependent promoter melting and clearance steps (34, 35, 41, 42). Curiously, only weak DNase I sensitivity is seen over the transcription initiation site on the template strand (Fig. 2A, label 3). This could indicate protection by transcription factors or rapid rebinding of Pol II to the highly efficient U snRNA promoters.
LM-PCR analysis of permanganate-sensitive sites.
To assay for single-stranded character within the U2 transcription unit, we treated HT1080 cells in vivo with the oxidizing reagent potassium permanganate and used LM-PCR to identify modified bases (1). Unlike DNase I, a large reagent that assays accessibility of the deoxyribose phosphate backbone, the small permanganate ion oxidizes the C4-5 bond of thymidines and, less efficiently, cytidines that are not protected by stacking within a DNA duplex (49). Nucleotides modified in vivo were compared to those in single-stranded and duplex DNA controls (Fig. 4). We included both controls because the highly structured U2 snRNA is known to assume alternative secondary structures (87), and it would not be surprising if this were also true for both the template and nontemplate strands of the U2 gene.
Overall, the permanganate reactivity appears to reflect the DNA conformation in vivo (Fig. 4, labels 2, 3, 5c, and 6), since little or no reactivity was seen on either strand over the DSE and PSE, consistent with binding of the known transcription factors, Sp1, Oct1, and SNAPc, to duplex DNA (8, 41, 42). Sequences downstream of the DSE are protected from permanganate on both the template and nontemplate strands, whereas sequences nearer the PSE are hypersensitive (Fig. 4). These data, in combination with the 10-bp DNase I ladder downstream of the DSE (Fig. 2A) and the observation that a positioned nucleosome can generate permanganate-hypersensitive sites (26), confirm the existence of a positioned nucleosome between the DSE and PSE (7, 112). The upstream half of the U2 coding region extending to position +90 appears to be mainly single stranded, with hyperreactive thymidines at positions +23 and +24 in the template strand. (The thymidine at position +2 is uninformative, since it reacts poorly in the single-strand control and in vivo.) The downstream half of the U2 coding region extending to +188 exhibits little or no permanganate oxidation in vivo, suggesting that this region is mainly duplex; most notably, all thymidines within the Sm site (ATTTTTG) at +98 are nonreactive.
Curiously, the 3′-end formation signal at +205 just downstream from the U2 coding region exhibits hypersensitive thymidines on the template strand and protected thymidines on the nontemplate strand (Fig. 4, labels 4 and 5b; summarized in Fig. 3), as well as two hypersensitive sites and one protected site beyond the signal on the nontemplate strand (Fig. 4, label 5b). DNA corresponding to an RNA processing signal would not be expected to exhibit nonduplex structure, but as discussed below, permanganate reactivity unaccompanied by DNase I sensitivity (Fig. 2A and 3) could reflect Pol II pausing during 3′-end formation or Pol II recycling from termination back to initiation.
The DNase I sensitivity (Fig. 2) and permanganate reactivity (Fig. 4) of the U2 coding and flanking sequences are summarized and compared in Fig. 1D. Although DNase I and permanganate both assay for single-stranded DNA character in vivo and should in principle provide complementary information, the two assays are not always easy to correlate. Nonetheless, the simplest interpretation of the permanganate data is consistent with the DNase I data: the first half of the U2 coding region is relatively open, the second half of the U2 coding region is mainly duplex or protected, and occasional hits in the 3′-end formation signal and immediately downstream may reflect transcriptionally coupled events, such as polymerase pausing, topological constraints, or release or gain of processing and/or termination factors.
U2 snRNA genes may be sites of preferential DNA damage.
CSB participates in global base excision repair of oxidative damage, as well as global and transcription-coupled nucleotide excision repair (105). The presence of persistently single-stranded DNA in the U2 genes of asynchronous cells (Fig. 1, 2, and 4) therefore suggested that preferential oxidative damage of the exposed bases might account for metaphase fragility of the tandemly repeated U1, U2, and 5S RNA genes (the RNU1, RNU2, and RN5S loci) in cells lacking functional CSB protein (111). The resulting accumulation of DNA damage and/or stalled RNA polymerases within the tandem arrays might then interfere locally with metaphase chromatin condensation.
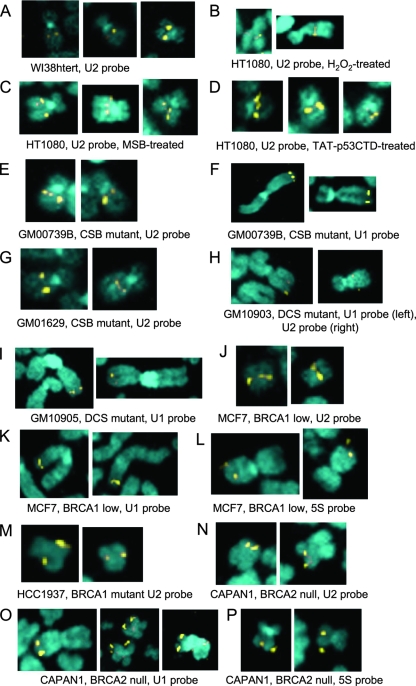
Consistent with this hypothesis, we found using our standard assay (111) that treatment of HT1080 cells with hydrogen peroxide in vivo, followed by 3 h in Colcemid, increased metaphase fragility of the RNU2 locus (Fig. 5B; Table 1); however, the effect was modest, presumably because all concentrations of peroxide tested inhibited progression into mitosis and caused considerable cell death (A. D. Bailey, unpublished observations). In contrast, the effect is maintained at higher concentrations of MSB, which also induces no more than 35% fragility (Fig. 5C; Table 1). Since both uninduced and induced levels of fragility depend on the genotype and age of the cell line, we suspect that the magnitude of the effect may depend on the stringency of mitotic checkpoints in each line. Similarly, protein transduction with residues 303 to 393 of the tetrameric p53 C-terminal domain (TAT-p53CTD), which is thought to mimic activation of p53 by DNA damage (110, 111), also induces fragility (Fig. 5D). Overall, these observations suggest that preferential oxidative DNA damage resulting from transient or persistently single-stranded DNA character during U snRNA transcription by Pol II (Fig. 2 and 4) could contribute to locus-specific metaphase fragility. The single-stranded DNA character could also explain the puzzling observation that tandemly repeated 5S RNA genes (the RN5S locus) are the other major site of inducible locus-specific metaphase chromosome fragility (111) (Fig. 5L and P): although 5S RNA genes are transcribed by Pol III not by Pol II, 5S genes are transiently single stranded when TFIIIA binds to the nontemplate strand of the 5S coding region during assembly of the Pol III preinitiation complex (18, 92).
FIG. 5.
Metaphase fragility of the RNU1, RNU2, and RN5S loci in normal and mutant cells either treated with oxidizing agents or transduced with TAT-p53CTD. (A) WI38 lung fibroblasts immortalized with htert and probed for U2 genes. The RNU2 signals never split but are occasionally unequal in this and all other cell lines. (B) HT1080 fibrosarcoma line treated with MSB and probed for U2 genes. RNU2 often incompletely condensed and split. (C) HT1080 treated with H2O2 and probed for U2 genes. RNU2 often split. (D) HT1080 transduced with TAT-p53CTD and probed for U2 genes. RNU2 often incompletely condensed and/or split. (E) GM00739B primary CSB fibroblasts from a patient with severe CS, probed for U2 genes. RNU2 often incompletely condensed and split. (F) GM00739B probed for U1 genes. RNU1 often split. (G) GM01629 primary CSB fibroblasts from a patient with severe CS, probed for U2 genes. RNU2 often incompletely condensed and/or split. (H) GM10903, a primary fibroblast from a patient with a CSB mutation causing severe DeSanctis-Cacchione syndrome (DCS) probed for U1 genes (left) and U2 genes (right). RNU1 and RNU2 often incompletely condensed and/or split. (I) GM10903 probed for U1 genes. RNU1 often incompletely condensed and/or split. (J) MCF7 breast adenocarcinoma line expressing low levels of BRCA1, probed for U2 genes. RNU2 often incompletely condensed and/or split. (K) MCF7 probed for U1 genes. RNU1 often incompletely condensed and/or split. (L) MCF7 probed for 5S genes. RN5S often incompletely condensed, split, or separated from the telomere. (M) HCC1937, a breast carcinoma line homozygous for a C-terminal deletion that abolishes ubiquitylation of known BRCA1 targets (95), probed for U2 genes. RNU2 often incompletely condensed and/or split. (N) CAPAN1 pancreatic carcinoma line, a compound heterozygote for loss of BRCA2, probed for U2 genes. RNU2 often incompletely condensed and/or split. (O) CAPAN1 probed for U1 genes. RNU1 often incompletely condensed, split, or separated from the telomere. The CAPAN1 karyotype is complex, and RNU1 loci are found on different derivative chromosomes. (P) CAPAN1 probed for 5S genes. RNU5 often separated from the telomere.
TABLE 1.
Hydrogen peroxide and MSB induce metaphase fragility of the RNU2 locus in HT1080 cells
| Treatment | % Fragilitya
|
|
|---|---|---|
| Per spread | Per locus | |
| H2O2 | ||
| 0 nM | 15 | 6.5 |
| 50 nM | 23 | 12 |
| 500 nM | 33 | 17 |
| 50 μM | 27 | 13 |
| 100 μM | 20 | 9.4 |
| MSB | ||
| 0 nM | 18 | 8.6 |
| 50 nM | 25 | 15 |
| 100 nM | 29 | 16 |
| 200 nM | 31 | 16 |
| 500 nM | 36 | 19 |
| 1 μM | 28 | 15 |
| 10 μM | 36 | 19 |
| 100 μM | 35 | 17 |
Per spread, percentage of nuclei with at least one fragile locus; per locus, percentage of total loci scored as fragile.
We next looked directly for preferential oxidative damage in the U1 or U2 snRNA genes using the modified FPG assay (63, 94). We treated HT1080 cells with three different oxidizing agents (hydrogen peroxide, potassium bromate, and menadione bisulfite) and prepared genomic DNA under conditions that inhibit in vitro oxidation. The DNA was cleaved with FPG, an N-glycosidase and AP lyase that releases damaged purine. The resulting single-strand breaks block PCR amplification so that interval-specific oxidative damage can be estimated from the ratio of products generated by quantitative PCR using cleaved and uncleaved DNA as templates. We were unable to detect any incremental damage in the U1 or U2 genes compared to genes for rRNA, glyceraldehyde-3-phosphate dehydrogenase, or the large subunit of RNA polymerase II (A. D. Bailey, data not shown); however, the FPG assay scores damage averaged over all 20 of the U2 snRNA genes in HT1080 cells (85, 86) and thus might fail to detect unrepaired damage to a minority of these genes (corresponding to less than a 50% decrease in the PCR signal). In addition, although the FPG assay scores any damaged purine, the major oxidation product is 8-oxoguanine and the single-stranded regions identified by DNase I and permanganate within the U2 genes are not GC rich.
BRCA1 and BRCA2 deficiency also cause RNU2 metaphase fragility.
If U2 genes are indeed sites of low but preferential oxidative damage, one would predict that loss of other remodeling or repair proteins might also cause locus-specific metaphase chromosome fragility. We therefore asked whether defects in BRCA1 or BRCA2, which like CSB are required for repair of oxidative damage (21, 52) and for regulation of Pol II transcription (75, 79, 103), can also cause metaphase fragility of the RNU1, RNU2, and RN5S loci (Table 2). Unsynchronized, subconfluent cultures of three cell lines—HCC1937 (BRCA1 mutant), MCF7 (BRCA1 low), and CAPAN-1 (BRCA2 null)—were treated with Colcemid for 3 h, and metaphase spreads were examined by fluorescent in situ hybridization using a biotinylated nonrepetitive probe for the U2 snRNA tandem repeat unit (111). As controls, we examined GM00739B (primary CSB fibroblasts from patient CS1AN with severe CS) for RNU2 and RNU1 fragility, GM01629 (another primary CSB fibroblast from a patient with severe CS) for RNU2 fragility, and GM10903 (a primary CSB fibroblast from a patient with severe DeSanctis-Cacchione syndrome) for RNU1 and RNU2 fragility (Fig. 5E to I). Whether calculated on a per-spread or per-locus basis (3), the level of metaphase RNU2 fragility observed in the BRCA1 and BRCA2 cell lines (Fig. 5J to P; Table 2) was significantly elevated relative to that for the controls (Fig. 5E to I; Table 2) and comparable to that seen previously for both CSB mutant fibroblasts (CS1AN) and normal fibroblasts (MRC5) transiently overexpressing the C-terminal domain of p53 (111). Similar levels of metaphase RNU2 fragility were observed both in primary CSB cells (GM00739B) and in these same cells after immortalization with an htert-expressing retroviral vector (GM00739Bhtert); moreover, RNU2 fragility was significantly reduced, as expected, by stable transfection of GM00739Bhtert cells with normal CSB cDNA (GM00739Bhtert CSBwt F1) and no RNU2 fragility was observed when normal primary WI38 fibroblasts were immortalized with the PG-13/htert retroviral vector alone (WI38htert) (Table 2), although expansion of one or both RNU2 loci is occasionally observed in these and other cell lines, perhaps reflecting overdenaturation during chromosome workup (Fig. 5A, middle and right panels).
TABLE 2.
Metaphase fragility of the RNU2 locus and other loci in BRCA1- and BRCA2-deficient cells
| Cell line (description) | % Fragilitya
|
|||||
|---|---|---|---|---|---|---|
|
RNU2
|
RNU1
|
RN5S
|
||||
| Per spread | Per locus | Per spread | Per locus | Per spread | Per locus | |
| MCF7 (BRCA1 low) | 79 | 41 | 100 | 49 | 73 | 32 |
| HCC1937 (BRCA1 mutant) | 74 | 36 | ND | ND | ND | ND |
| CAPAN-1 (BRCA2 null) | 70 | 23 | 84 | 60 | 76 | 40 |
| GM00739Bhtert (CSB mutant) | 68 | 51 | ND | ND | ND | ND |
| GM00739Bhtert CSBwt F1 (rescued) | 31 | 17 | ND | ND | ND | ND |
| WI38htert | 12 | 6 | ND | ND | ND | ND |
Per spread, percentage of nuclei with at least one fragile locus; per locus, percentage of total loci scored as fragile; ND, not determined.
Specific U2 snRNA coding sequences are not required for metaphase chromosome fragility.
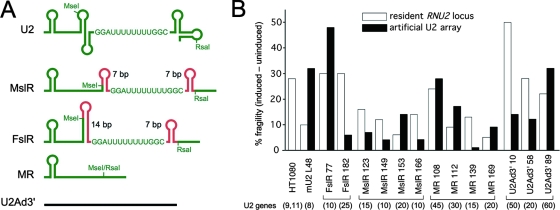
Mutation or loss of CSB causes locus-specific metaphase fragility of the tandemly repeated genes encoding U1 snRNA (the RNU1 locus), U2 snRNA (the RNU2 locus), and 5S rRNA (the RN5S locus) (111). All three small RNAs are highly structured, suggesting an alternative or complementary explanation for locus-specific metaphase fragility: newly formed secondary structure in the nascent RNA (96) or related structures in the template or nontemplate strand of the DNA could potentially stall or arrest Pol II by displacing the protein “lid” or “clamp” that helps to encircle the nascent transcript. To test this hypothesis, we altered the structure of the nascent U2 snRNA coding region by deleting or replacing U2 snRNA stem-loops IIa, IIb, III, and IV; we also expanded the oligo(U) tract of the natural Sm site from U5 to U8 in an attempt to mimic a eubacterial rho-independent transcription terminator (Fig. 6A). Although deletion or replacement of these stem-loops had no consistent effect on metaphase fragility, we found to our surprise that fragility was similarly unaffected when the U2 snRNA coding region was completely replaced by unstructured “stuffer” sequences from the adenovirus 2 major late transcription unit (36) (Fig. 6A). These data suggest that the U2 promoter and possibly the promoter-dependent U snRNA 3′ end formation signal (20, 36) are the essential determinants of metaphase fragility. This would explain why U2 transcription is essential for fragility (Fig. 1C) and would suggest that the CSB chromatin remodeling function (17, 79) required to prevent locus-specific metaphase fragility (111) acts directly on factors bound to the U2 promoter or on the dedicated transcription unit established jointly by the U snRNA promoter and 3′-end formation signal.
FIG. 6.
Effect of U2 snRNA structure on fragility of artificial U2 tandem arrays. (A) Secondary structure of altered U2 snRNAs. The expanded Sm site AUUUUUUUUG between U2 stem-loops IIb and III is shown explicitly. Green, normal U2 sequence; red, artificial sequence, with stem length in base pairs, and tetraloops for stability. (B) Fragility of artificial U2 arrays. Note that the observed level of fragility varies from one independently derived line to another, presumably reflecting position effects, copy number, and/or the relative (and heritable) activity of resident and artificial arrays (3, 68). Tandem U2 gene copy numbers are in parentheses.
SNAPc remains bound to the U2 promoter under conditions that induce locus-specific metaphase fragility.
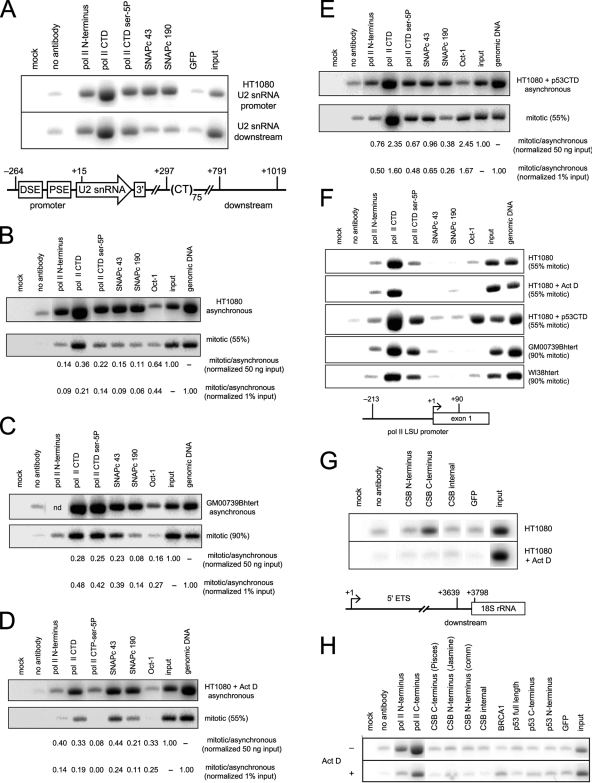
The requirement for U2 snRNA promoter sequences (Fig. 6) and the existence of partially single-stranded DNA within the U2 snRNA coding region (Fig. 1, 2, and 4) suggested that RNU2 metaphase fragility observed in CSB-deficient cells (111) might reflect persistent binding of transcription, chromatin remodeling, or DNA repair factors to the U2 genes during metaphase. We therefore used ChIP to examine the U2 genes for the presence of factors that are known to be involved in either U2 snRNA transcription (Pol II, SNAPc, and Oct-1) (35, 41, 42) or metaphase fragility of the RNU2, RNU1, and RN5S loci (CSB and p53) (111).
As shown in Fig. 7, we found that the SNAPc 43 and SNAPc 190 subunits of the U snRNA-specific transcription factor SNAPc (65)—also known as PTFγ and PTFα, respectively (109)—are easily detected on the U2 promoter (Fig. 7A) but are removed (or become inaccessible to antibody) as the cells reach metaphase (Fig. 7B). Most importantly, SNAPc 43 (but not SNAPc 190) is preferentially retained on the U2 promoter under three very different conditions that induce metaphase fragility of the RNU2 locus: loss of CSB function in GM00739Bhtert cells derived from the severely affected CS patient CS1AN (79, 97) (Fig. 7C), treatment with low doses of actinomycin D (110, 111) (Fig. 7D), and transfection (110, 111) or transduction (Fig. 6E) with the tetrameric C-terminal domain of p53 (TAT-p53CTD). These data demonstrate that SNAPc 43 is normally removed from the DNA before metaphase chromatin condensation takes place but remains on the genes under conditions that induce locus-specific metaphase fragility. Moreover, since both the PSE and DSE footprints are substantially reduced at metaphase (8) and SNAPc is recruited to the PSE through an interaction of SNAPc 190 with Oct-1 bound to the DSE (41, 42, 112), the data suggest that the U2 promoter complex is disassembled at metaphase and that failure to do so causes localized failure of chromatin condensation. Interestingly, all three conditions that induce RNU2 metaphase fragility appear to cause greater retention of SNAPc 43 than of SNAPc 190 (Fig. 7C, D, and E), suggesting that SNAPc may be removed piecewise during promoter disassembly. Alternatively, the fragile state may render SNAPc 190 inaccessible to our antibody, an interpretation that is far more easily reconciled with previous observations that SNAPc 190 interacts with all the other SNAPc subunits (41) and that U2 transcription can be activated by a minimal complex of SNAPc 43, 50, and 190 in which only SNAPc 50 and 190 can be UV cross-linked to the DNA (34).
FIG. 7.
ChIP assays for factors bound to U2 genes at interphase and metaphase. (A) SNAPc and Pol II CTD phosphorylation on the U2 snRNA promoter (−264 to +15) and downstream flanking sequence (+791 to +1019) in HT1080 cells. The abundance of SNAPc on the promoter compared to the downstream fragment demonstrates that ChIP has sufficient resolution for these experiments, although chromatin shearing to the limit size of 500 bp leaves many fragments of >2 kb which reduce resolution on the small U snRNA genes. (B) SNAPc lost from the U2 promoter at metaphase in HT1080 cells. Quantification below ChIP panels is the ratio of mitotic to asynchronous signals when normalized to 50 ng input (upper row) or 1% input DNA (lower row). (C) Some SNAPc 43 but little SNAPc 190 remains on the U2 promoter in the CSB null cell line GM00739Bhtert, derived from patient CS1AN. Quantification is as in panel B. (D) Some SNAPc 43 but little SNAPc 190 remains on the U2 promoter at metaphase in HT1080 cells treated with a low concentration of actinomycin D to induce RNU2 fragility. Quantification is as in panel B. (E) Most SNAPc 43 but little SNAPc 190 remains on the U2 promoter at metaphase in HT1080 cells transduced with the tetrameric C-terminal domain of p53 (TAT-p53CTD) to induce RNU2 fragility. Quantification is as in panel B. (F) Neither SNAPc 43 nor SNAPc 190 is retained at metaphase on the promoter of the largest subunit of RNA polymerase II in untreated HT1080 cells, HT1080 treated with actinomycin D or transduced with TAT-p53CTD, or in mitotic GM00739Bhtert or WI38htert cells. (G) CSB can be detected on active rRNA transcription units (PCR primers amplify +3639 to +3798), but only the CSB N terminus is accessible to our antibody. (H) Neither CSB nor BRCA1 can be detected on the U2 snRNA promoter before or after treatment of asynchronous HT1080 cells with actinomycin D. Pisces is a polyclonal antibody directed against the C terminus of CSB, and Jasmine is a polyclonal antibody against the N terminus (80); “comm” is a commercial polyclonal antibody against CSB (Santa Cruz).
Pol II CTD.
Although SNAPc 43, but possibly not SNAPc 190, remains on the U2 snRNA promoter under all three conditions that induce RNU2 metaphase fragility (loss of CSB [Fig. 7C], treatment with actinomycin D [Fig. 7D], and transduction with TAT-p53CTD [Fig. 7E]), Pol II occupancy is more complex: In CSB null cells with a mitotic index of >90%, the Pol II C-terminal domain (CTD) and serine-5P signals appear undiminished, although SNAPc 190 is low or lost (Fig. 7C); since SNAPc 190 is thought to form the backbone of the SNAPc complex and is required for initiation in vitro (34), this is consistent with our earlier hypothesis (111) that CSB may play a role in removing Pol II from the U2 transcription unit as metaphase approaches. Transduction with TAT-p53CTD has the same effect on SNAPc 43 and 190 as loss of CSB function, and the CTD and serine-5P signals also remain high (Fig. 7E). However, treatment with low concentrations of actinomycin D, which has the same effect on SNAPc 43 and 190 as loss of CSB or transduction with TAT-p53CTD, causes both the Pol II CTD and serine-5P signals to drop dramatically (Fig. 7D). Thus, metaphase fragility correlates with U2 promoter occupancy by SNAPc, not by Pol II, consistent with evidence that the U2 promoter recruits SNAPc independently of Pol II (24). Finally, the CTD, serine-5P, and Oct-1 signals observed on the promoter for the largest subunit of Pol II resemble those seen on U2 genes in both normal cells (HT1080 and WI38htert) and CSB null cells (GM00739htert) (Fig. 7F). This control confirms that the SNAPc signal is independent of the Pol II and Oct-1 signals in the cell types studied and that Pol II occupancy can be uncoupled from locus-specific metaphase fragility.
CSB and BRCA1.
We easily detected CSB on active rRNA transcription units (Fig. 7G), as previously observed (11), but little or no CSB could be detected on U2 genes either in unsynchronized cells or when RNU2 fragility was induced by actinomycin D (Fig. 7H). This could mean that CSB is inaccessible to our antibody when bound to U2 snRNA genes (only the N terminus of CSB is accessible on rRNA genes) (Fig. 7H), that binding of CSB is weak or transient, or that CSB acts indirectly to induce metaphase RNU2 fragility. Similarly, although BRCA1 deficiencies also cause metaphase fragility of the RNU2 locus (Fig. 5B; Table 2), very little BRCA1 can be detected on U2 snRNA genes even under conditions that induce fragility (Fig. 7H).
DISCUSSION
Using DNase I sensitivity and permanganate modification, we present evidence that sequences within the U2 snRNA coding region from +15 to +56 exhibit significant single-stranded character in unsynchronized cells (Fig. 1, 2, and 3). The single-strandedness is unlikely to reflect stalled or paused Pol II (60), because U2 snRNA genes are as efficiently transcribed by Pol II as 5S rRNA genes are by Pol III or 45S rRNA genes by Pol I. Single-strandedness is also unlikely to reflect direct binding of transcription factors to the RNA coding region as observed during assembly of the eukaryotic 5S rRNA transcription complex (18, 31, 44, 92), because complete replacement of the U2 coding region by “stuffer” sequences does not affect the efficiency of transcription (36). We speculate that the unusual architecture of the dedicated U snRNA promoter establishes a persistently open transcriptional state in order to facilitate polymerase binding, initiation, or promoter clearance, potentially by circumventing a TFIIH-dependent slow step.
Components of the 10 subunit TFIIH complex perform two separable functions during initiation by Pol II: the XPB and to a lesser extent XPD helicase subunits facilitate initiation and promoter escape, and the CDK7 cyclin-dependent kinase subunit phosphorylates serine 5 of the Pol II CTD (12), which subsequently recruits and activates the capping complex (38). Although TFIIH may not be required for U snRNA transcription in vitro (48), the observation that U2 transcripts continue to be capped when serine 2 phosphorylation by P-TEFb is inhibited by KM05283 (72) suggests that serine 5 is phosphorylated by the CDK7 component of TFIIH in vivo (72). However, the intriguing possibility remains that serine 5 phosphorylation may be the only TFIIH activity required for U snRNA transcription; the open U2 chromatin conformation could facilitate transcription by circumventing the need for helicase activity during initiation and/or promoter escape. In fact, promoter escape and clearance are rate limiting for reconstituted Pol I (84) and Pol II (47) transcription, and some activators regulate Pol II at both initiation and promoter escape (28, 62). The persistently open transcriptional state could also prevent nucleosome binding, thus avoiding nucleosome-induced backtracking and arrest (45), especially in these unusual transcription units where the nascent RNA (and presumably both strands of the corresponding DNA) have the potential for secondary structure. Although the U2 transcription initiation site itself exhibits little or no single-strandedness (Fig. 2 and 4), this could indicate protection by the same promoter architecture that maintains single-stranded character downstream of +15 or such rapid rebinding of Pol II that the initiation site appears closed because it is in continual use.
An open chromatin state might also allow functional looping or even physical juxtaposition of the 5′ and 3′ ends of the U2 genes in order to facilitate local recycling of Pol II as documented for yeast Pol III (22) and human mitochondrial RNA polymerase (69) and as suggested for human Pol II based on recent evidence that the general transcription factor TFIIB is associated with both the promoter and the polyadenylation site (93). Indeed, just as components of the polyadenylation apparatus are loaded onto promoter-bound factors and then transferred to the Pol II CTD (15), the promoter dependence of U1 and U2 snRNA 3′ end formation (20, 36) suggests that Integrator, the endonuclease complex that generates the 3′ end of the U2 precursor (5, 66), may first bind to the Pol II CTD at the promoter (24). This scenario could also explain the unexpected permanganate reactivity of the 3′ end formation signal and sequences immediately downstream (Fig. 3 and 4), which is not accompanied by DNase I sensitivity (Fig. 2A and 3): although U2 transcription can continue 800 bp past the 3′ end formation signal (19, 40, 99), Pol II may pause on these sequences, awaiting endonucleolytic cleavage of the nascent U2 precursor by Integrator (5, 66) or facilitated recycling back onto the promoter.
We also show by ChIP that the dedicated SNAPc transcription factor which binds to the PSE of the U2 snRNA promoter is normally removed (or becomes inaccessible to antibody) during metaphase (Fig. 7B) but is retained when locus-specific metaphase fragility of the U1, U2, and 5S genes is induced (110, 111) by loss of CSB function (Fig. 7C), low doses of actinomycin D (Fig. 7D), or by transduction with the tetrameric C-terminal domain of p53 (Fig. 7E). Although there are subtle differences between the human U1 and U2 snRNA transcription units (41, 42), the overall architecture of these genes is very similar, and it seems likely that retention of SNAPc during metaphase also explains RNU1 fragility. Since retention of SNAPc correlates with locus-specific chromosomal fragility, we speculate that the persistently open transcriptional state established by the dedicated U2 snRNA promoter must be disassembled to allow proper metaphase chromatin condensation. SNAPc also recruits TATA binding protein (TBP) to TATA-less U snRNA genes (41); this suggests that loss of TBP may contribute to disassembly and is consistent with previous observations that the U2 promoter binds transcription factors during interphase but not metaphase (8). In contrast, the TBP-containing transcription factor TFIID remains on many if not most TATA-containing mRNA promoters through mitosis, although Pol II itself is removed (16, 74).
Although loss of functional CSB protein induces locus-specific metaphase fragility of the U1, U2, and 5S genes (111), we were unable to detect CSB on U2 genes in either unsynchronized or metaphase cells (Fig. 7H) under conditions where we readily detected CSB on rRNA genes (Fig. 7G), as previously observed (11). Conceivably, transient binding of CSB to U2 genes may suffice to remove SNAPc and/or disassemble the open transcriptional state as metaphase approaches; some remodeling proteins, like mammalian BRG1, bind stably to target genes (98), whereas others, like yeast Isw2, have brief dwell times (32). Alternatively, CSB could bind U2 genes through an intermediary factor: CSB binds p53 (111), p53 is known to bind U1 snRNA genes which strongly resemble U2 genes (41), and binding of SWI/SNF chromatin remodeling factors, such as BRG1, to p53 is required for p53-driven activation of the p21 promoter (51). Indeed, many p53-driven promoters exist in a constitutively DNase I-hypersensitive state (10), not unlike the upstream region of the U2 coding region (Fig. 1, 2, and 4). A third scenario to explain why CSB and BRCA1 proteins cannot be detected by ChIP (Fig. 7H) is that loss of CSB (111), BRCA1, or BRCA2 may cause locus-specific fragility (Fig. 5A; Table 2) indirectly by sequestering chromatin remodeling or DNA repair factors elsewhere in the genome.
We found previously that at least five tandem copies of the U2 repeat unit are required to exhibit cytologically detectable locus-specific metaphase fragility (3); however, it is still unclear whether fragility is a cooperative property requiring at least five clustered genes or a stochastic property of individual genes in which experimentally detectable fragility increases with local gene density. A related possibility is that locus-specific fragility might be an intrinsic liability of tandemly repeated genes, not unlike the intrinsic recombinogenic properties of tandem arrays, which can undergo homologous recombination either by looping back on themselves in different registers or by aligning in or out of register with an identical array on the sister chromatid. In fact, the danger of expansion or contraction of tandem arrays appears to be so great that special mechanisms have evolved for suppression of transcription and recombination within ribosomal DNA arrays (13). On the other hand, a solitary uncondensed gene seems to have no cytologically detectable effect on chromatin condensation: for example, active genes are scattered throughout the inactive X chromosome (108), and the heat shock gene hsp70i maintains an open chromatin conformation throughout mitosis by recruiting protein phosphatase 2A locally to inactivate condensin complexes (107). The U1, U2, and 5S tandem arrays may simply increase the local concentration of independent transcription units, thereby magnifying the effect of any impediment to condensation throughout the array. For example, if locus-specific fragility reflects occasional damage to U2 genes or occasional stalling of Pol II by secondary structure in the nascent U2 snRNA or DNA template (Fig. 6), loss of BRCA1 might cause fragility by impairing degradation or disassembly of the stalled transcription or repair complexes (46, 106). Thus, the persistently open chromatin structure of the U2 snRNA genes may reflect a compromise between the benefits of rapid reinitiation and/or promoter clearance and the dangers of metaphase fragility.
Acknowledgments
We thank Michelle Baranski and Steve Hauschka for help with quantitative PCR; Denise Galloway for the PG-13/htert retrovirus; Brian Kennedy for WI38 cells; Piri Welcsh and Mary-Claire King for BRCA1- and BRCA2-deficient cell lines; and Nouria Hernandez for SNAPc 43 and 190 antibodies.
This work was supported by Public Health Service award GM-41624 from the National Institutes of Health.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Akman, S. A., J. H. Doroshow, and M. Dizdaroglu. 1990. Base modifications in plasmid DNA caused by potassium permanganate. Arch. Biochem. Biophys. 282202-205. [DOI] [PubMed] [Google Scholar]
- 2.Ares, M., Jr., J. S. Chung, L. Giglio, and A. M. Weiner. 1987. Distinct factors with Sp1 and NF-A specificities bind to adjacent functional elements of the human U2 snRNA gene enhancer. Genes Dev. 1808-817. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, A. D., Z. Li, T. Pavelitz, and A. M. Weiner. 1995. Adenovirus type 12-induced fragility of the human RNU2 locus requires U2 small nuclear RNA transcriptional regulatory elements. Mol. Cell. Biol. 156246-6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, A. D., T. Pavelitz, and A. M. Weiner. 1998. The microsatellite sequence (CT)n·(GA)n promotes stable chromosomal integration of large tandem arrays of functional human U2 small nuclear RNA genes. Mol. Cell. Biol. 182262-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baillat, D., M. A. Hakimi, A. M. Naar, A. Shilatifard, N. Cooch, and R. Shiekhattar. 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123265-276. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, L. B., T. Manser, and A. M. Weiner. 1985. Human U1 small nuclear RNA genes: extensive conservation of flanking sequences suggests cycles of gene amplification and transposition. Mol. Cell. Biol. 52159-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, D. C., I. H. Greger, and S. Murphy. 2000. In vivo footprinting studies suggest a role for chromatin in transcription of the human 7SK gene. Gene 24733-44. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, D. C., A. Pombo, and S. Murphy. 2003. Interaction of proteins with promoter elements of the human U2 snRNA genes in vivo. Gene. 315103-112. [DOI] [PubMed] [Google Scholar]
- 9.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl. Acad. Sci. USA 9513887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braastad, C. D., Z. Han, and E. A. Hendrickson. 2003. Constitutive DNase I hypersensitivity of p53-regulated promoters. J. Biol. Chem. 2788261-8268. [DOI] [PubMed] [Google Scholar]
- 11.Bradsher, J., J. Auriol, L. Proietti de Santis, S. Iben, J. L. Vonesch, I. Grummt, and J. M. Egly. 2002. CSB is a component of RNA pol I transcription. Mol. Cell 10819-829. [DOI] [PubMed] [Google Scholar]
- 12.Bradsher, J., F. Coin, and J. M. Egly. 2000. Distinct roles for the helicases of TFIIH in transcript initiation and promoter escape. J. Biol. Chem. 2752532-2538. [DOI] [PubMed] [Google Scholar]
- 13.Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis, and F. Winston. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12165-170. [DOI] [PubMed] [Google Scholar]
- 14.Bushnell, D. A., P. Cramer, and R. D. Kornberg. 2002. Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc. Natl. Acad. Sci. USA 991218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calvo, O., and J. L. Manley. 2003. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 171321-1327. [DOI] [PubMed] [Google Scholar]
- 16.Christova, R., and T. Oelgeschlager. 2002. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 479-82. [DOI] [PubMed] [Google Scholar]
- 17.Citterio, E., V. Van Den Boom, G. Schnitzler, R. Kanaar, E. Bonte, R. E. Kingston, J. H. Hoeijmakers, and W. Vermeulen. 2000. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 207643-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costanzo, G., S. Camier, P. Carlucci, L. Burderi, and R. Negri. 2001. RNA polymerase III transcription complexes on chromosomal 5S rRNA genes in vivo: TFIIIB occupancy and promoter opening. Mol. Cell. Biol. 213166-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuello, P., D. C. Boyd, M. J. Dye, N. J. Proudfoot, and S. Murphy. 1999. Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. EMBO J. 182867-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Vegvar, H. E., E. Lund, and J. E. Dahlberg. 1986. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell 47259-266. [DOI] [PubMed] [Google Scholar]
- 21.de Waard, H., J. de Wit, J. O. Andressoo, C. T. van Oostrom, B. Riis, A. Weimann, H. E. Poulsen, H. van Steeg, J. H. Hoeijmakers, and G. T. van der Horst. 2004. Different effects of CSA and CSB deficiency on sensitivity to oxidative DNA damage. Mol. Cell. Biol. 247941-7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dieci, G., and A. Sentenac. 1996. Facilitated recycling pathway for RNA polymerase III. Cell 84245-252. [DOI] [PubMed] [Google Scholar]
- 23.Eberhardy, S. R., and P. J. Farnham. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 27648562-48571. [DOI] [PubMed] [Google Scholar]
- 24.Egloff, S., D. O'Reilly, R. D. Chapman, A. Taylor, K. Tanzhaus, L. Pitts, D. Eick, and S. Murphy. 2007. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 3181777-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elco, C. P. 1998. Senior thesis. Yale University, New Haven, CT.
- 26.Fitzgerald, D. J., and J. N. Anderson. 1999. DNA distortion as a factor in nucleosome positioning. J. Mol. Biol. 293477-491. [DOI] [PubMed] [Google Scholar]
- 27.Frey, M. R., A. D. Bailey, A. M. Weiner, and A. G. Matera. 1999. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 9126-135. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda, A., T. Nakadai, M. Shimada, T. Tsukui, M. Matsumoto, Y. Nogi, M. Meisterernst, and K. Hisatake. 2004. Transcriptional coactivator PC4 stimulates promoter escape and facilitates transcriptional synergy by GAL4-VP16. Mol. Cell. Biol. 246525-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrity, P. A., and B. J. Wold. 1992. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc. Natl. Acad. Sci. USA 891021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehring, N. H., M. W. Hentze, and K. Pantopoulos. 1999. Inactivation of both RNA binding and aconitase activities of iron regulatory protein-1 by quinone-induced oxidative stress. J. Biol. Chem. 2746219-6225. [DOI] [PubMed] [Google Scholar]
- 31.Geiduschek, E. P., and G. A. Kassavetis. 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 3101-26. [DOI] [PubMed] [Google Scholar]
- 32.Gelbart, M. E., N. Bachman, J. Delrow, J. D. Boeke, and T. Tsukiyama. 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19942-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Granneman, S., and S. J. Baserga. 2005. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr. Opin. Cell Biol. 17281-286. [DOI] [PubMed] [Google Scholar]
- 34.Hanzlowsky, A., B. Jelencic, G. Jawdekar, C. S. Hinkley, J. H. Geiger, and R. W. Henry. 2006. Co-expression of multiple subunits enables recombinant SNAPC assembly and function for transcription by human RNA polymerases II and III. Protein Expr. Purif. 48215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez, N. 2001. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 27626733-26736. [DOI] [PubMed] [Google Scholar]
- 36.Hernandez, N., and A. M. Weiner. 1986. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell 47249-258. [DOI] [PubMed] [Google Scholar]
- 37.Hershkovitz, M., and A. D. Riggs. 1997. Ligation-mediated PCR for chromatin-structure analysis of interphase and metaphase chromatin. Methods 11253-263. [DOI] [PubMed] [Google Scholar]
- 38.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3405-411. [DOI] [PubMed] [Google Scholar]
- 39.Holstege, F. C., and H. T. Timmers. 1997. Analysis of open complex formation during RNA polymerase II transcription initiation using heteroduplex templates and potassium permanganate probing. Methods 12203-211. [DOI] [PubMed] [Google Scholar]
- 40.Jacobs, E. Y., I. Ogiwara, and A. M. Weiner. 2004. Role of the C-terminal domain of RNA polymerase II in U2 snRNA transcription and 3′ processing. Mol. Cell. Biol. 24846-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jawdekar, G. W., A. Hanzlowsky, S. L. Hovde, B. Jelencic, M. Feig, J. H. Geiger, and R. W. Henry. 2006. The unorthodox SNAP50 zinc finger domain contributes to cooperative promoter recognition by human SNAPC. J. Biol. Chem. 28131050-31060. [DOI] [PubMed] [Google Scholar]
- 42.Jawdekar, G. W., and R. W. Henry. Transcriptional regulation of human small nuclear RNA genes. Biochim. Biophys. Acta Gene Reg. Mech., in press. [DOI] [PMC free article] [PubMed]
- 43.Jiang, C., and D. Liao. 1999. Striking bimodal methylation of the repeat unit of the tandem array encoding human U2 snRNA (the RNU2 locus). Genomics 62508-518. [DOI] [PubMed] [Google Scholar]
- 44.Kassavetis, G. A., B. R. Braun, L. H. Nguyen, and E. P. Geiduschek. 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60235-245. [DOI] [PubMed] [Google Scholar]
- 45.Kireeva, M. L., B. Hancock, G. H. Cremona, W. Walter, V. M. Studitsky, and M. Kashlev. 2005. Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell 1897-108. [DOI] [PubMed] [Google Scholar]
- 46.Kleiman, F. E., F. Wu-Baer, D. Fonseca, S. Kaneko, R. Baer, and J. L. Manley. 2005. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev. 191227-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kugel, J. F., and J. A. Goodrich. 1998. Promoter escape limits the rate of RNA polymerase II transcription and is enhanced by TFIIE, TFIIH, and ATP on negatively supercoiled DNA. Proc. Natl. Acad. Sci. USA 959232-9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuhlman, T. C., H. Cho, D. Reinberg, and N. Hernandez. 1999. The general transcription factors IIA, IIB, IIF, and IIE are required for RNA polymerase II transcription from the human U1 small nuclear RNA promoter. Mol. Cell. Biol. 192130-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambrinakos, A., K. E. Humphrey, J. J. Babon, T. P. Ellis, and R. G. Cotton. 1999. Reactivity of potassium permanganate and tetraethylammonium chloride with mismatched bases and a simple mutation detection protocol. Nucleic Acids Res. 271866-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langst, G., T. Schatz, J. Langowski, and I. Grummt. 1997. Structural analysis of mouse rDNA: coincidence between nuclease hypersensitive sites, DNA curvature and regulatory elements in the intergenic spacer. Nucleic Acids Res. 25511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee, D., J. W. Kim, T. Seo, S. G. Hwang, E. J. Choi, and J. Choe. 2002. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 27722330-22337. [DOI] [PubMed] [Google Scholar]
- 52.Le Page, F., V. Randrianarison, D. Marot, J. Cabannes, M. Perricaudet, J. Feunteun, and A. Sarasin. 2000. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res. 605548-5552. [PubMed] [Google Scholar]
- 53.Li, Z., A. Yu, and A. M. Weiner. 1998. Adenovirus type 12-induced fragility of the human RNU2 locus requires p53 function. J. Virol. 724183-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao, D. 1999. Concerted evolution: molecular mechanism and biological implications. Am. J. Hum. Genet. 6424-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao, D., T. Pavelitz, J. R. Kidd, K. K. Kidd, and A. M. Weiner. 1997. Concerted evolution of the tandemly repeated genes encoding human U2 snRNA (the RNU2 locus) involves rapid intrachromosomal homogenization and rare interchromosomal gene conversion. EMBO J. 16588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao, D., T. Pavelitz, and A. M. Weiner. 1998. Characterization of a novel class of interspersed LTR elements in primate genomes: structure, genomic distribution, and evolution. J. Mol. Evol. 46649-660. [DOI] [PubMed] [Google Scholar]
- 57.Liao, D., and A. M. Weiner. 1995. Concerted evolution of the tandemly repeated genes encoding primate U2 small nuclear RNA (the RNU2 locus) does not prevent rapid diversification of the (CT)n·(GA)n microsatellite embedded within the U2 repeat unit. Genomics 30583-593. [DOI] [PubMed] [Google Scholar]
- 58.Liao, D., A. Yu, and A. M. Weiner. 1999. Coexpression of the adenovirus 12 E1B 55 kDa oncoprotein and cellular tumor suppressor p53 is sufficient to induce metaphase fragility of the human RNU2 locus. Virology 25411-23. [DOI] [PubMed] [Google Scholar]
- 59.Lindgren, V., M. Ares, Jr., A. M. Weiner, and U. Francke. 1985. Human genes for U2 small nuclear RNA map to a major adenovirus 12 modification site on chromosome 17. Nature 314115-116. [DOI] [PubMed] [Google Scholar]
- 60.Lis, J. T. 2007. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature 450198-202. [DOI] [PubMed] [Google Scholar]
- 61.Little, R. D., and D. C. Braaten. 1989. Genomic organization of human 5 S rDNA and sequence of one tandem repeat. Genomics 4376-383. [DOI] [PubMed] [Google Scholar]
- 62.Liu, J., S. Akoulitchev, A. Weber, H. Ge, S. Chuikov, D. Libutti, X. W. Wang, J. W. Conaway, C. C. Harris, R. C. Conaway, D. Reinberg, and D. Levens. 2001. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell 104353-363. [DOI] [PubMed] [Google Scholar]
- 63.Lu, T., Y. Pan, S. Y. Kao, C. Li, I. Kohane, J. Chan, and B. A. Yankner. 2004. Gene regulation and DNA damage in the ageing human brain. Nature 429883-891. [DOI] [PubMed] [Google Scholar]
- 64.Lund, E., and J. E. Dahlberg. 1984. True genes for human U1 small nuclear RNA. Copy number, polymorphism, and methylation. J. Biol. Chem. 2592013-2021. [PubMed] [Google Scholar]
- 65.Ma, B., and N. Hernandez. 2001. A map of protein-protein contacts within the small nuclear RNA-activating protein complex SNAPc. J. Biol. Chem. 2765027-5035. [DOI] [PubMed] [Google Scholar]
- 66.Mandel, C. R., S. Kaneko, H. Zhang, D. Gebauer, V. Vethantham, J. L. Manley, and L. Tong. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mangin, M., M. Ares, Jr., and A. M. Weiner. 1986. Human U2 small nuclear RNA genes contain an upstream enhancer. EMBO J. 5987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mangin, M., M. Ares, Jr., and A. M. Weiner. 1985. U1 small nuclear RNA genes are subject to dosage compensation in mouse cells. Science 229272-275. [DOI] [PubMed] [Google Scholar]
- 69.Martin, M., J. Cho, A. J. Cesare, J. D. Griffith, and G. Attardi. 2005. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell 1231227-1240. [DOI] [PubMed] [Google Scholar]
- 70.Matera, A. G., A. M. Weiner, and C. W. Schmid. 1990. Structure and evolution of the U2 small nuclear RNA multigene family in primates: gene amplification under natural selection? Mol. Cell. Biol. 105876-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mayer, C., and I. Grummt. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 256384-6391. [DOI] [PubMed] [Google Scholar]
- 72.Medlin, J., A. Scurry, A. Taylor, F. Zhang, B. M. Peterlin, and S. Murphy. 2005. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 244154-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Medlin, J. E., P. Uguen, A. Taylor, D. L. Bentley, and S. Murphy. 2003. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 22925-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Michelotti, E. F., S. Sanford, and D. Levens. 1997. Marking of active genes on mitotic chromosomes. Nature 388895-899. [DOI] [PubMed] [Google Scholar]
- 75.Moisan, A., C. Larochelle, B. Guillemette, and L. Gaudreau. 2004. BRCA1 can modulate RNA polymerase II carboxy-terminal domain phosphorylation levels. Mol. Cell. Biol. 246947-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mosesso, P., S. Penna, G. Pepe, C. Lorenti-Garcia, and F. Palitti. 2004. Potassium bromate but not X-rays cause unexpectedly elevated levels of DNA breakage similar to those induced by ultraviolet light in Cockayne syndrome (CS-B) fibroblasts. Cytogenet. Genome Res. 104178-181. [DOI] [PubMed] [Google Scholar]
- 77.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246780-786. [DOI] [PubMed] [Google Scholar]
- 78.Nagahara, H., A. M. Vocero-Akbani, E. L. Snyder, A. Ho, D. G. Latham, N. A. Lissy, M. Becker-Hapak, S. A. Ezhevsky, and S. F. Dowdy. 1998. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat. Med. 41449-1452. [DOI] [PubMed] [Google Scholar]
- 79.Newman, J. C., A. D. Bailey, and A. M. Weiner. 2006. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc. Natl. Acad. Sci. USA 1039613-9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Newman, J. C., A. D. Bailey, H.-Y. Fan, T. Pavelitz, and A. M. Weiner. 2008. An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 4e1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nilsen, T. W. 2003. The spliceosome: the most complex macromolecular machine in the cell? Bioessays 251147-1149. [DOI] [PubMed] [Google Scholar]
- 82.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2599-104. [DOI] [PubMed] [Google Scholar]
- 83.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11205-214. [DOI] [PubMed] [Google Scholar]
- 84.Panov, K. I., J. K. Friedrich, J. Russell, and J. C. Zomerdijk. 2006. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 253310-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pavelitz, T., D. Liao, and A. M. Weiner. 1999. Concerted evolution of the tandem array encoding primate U2 snRNA (the RNU2 locus) is accompanied by dramatic remodeling of the junctions with flanking chromosomal sequences. EMBO J. 183783-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pavelitz, T., L. Rusche, A. G. Matera, J. M. Scharf, and A. M. Weiner. 1995. Concerted evolution of the tandem array encoding primate U2 snRNA occurs in situ, without changing the cytological context of the RNU2 locus. EMBO J. 14169-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perriman, R. J., and M. Ares, Jr. 2007. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 21811-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Preuss, S., and C. S. Pikaard. 2007. rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochim. Biophys. Acta 1769383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rapin, I., Y. Lindenbaum, D. W. Dickson, K. H. Kraemer, and J. H. Robbins. 2000. Cockayne syndrome and xeroderma pigmentosum. Neurology 551442-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rincon, J. C., S. K. Engler, B. W. Hargrove, and G. R. Kunkel. 1998. Molecular cloning of a cDNA encoding human SPH-binding factor, a conserved protein that binds to the enhancer-like region of the U6 small nuclear RNA gene promoter. Nucleic Acids Res. 264846-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sandmeier, J. J., S. French, Y. Osheim, W. L. Cheung, C. M. Gallo, A. L. Beyer, and J. S. Smith. 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 214959-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schroder, O., E. P. Geiduschek, and G. A. Kassavetis. 2003. A single-stranded promoter for RNA polymerase III. Proc. Natl. Acad. Sci. USA 100934-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Singh, B. N., and M. Hampsey. 2007. A transcription-independent role for TFIIB in gene looping. Mol. Cell 27806-816. [DOI] [PubMed] [Google Scholar]
- 94.Tchou, J., V. Bodepudi, S. Shibutani, I. Antoshechkin, J. Miller, A. P. Grollman, and F. Johnson. 1994. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J. Biol. Chem. 26915318-15324. [PubMed] [Google Scholar]
- 95.Tomlinson, G. E., T. T. Chen, V. A. Stastny, A. K. Virmani, M. A. Spillman, V. Tonk, J. L. Blum, N. R. Schneider, I. I. Wistuba, J. W. Shay, J. D. Minna, and A. F. Gazdar. 1998. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 583237-3242. [PubMed] [Google Scholar]
- 96.Toulokhonov, I., J. Zhang, M. Palangat, and R. Landick. 2007. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell 27406-419. [DOI] [PubMed] [Google Scholar]
- 97.Troelstra, C., A. van Gool, J. de Wit, W. Vermeulen, D. Bootsma, and J. H. Hoeijmakers. 1992. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell 71939-953. [DOI] [PubMed] [Google Scholar]
- 98.Tu, N., Y. Hu, and N. F. Mivechi. 2006. Heat shock transcription factor (Hsf)-4b recruits Brg1 during the G1 phase of the cell cycle and regulates the expression of heat shock proteins. J. Cell Biochem. 981528-1542. [DOI] [PubMed] [Google Scholar]
- 99.Uguen, P., and S. Murphy. 2003. The 3′ ends of human pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. EMBO J. 224544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Van Arsdell, S. W., and A. M. Weiner. 1984. Human genes for U2 small nuclear RNA are tandemly repeated. Mol. Cell. Biol. 4492-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van der Drift, P., A. Chan, G. Laureys, N. van Roy, G. Sickmann, J. den Dunnen, A. Westerveld, F. Speleman, and R. Versteeg. 1995. Balanced translocation in a neuroblastoma patient disrupts a cluster of small nuclear RNA U1 and tRNA genes in chromosomal band 1p36. Genes Chromosomes Cancer 1435-42. [DOI] [PubMed] [Google Scholar]
- 102.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 216820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Welcsh, P. L., M. K. Lee, R. M. Gonzalez-Hernandez, D. J. Black, M. Mahadevappa, E. M. Swisher, J. A. Warrington, and M. C. King. 2002. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc. Natl. Acad. Sci. USA 997560-7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Westin, G., J. Zabielski, K. Hammarstrom, H. J. Monstein, C. Bark, and U. Pettersson. 1984. Clustered genes for human U2 RNA. Proc. Natl. Acad. Sci. USA 813811-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wong, H. K., M. Muftuoglu, G. Beck, S. Z. Imam, V. A. Bohr, and D. M. Wilson III. 2007. Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 354103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu, W., H. Nishikawa, R. Hayami, K. Sato, A. Honda, S. Aratani, T. Nakajima, M. Fukuda, and T. Ohta. 2007. BRCA1 ubiquitinates RPB8 in response to DNA damage. Cancer Res. 67951-958. [DOI] [PubMed] [Google Scholar]
- 107.Xing, H., D. C. Wilkerson, C. N. Mayhew, E. J. Lubert, H. S. Skaggs, M. L. Goodson, Y. Hong, O. K. Park-Sarge, and K. D. Sarge. 2005. Mechanism of hsp70i gene bookmarking. Science 307421-423. [DOI] [PubMed] [Google Scholar]
- 108.Yasuhara, J. C., and B. T. Wakimoto. 2006. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 22330-338. [DOI] [PubMed] [Google Scholar]
- 109.Yoon, J. B., S. Murphy, L. Bai, Z. Wang, and R. G. Roeder. 1995. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol. Cell. Biol. 152019-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu, A., A. D. Bailey, and A. M. Weiner. 1998. Metaphase fragility of the human RNU1 and RNU2 loci is induced by actinomycin D through a p53-dependent pathway. Hum. Mol. Genet. 7609-617. [DOI] [PubMed] [Google Scholar]
- 111.Yu, A., H. Y. Fan, D. Liao, A. D. Bailey, and A. M. Weiner. 2000. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol. Cell 5801-810. [DOI] [PubMed] [Google Scholar]
- 112.Zhao, X., P. S. Pendergrast, and N. Hernandez. 2001. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol. Cell 7539-549. [DOI] [PubMed] [Google Scholar]
- 113.zur Hausen, H. 1967. Induction of specific chromosomal aberrations by adenovirus type 12 in human embryonic kidney cells. J. Virol. 11174-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]