Abstract
Cell proliferation is regulated by the induction of growth promoting genes and the suppression of growth inhibitory genes. Malignant growth can result from the altered balance of expression of these genes in favor of cell proliferation. Induction of the transcription factor, c-Myc, promotes cell proliferation and transformation by activating growth promoting genes, including the ODC and cdc25A genes. We show that c-Myc transcriptionally represses the expression of a growth arrest gene, gas1. A conserved Myc structure, Myc box 2, is required for repression of gas1, and for Myc induction of proliferation and transformation, but not for activation of ODC. Activation of a Myc-estrogen receptor fusion protein by 4-hydroxytamoxifen was sufficient to repress gas1 gene transcription. These findings suggest that transcriptional repression of growth arrest genes, including gas1, is one step in promotion of cell growth by Myc.
c-Myc is a strong potentiator of proliferation and tumorigenesis. Its expression results in a reduced capacity for cells to differentiate and an increased potential to cycle (1–4). Cells with deregulated Myc expression are good targets for additional mutations that lead to tumor formation. Although c-myc is able to immortalize primary cells in culture, its deregulated expression alone is not sufficient to produce the malignant phenotype. c-myc cooperates with an activated ras oncogene in the transformation of rat embryo fibroblasts (REFs) (5). By using ras cooperation as an assay, Myc’s transforming activity has been found to be dependent on its N-terminal transactivation domain as well as its C-terminal basic region/helix-loop-helix/leucine zipper (LZ) domain (6–10).
Myc is a transcription factor, and its role in transformation has thus far been mainly ascribed to the enhancement of expression of growth promoting genes. A number of Myc-inducible genes have been identified (11–19). Among these, the ornithine decarboxylase (ODC) and cdc25A genes stand out as targets for Myc activation because of their involvement in cell cycle progression. We and others have reported (9, 20, 21) that Myc is also capable of repressing gene expression through actions dependent upon initiator sequences in the regulated gene promoter. In the cases of the adenovirus-2 major late and C/EBPα promoters, Myc repression has also been shown to be dependent upon a small region in the Myc N terminus known as Myc box 2 (MB2; amino acids 129–143) which is highly conserved among Myc family members, and upon the basic region/helix-loop-helix/LZ domain, as well as upon the integrity of the core promoter initiator sequence (9, 20, 21). Deletion of Myc box 2 not only abrogates repression of transcription, but also results in failure of Myc to cooperate with activated Ras in REF transformation (6–10). A box 2 deletion mutant of Myc that does not repress (ΔMB2), however, retained its ability to transactivate the ODC promoter (10, 14). These observations raise the possibility that a function dependent upon box 2, such as transcription repression, is required for Myc induction of growth and for Ras cooperation.
When quiescent cells are stimulated by serum, expression of growth arrest-specific (gas) mRNAs is suppressed and cells transit from G0 to G1 (22, 23). Repression of gas gene expression coincides with the induction of immediate early mitogenic response genes, such as c-myc. Gas1 is a membrane-associated protein that blocks the G0-to-S phase transition of quiescent fibroblasts when ectopically expressed, without affecting the early serum response (23). Gas1 also blocks cell proliferation in several transformed cell lines except those transformed by simian virus 40 or adenovirus (23, 24). Gas1 activates a transactivation-independent p53-mediated growth arrest function (25, 26). We report here that c-Myc represses transcription of gas1 and suggest that Myc’s promotion of cell growth depends, in part, on repression of growth arrest genes, as well as upon inducing genes for growth stimulatory functions.
MATERIALS AND METHODS
Plasmids.
Plamids expressing the wild-type human c-myc gene regulated by the long terminal repeat of the Moloney murine leukemia virus (pLTR-Hm) or the cytomegalovirus major immediate early promoter (pCMV-Hm) and plasmids pLTR-HmΔMB2 (c-myc-deleted of sequences encoding aa 132–143 of MB2) and pCMV-HmΔLZ (c-myc-deleted of sequences encoding aa 411–439) have been described (9, 10, 32). pCMV-HmΔMB2 was constructed by subcloning the HindIII/EcoRI fragment of pLTR-HmΔMB2 into pcDNAI (Invitrogen). Reporter constructs pGas1-Luc and pODC-chloramphenicol acetyltransferase (CAT) were the kind gifts of R. de Martin and J. Cleveland, respectively.
Transfections.
Transformation assays in Rat-1a cells were performed by cotransfection of 1 μg of plasmid Rous sarcoma virus-neo (Invitrogen) with 5 μg of pLTR-Hm or pLTR-HmΔMB2 by the calcium phosphate DNA precipitation method followed by G418 (1,500 μg/ml) selection. Photomicrographs were taken with 320× magnification after 4 weeks of selection. Transient transfections were performed in Rat-1 or NIH 3T3 cells as described (9). pGas1-luc (5 μg) or pODC-CAT (4 μg) was cotransfected with 1 μg of pCMV-Hm, pCMV-Hm ΔMB2, pCMV-Hm ΔLZ, or empty vector pcDNA. Transfection efficiency was controlled by using a cotransfected pCMV-β-galactosidase (0.1 μg) plasmid. Luciferase or CAT activity was determined 48 hr later and normalized for β-galactosidase activity. All relative luciferase or CAT activities were then normalized to that of the empty vector (pcDNA) alone.
Northern Blot Analysis.
RNA was isolated by a modification (Ultraspec, Biotecx Laboratories, Houston) of the guanidinium acid-phenol extraction method (27), fractionated on 1% agarose-6% formaldehyde gels and transferred to Duralon-UV membranes (Stratagene) by capillary blotting. After UV cross-linking, membranes were hybridized sequentially after stripping to cDNA probes to gas1, c-myc, and glyceraldehyde-3-phosphate dehydrogenase (gapdh) labeled with [α-32P]dCTP by random priming (Boehringer Mannheim). Northern blot analysis for gas1 mRNA was performed by using a 2-kb EcoRI/BamHI fragment from pA1 (23), and for c-myc mRNA by using a HindIII/EcoRI fragment of pCMV-Hm. The gapdh cDNA probe was obtained from CLONTECH. Quantitation of autoradiographs was performed by using the nih image software.
Western Blot Analysis.
For detection of Gas1 or c-Myc, 50 μg of whole cell lysate (100 μg for transiently transfected cells) were fractionated by SDS/PAGE (8% acrylamide), transferred to a nitrocellulose membrane, and immunoblotted with a polyclonal antiserum to murine Gas1 (23) or with a mAb to c-Myc (Santa Cruz Biotechnology). Myc-estrogen receptor fusion protein (MycER) and ΔMycER fusion proteins were immunoprecipitated from whole cell extracts prepared from ≈5 × 106 cells [Rat-1 control, MycER, and ΔMycER cells before and after a 4 hr 4-hydroxytamoxifen (4-OHT) treatment] with 2 μg of a mAb raised against a C-terminal peptide of the ER (Santa Cruz Biotechnology). Western blot analysis of the fusion proteins was performed by using a mAb to human c-Myc (9E10, Santa Cruz Biotechnology).
Flow Cytometric Analysis.
Cell cycle distribution experiments were performed as described (28) by using a FACScan flow cytometer (Becton Dickinson). Briefly, 5 × 105 cells were pelleted by centrifugation and resuspended in 0.5 ml PBS supplemented with 2% (vol/vol) fetal calf serum, and fixed in 100% ethanol. The fixed cells were pelleted and resuspended in 0.75 ml of PBS (with 2% serum), stained with propidium iodide, and treated with RNase A. These cells were then analyzed by flow cytometry for DNA content.
Nuclear Run-Off Assay.
Nuclei were prepared and nuclear run-off transcription assays were performed as described (29) with the following modifications. Five μg of linearized gas1 or gapdh plasmid DNA or empty Bluescript (pBS) plasmid DNA were immobilized onto nitrocellulose membrane. 32P-labeled transcripts (15 × 106 cpm) were hybridized to DNA probes at 62°C for 6 hr in QuikHyb solution (Stratagene) in a Hybaid (Middlesex, U.K.) hybridization oven. Hybridized membranes were washed twice in 2× standard saline citrate (SSC)/0.1% SDS for 15 min at room temperature, followed by 0.1× SSC/0.1% SDS for 30 min at 57°C, followed by incubation with 10 mg/ml RNase A at 37°C for 30 min. The filters were rinsed in 0.1× SSC/0.1% SDS, air dried, and exposed to autoradiographic film at −80°C.
RESULTS
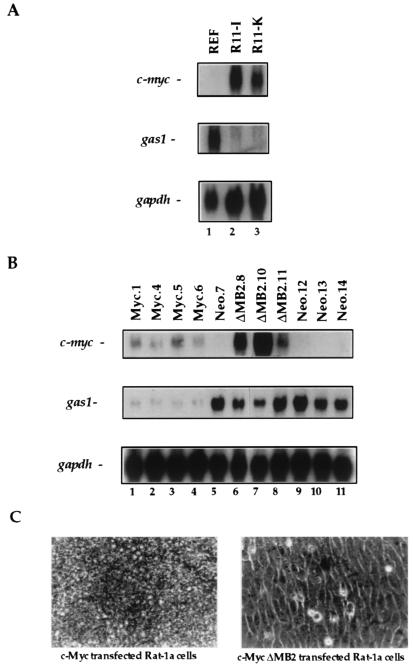
To establish the role of Myc in gas1 regulation, we examined the expression of gas1 in Myc-transformed cells. REFs were stably transformed by cotransfection of c-myc and activated H-ras plasmids. RNA was prepared from untransfected REFs as well as from two clones of Myc-Ras transformed REFs. Northern blot analysis was performed by using cDNA probes for gas1 and c-myc. The Myc-Ras transformed REFs (lanes 2 and 3, Fig. 1A) expressed high levels of c-myc and barely detectable levels of gas1. In contrast, the untransformed control REFs (lane 1) demonstrated no detectable c-myc expression, but very high gas1 mRNA expression. Thus, gas1 expression was suppressed in the presence of myc overexpression. Because gas1 mRNA levels are significantly lower in K-ras-transformed NIH 3T3 cells than in NIH 3T3 cells (30), it was not clear from our results whether suppression of gas1 was an effect of expression of Myc, Ras, or both proteins. Because c-Myc overexpression alone (in the absence of mutant Ras) is able to transform Rat-1a fibroblasts (12, 31), we generated Myc-transformed Rat-1a cell lines. In addition, we generated stable Rat-1a cell lines constitutively expressing the box 2 deletion mutant, ΔMB2. The latter cell lines were not morphologically transformed, in contrast to those stably expressing exogenous wild-type c-Myc (Fig. 1C), in agreement with earlier reports that cooperation of Myc with Ras in REF cell transformation depends on MB2 (9, 10, 12, 31). Expression of c-myc resulted in massive apoptosis making isolation of cell lines with high c-myc levels difficult. However, even in the Myc-transformed Rat-1a cell lines that expressed only low levels of exogenous c-myc, gas1 mRNA expression was strongly repressed (lanes 1–4, Fig. 1B). In contrast, cell lines stably expressing ΔMB2, even at extremely high levels, continued to express abundant gas1 mRNA (lanes 6–8, Fig. 1B). Control Rat-1a cell lines stably transfected with the neomycin-containing empty vector (lanes 5, 9–11, Fig. 1B) demonstrated no detectable c-myc expression, and abundant gas1 expression. Thus, repression of the gas1 gene correlated with Myc expression and box 2 function.
Figure 1.
Gas1 expression is repressed in Myc-transformed cells. (A) REFs were cotransfected with c-myc and activated H-ras expression plasmids and transformed foci were isolated (9, 10). Randomly selected clones or untransfected REFs were grown to confluence, then cultured in 0.5% fetal calf serum/DMEM for 20 hr. RNA was analyzed by Northern blotting with gas1 and c-myc cDNA probes. A gapdh probe was used to control for RNA loading. (B) Rat-1a cells were stably transfected with expression vectors containing wild-type c-myc (Myc), c-myc deleted of sequences encoding amino acids 132–143 (ΔMB2), or the empty vector; and cell lines were generated. Cells from randomly selected clones were grown to confluence, then cultured in serum-deprived medium as above. RNA was analyzed as in A. (C) Photomicrographs of representative Rat-1a/Myc and Rat-1a/ΔMB2 clones. Magnification, ×320.
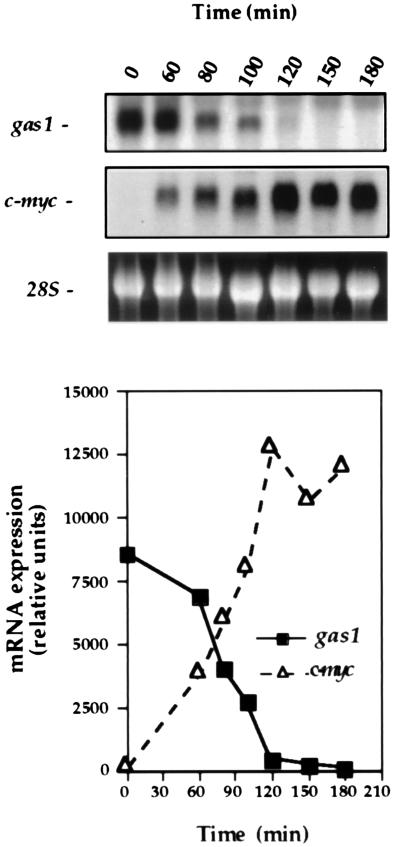
To establish further the role of Myc in gas1 regulation, we determined the kinetics of gas1 expression relative to the immediate early induction of c-myc by serum. Rat-1 fibroblasts were grown to confluence, made quiescent by serum-deprivation through culture in 0.1% fetal bovine serum (FBS)/DMEM for 72 hr (93% of cells were in G0/G1 by fluorescence-activated cell sorter analysis), and then stimulated with FBS (20% final concentration). RNA was harvested at the indicated times over the course of 180 min and underwent Northern blot analysis (Fig. 2). c-myc RNA expression was clearly elevated within 60 min of serum treatment. A decline of gas1 mRNA levels was detected at 80 min, immediately following appearance of c-myc mRNA, and gas1 mRNA levels continued to decline progressively such that they were undetectable by 150–180 min postserum treatment. A similar suppression of gas1 has been observed in NIH 3T3 cells following exposure to serum (22–25). These data show that gas1 mRNA repression closely follows c-myc expression in a serum-stimulated cell.
Figure 2.
Induction of Myc expression is followed by repression of gas1 mRNA expression. Rat-1 cells were made quiescent by growing to confluence followed by serum-deprivation in 0.1% FBS/DMEM for 72 hr, and then were stimulated with serum (20% FBS, final concentration) for the indicated times. RNA underwent Northern blot analysis for gas1 and c-myc expression. The 28S rRNA band in the ethidium bromide stained gel is shown as a control for RNA loading. Hybridization signals were quantitated by using nih image software.
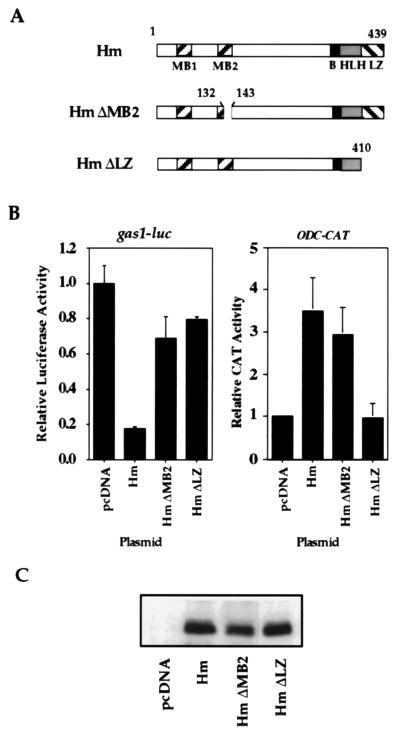
To examine the functional relationship between the Myc protein and gas1 transcription, we measured changes in gas1 promoter activity upon exogenous expression of Hm, or Myc deleted of amino acids 132–143 of MB2 (Hm ΔMB2), or Myc deleted of amino acids distal to position 410 (32), a mutation that removes the entire leucine repeat (Hm ΔLZ, Fig. 3A). Both Hm ΔMB2 and Hm ΔLZ fail to cooperate with activated Ras in REF transformation assays (6–10). A luciferase reporter gene dependent on 3.5 kb of the 5′ flanking region of the gas1 promoter (33) was cotransfected into Rat-1 cells together with the above Myc expression plasmids (Fig. 3B). Wild-type Myc repressed gas1 promoter activity ≈6-fold, whereas ΔMB2 repressed gas1 promoter activity only 1.4-fold. Similar results were observed in NIH 3T3 cells (T.C.L. and E.B.Z., unpublished observations). Although ΔMB2 was severely defective in gas1 transcriptional repression, its transactivation of the ODC promoter was comparable to that of wild-type Myc (Fig. 3B) as reported (10, 14). As a control, ΔLZ, which is defective in dimerization with Max (32), and which is unable to cooperate with activated Ras to transform REFs, failed to repress the gas1 promoter (1.3-fold repression), and did not induce transcription of the ODC promoter. The effect on these promoters of a double mutant with deletion of MB2 and the LZ was not tested. Western blot analysis shows that the ectopic proteins were all expressed at similar levels (Fig. 3C). These experiments show that Myc transcriptionally represses the gas1 promoter by a mechanism dependent on MB2 and the LZ.
Figure 3.
Myc represses gas1 promoter activity. (A) Schematic representation of c-Myc and mutants used. (B) Gas1 and ODC reporter plasmids (14, 33) were cotransfected into Rat-1 cells together with the c-myc expression vector pCMV-Hm (32), or with plasmids expressing the MB2 or LZ deletion mutants of myc, or with empty vector. All transfections included a pCMV-β-galactosidase plasmid to normalize for transfection efficiency. (C) Western blot analysis of wild-type and mutant Myc proteins following transient transfection.
The findings that expression of c-myc in vivo is followed closely by repression of gas1, and that ectopically expressed Myc represses gas1 promoter activity, both suggest that Myc itself is a negative regulator of the gas1 gene. To determine whether Myc protein activation is sufficient to repress gas1, we employed a 4-OHT-specific inducible MycER protein in nontransformed Rat-1 cells (34, 35). In this system, the human c-myc coding sequence is fused in frame to sequences encoding the hormone binding domain of the murine ER. The system employs a mutant of the hormone binding domain that selectively binds 4-OHT, a synthetic analog of 17-β-estradiol, but not 17-β-estradiol itself, which is present in serum (34, 35), thus facilitating the culture of these cells during the experiment. In addition, the mutation of the ER hormone binding domain abolishes the inherent ligand-dependent transactivation activity of the ER, thereby permitting study of transcriptional control by exogenous domains in hormone binding domain fusion proteins (35). The MycER fusion protein is expressed constitutively in Rat1/MycER cells, but is inactive until stimulation with 4-OHT derepresses the hormone-binding domain of the fusion protein. This induction, which occurs within minutes of treatment, enables the Myc portion to function (12, 31, 34–37). Parental Rat-1 cells and Rat-1 cells expressing a mutant, ΔMycER (Rat1/ΔMycER), which has a deletion of amino acids 106–143 spanning the MB2 region, were used as controls. The amino acids 106–143 deletion mutant transactivates the ODC promoter (10, 14), but in the ΔMycER fusion, it is defective in transformation (12, 31), transactivation of the cdc25A cell-cycle phosphatase gene (19), autosuppression (37), and stimulation of apoptosis (34, 35).
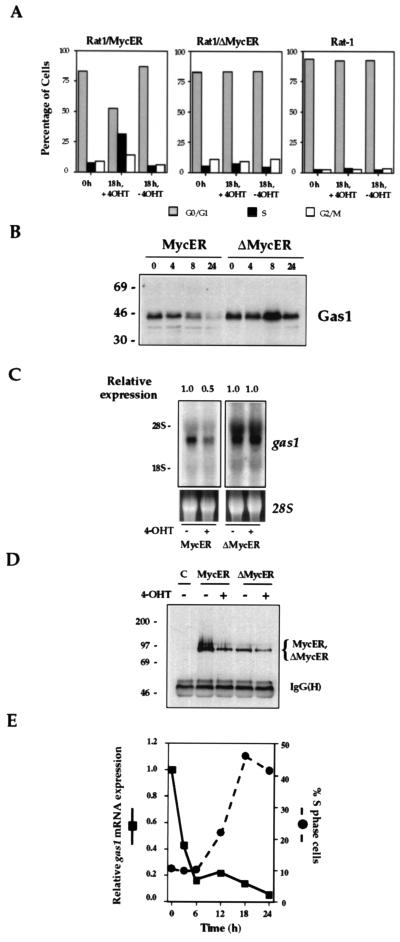
Rat1/MycER cells and control Rat1/ΔMycER or Rat-1 cells were grown to confluence and deprived of serum for 72 hr. Quiescent cells were then treated with 4-OHT for 18 hr and analyzed for cell cycle distribution by fluorescence activated cytometric scanning. Also, cells were treated for 0, 4, 8, and 24 hr to activate MycER and assayed for gas1 expression. Cells made quiescent by serum-starvation or by high density growth conditions express high levels of gas1 (22–24). Activation of MycER resulted in a transition of the cells from G0/G1 to S phase (Fig. 4A), and a decrease of Gas1 protein levels (Fig. 4B). The increase in Gas1 protein levels at 8 hr following 4-OHT treatment in ΔMycER cells was not observed in repeat experiments. gas1 mRNA levels were clearly suppressed by 4 hr following 4-OHT treatment (Fig. 4C). 4-OHT treatment of control Rat1/ΔMycER or Rat-1 did not elicit these effects (Figs. 4 A–C). MycER and ΔMycER fusion proteins were expressed at similar levels both before and after 4-OHT treatment (Fig. 4D). These observations indicate that induction of Myc in the absence of other cellular treatments, such as serum stimulation, is sufficient to repress gas1 expression in vivo and that this repression is dependent on a region in Myc that spans MB2. Also, repression of gas1 expression by activated MycER is associated with a G1-to-S phase transition.
Figure 4.
Activation of MycER is sufficient to cause entry into S phase and to repress gas1 expression. (A) Cell cycle distribution of 0 and 18 hr 4-OHT-treated and -untreated Rat1/MycER, Rat1/ΔMycER, and control Rat-1 cells. Cells were made quiescent as in Fig. 2, then stimulated with 4-OHT (200 nM) or ethanol carrier for 18 hr. Cells were harvested, fixed in ethanol, and stained with propidium iodide. DNA content was measured to assess cell cycle distribution. (B) Western blot analysis of Gas1 expression following 4-OHT treatment (0, 4, 8, and 24 hr) of quiescent MycER and ΔMycER cells. (C) Northern blot analysis and quantitation of gas1 mRNA expression in quiescent MycER and ΔMycER cells either untreated or following 4-OHT treatment (4 hr). (D) Western detection of immunoprecipitated MycER and ΔMycER fusion proteins (see brace) from whole cell extracts before and after a 4 hr 4-OHT treatment of MycER and ΔMycER cells. Rat-1 cells were used as a negative control. The lower band represents the Ig heavy chain [IgG(H)]. (E) Correlation of entry into S phase with repression of gas1 expression upon MycER activation. Quiescent MycER cells were treated with 4-OHT (200 nM) for the indicated times. For each time point, RNA was extracted for Northern blot analysis of gas1 and gapdh expression and fluorescence-activated cell sorter analysis was performed. Shown is the relative gas1 mRNA expression (defined as the ratio of quantitated gas1/gapdh band intensities, normalized to the ratio at time 0 hr) and percentage of cells in S phase plotted as a function of time following 4-OHT treatment.
To determine the kinetic relationship between Myc repression of gas1 expression and progression of cells through G1 into S, we measured the decline of gas1 mRNA and the change in cell cycle distribution of quiescent Rat1/MycER cells following treatment with 4-OHT over a 24-hr period. The high level of gas1 mRNA in quiescent cells fell ≈60% within 3 hr of 4-OHT treatment, 80% by 6 hr, and decreased to only 4% of its initial level by 24 hr (Fig. 4E). Fluorescence-activated cell sorter analysis of these cells, however, indicated that the S phase population did not increase until 12 hr following 4-OHT treatment, whereupon it started to rise. Because Myc repression of gas1 expression in vivo commenced well in advance of the increase in S phase cells, gas1 repression is unlikely to be simply a consequence of changes in cell cycle distribution.
To determine whether Myc represses gas1 at the transcriptional level, we analyzed the Myc-induced changes in gas1 transcription by nuclear run-off assays. Rat1/MycER cells were grown to confluence, deprived of serum by incubation in 0.1% FBS/DMEM for 72 hr, and then treated with 4-OHT for 0, 1, and 3 hr. Nuclei were isolated, and transcription run-off assays were performed. At 1 hr following 4-OHT treatment, gas1 transcription was reduced to 76% of the initial rate (Fig. 5 A and B). At 3 hr, gas1 transcription dropped to 17% of the initial rate. Transcription of the gapdh gene, a positive control, did not change during this period of 4-OHT treatment. Fig. 5B shows that the decrease in the rate of gas1 transcription over 3 hr is linear and suggests that repression of gas1 transcription begins almost immediately following MycER activation. These results indicate that negative regulation of gas1 gene expression by 4-OHT activation of MycER is at the level of transcription, consistent with Myc action at the gas1 promoter.
Figure 5.
Rate of gas1 gene transcription following activation of MycER. (A) Rat1/MycER cells were made quiescent as described in Figs. 2 and 4, and treated for 0, 1, and 3 hr with 4-OHT (200 nM). Nuclei were prepared and nuclear run-off transcription assays were performed as described in Materials and Methods. 32P-labeled transcripts were hybridized to DNA probes immobilized on nitrocellulose membranes. Blots were visualized by autoradiography. (B) Quantitation of blots was performed by using hih image software. gas1 band intensities were normalized to that of gapdh, and the amounts relative to that at 0 hr are shown.
DISCUSSION
Constitutively high Myc expression can override growth arrest and results in decreased growth factor requirements, increased growth rate, and a shorter G1 phase (4). Activation of c-Myc by a MycER system similar to the one used in our studies is sufficient to induce DNA synthesis in quiescent cells (12). c-Myc, therefore, makes essential contributions to cellular progression through the G1 phase of the cell cycle.
The role that Myc plays in the cell cycle, particularly in G1, remains unclear. However, evidence to date suggests that Myc may regulate diverse cellular pathways. In several cases, Myc is working as a gene activator. Two genes, the ODC and cdc25A genes, both with oncogenic properties (38–40), are cell cycle targets under direct positive regulation by Myc. ODC encodes the rate-limiting enzyme for polyamine biosynthesis essential for progression into S phase (14). Cdc25A encodes a cell cycle cyclin-dependent kinase activating phosphatase (19). Both genes can be induced in serum-starved MycER cells by estrogen (16, 19), and respond to c-Myc by an E-box-dependent mechanism (14, 19). Other cell cycle regulators, including cyclin A and cyclin E, as well as the cad gene that encodes an enzyme involved in pyrimidine biosynthesis, are also induced in response to Myc (15, 18). In addition, Myc induces expression of the α-prothymosin (12, 41) and ECA39 (13) genes although their functions are not known.
We present the finding that Myc activation can repress transcription of the gas1 gene, a cell cycle inhibitory gene. By using the 4-OHT regulated MycER system to activate Myc in the absence of other external stimuli, we show that repression is rapid and commences almost immediately following 4-OHT treatment, well in advance of any measurable changes in the cell cycle distribution of the 4-OHT stimulated Rat1/MycER cells. This indicates that suppression of gas1 expression is not an indirect effect of entry of cells into S phase. Repression of gas1 transcription is followed by a decline in the levels of gas1 mRNA and Gas1 protein. Because Gas1 arrests cell proliferation (23), transcriptional repression of gas1 by Myc decreases the levels of a protein that inhibits cell cycling. Other inhibitors of cell cycling that are repressed by Myc include C/EBPα (9, 42) and p21WAF1/CIP1 (G. Y. Yan and E.B.Z., unpublished observation). Myc also represses the cyclin-dependent kinase inhibitor p27 at the protein level, but not the mRNA level; however the regulatory mechanism is not known (43).
Myc appears to be a protein with many targets, and Myc may therefore serve as a master regulator of expression of cell cycle-related genes. Some targets of Myc, such as ODC and cdc25A, function on growth stimulatory pathways. Here we show a second activity of Myc, repression of a gene that functions on a growth inhibitory pathway. Other oncoproteins including Myb (44, 45), E1A (46–49), Fos (50, 51), and Jun (50) also have dual regulatory functions and work both as transcriptional activators and repressors.
Repression of gas1 transcription by serum in NIH 3T3 cells was reported to be independent of de novo protein synthesis (30). This suggests that a Myc-independent pathway for regulation may exist. Because in our experiments cycloheximide on its own repressed gas1 (T.C.L. and E.B.Z., unpublished observation) partially in contrast to an earlier report (30), it was not possible to determine in the current work whether Myc can repress gas1 in the absence of new protein synthesis in the MycER cells. The specific mechanism by which Myc represses gas1 transcription remains to be established. However, the repression is rapid and begins almost immediately following activation of MycER, consistent with a direct effect of Myc on the gas1 promoter. Recently, Miz-1, an initiator binding protein, that interacts with Myc and that may mediate Myc’s repressive effects has been cloned (52). Such a protein could tether Myc to initiator sites of promoters and enable Myc to block initiation of transcription directly, resulting in gene repression.
Although c-Myc expression has been closely associated with cell proliferation and neoplasia, and Myc functions as a transcriptional activator of the cdc25A and ODC genes, the molecular details of how Myc regulates cell cycling remain to be determined. Taken together, our findings support a model wherein Myc induces cell proliferation, at least in part, by repressing genes with growth arrest activity (53). The mutant ΔMB2 retains the ability to activate ODC gene transcription, but fails to repress gas1. When viewed in the light of ΔMB2’s inability to transform cells or to induce cell cycle progression, the phenotype raises the possibility that MB2 contributes a transcriptional repressor function necessary for Myc’s growth stimulatory effects. MB2 may also contribute to the induction of a class of genes with E boxes distal to the promoter that stimulate growth (41, 54). By simultaneously stimulating expression of growth inducers and repressing growth inhibitors, Myc may exert a powerful regulation of cell growth. We are currently identifying other classes of growth suppressive genes, in addition to gas1, which are targeted by Myc for repression.
Acknowledgments
The authors thank R. de Martin for providing the pGas1-luc reporter construct, J. Cleveland for providing the pODC-CAT reporter construct, L. Penn and G. Evan for providing the MycER-expressing cells, C. Schneider for providing Gas1 antisera, M. Eilers for discussing results before publication, T. Serra for preparation of the manuscript, J. Perez for critical reading of the manuscript, and members of the Ziff lab for helpful discussions. This work was supported by a grant to E.B.Z from the National Institutes of Health (R01 AG13620). T.C.L. was supported by National institutes of Health Grant CA-01713, American Cancer Society Institutional Grant IRG-14-39, and a Skirball Institute Award. Support from the Beatrice and Samuel A. Seaver Foundation is gratefully acknowledged. E.B.Z. is an Investigator of the Howard Hughes Medical Institute. T.C.L. is a Physician-Scientist Scholar of the Skirball Institute of Biomolecular Medicine.
ABBREVIATIONS
- ODC
ornithine decarboxylase
- MB2
Myc box 2
- Hm
wild-type human c-myc
- 4-OHT
4-hydroxytamoxifen
- ER
estrogen receptor
- MycER
Myc-ER fusion protein
- LZ
leucine zipper
- REFs
rat embryo fibroblasts
- CAT
chloramphenicol acetyltransferase
- FBS
fetal bovine serum
- gas
growth arrest-specific
- CMV
cytomegalovirus
- LTR
long terminal repeat of the Moloney murine leukemia virus
- gapdh
glyceraldehyde-3-phosphate dehydrogenase
References
- 1.DePinho R A, Schreiber-Agus N, Alt F W. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- 2.Spencer C A, Groudine M. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 3.Marcu K B, Bossone S A, Patel A J. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 4.Henriksson M, Luscher B. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 5.Land H, Parada L F, Weinberg R A. Nature (London) 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 6.Stone J, de L T, Ramsay G, Jakobovits E, Bishop J M, Varmus H, Lee W. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang C V, McGuire M, Buckmire M, Lee W M. Nature (London) 1989;337:664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- 8.Kato G J, Barrett J, Villa G M, Dang C V. Mol Cell Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L H, Nerlov C, Prendergast G, MacGregor D, Ziff E B. EMBO J. 1994;13:4070–4079. doi: 10.1002/j.1460-2075.1994.tb06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacGregor D, Li L H, Ziff E B. J Cell Physiol. 1996;167:95–105. doi: 10.1002/(SICI)1097-4652(199604)167:1<95::AID-JCP11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 11.Prendergast G C, Diamond L E, Dahl D, Cole M D. Mol Cell Biol. 1989;10:1265–1269. doi: 10.1128/mcb.10.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eilers M, Schirm S, Bishop J M. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benvenisty N, Leder A, Kuo A, Leder P. Genes Dev. 1992;6:2513–2523. doi: 10.1101/gad.6.12b.2513. [DOI] [PubMed] [Google Scholar]
- 14.Bello-Fernandez C, Packham G, Cleveland J L. Proc Natl Acad Sci USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen-Durr P, Meichle A, Steiner P, Pagano M, Finke K, Botz J, Wessbecher J, Draetta G, Eilers M. Proc Natl Acad Sci USA. 1993;90:3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner A J, Meyers C, Laimins L A, Hay N. Cell Growth Diff. 1993;4:879–883. [PubMed] [Google Scholar]
- 17.Roy B, Beamon J, Balint E, Reisman D. Mol Cell Biol. 1994;14:7805–7815. doi: 10.1128/mcb.14.12.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miltenberger R J, Sukow K A, Farnham P J. Mol Cell Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galaktionov K, Chen X, Beach D. Nature (London) 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 20.Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, Alitalo K, Eilers M. Mol Cell Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy A L, Carruthers C, Gutjahr T, Roeder R G. Nature (London) 1993;365:359–361. doi: 10.1038/365359a0. [DOI] [PubMed] [Google Scholar]
- 22.Schneider C, King R M, Philipson L. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 23.Del Sal G, Ruaro M E, Philipson L, Schneider C. Cell. 1992;70:595–607. doi: 10.1016/0092-8674(92)90429-g. [DOI] [PubMed] [Google Scholar]
- 24.Del Sal G, Collavin L, Ruaro M E, Edomi P, Saccone S, Valle G D, Schneider C. Proc Natl Acad Sci USA. 1994;91:1848–1852. doi: 10.1073/pnas.91.5.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Sal G, Ruaro E M, Utrera R, Cole C N, Levine A J, Schneider C. Mol Cell Biol. 1995;15:7152–7160. doi: 10.1128/mcb.15.12.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruaro E M, Collavin L, Del Sal G, Haffner R, Oren M, Levine A J, Schneider C. Proc Natl Acad Sci USA. 1997;94:4675–4680. doi: 10.1073/pnas.94.9.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 28.Kerkhoff E, Ziff E B. EMBO J. 1995;14:1892–903. doi: 10.1002/j.1460-2075.1995.tb07181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenberg M E, Ziff E B. Nature (London) 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 30.Ciccarelli C, Philipson L, Sorrentino V. Mol Cell Biol. 1990;10:1525–1529. doi: 10.1128/mcb.10.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eilers M, Picard D, Yamamoto K R, Bishop J M. Nature (London) 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- 32.Prendergast G C, Lawe D, Ziff E B. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 33.de Martin R, Cowled P A, Smith S E, Papavassiliou A G, Sorrentino V, Philipson L, Bohmann D. J Biol Chem. 1993;268:22788–22793. [PubMed] [Google Scholar]
- 34.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 35.Littlewood T D, Hancock D C, Danielian P S, Parker M G, Evan G I. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daksis J I, Lu R Y, Facchini L M, Marhin W W, Penn L J. Oncogene. 1994;9:3635–3645. [PubMed] [Google Scholar]
- 37.Facchini L M, Chen S, Marhin W W, Lear J N, Penn L Z. Mol Cell Biol. 1997;17:100–114. doi: 10.1128/mcb.17.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auvinen M, Paasinen A, Andersson L D, Hollta E. Nature (London) 1992;360:355–358. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- 39.Moshier J A, Dosicu J, Skunca M, Luk G D. Cancer Res. 1993;55:5358–5365. [Google Scholar]
- 40.Galaktionov K, Le A K, Eckstein J, Draetta G, Meckler J, Loda M, Beach D. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 41.Gaubatz S, Meichle A, Eilers M. Mol Cell Biol. 1994;14:3853–3862. doi: 10.1128/mcb.14.6.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freytag S O, Geddes T J. Science. 1992;256:379–382. doi: 10.1126/science.256.5055.379. [DOI] [PubMed] [Google Scholar]
- 43.Steiner P, Philipp A, Lukas J, Godden-Kent D, Pagano M, Mittnacht S, Bartek J, Eilers M. EMBO J. 1995;14:4814–4826. doi: 10.1002/j.1460-2075.1995.tb00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson M A, Ransay R G. BioEssays. 1995;17:341–350. doi: 10.1002/bies.950170410. [DOI] [PubMed] [Google Scholar]
- 45.Mizuguchi G, Kanei-Ishii C, Takahashi T, Yasukawa T, Nagase T, Horikoshi M, Yamamoto T, Ishii S. J Biol Chem. 1995;270:9384–9389. doi: 10.1074/jbc.270.16.9384. [DOI] [PubMed] [Google Scholar]
- 46.Velcich A, Ziff E B. Mol Cell Biol. 1990;10:6273–6282. doi: 10.1128/mcb.10.12.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nevins J R. Curr Top Microbiol Immunol. 1995;199:25–32. doi: 10.1007/978-3-642-79586-2_2. [DOI] [PubMed] [Google Scholar]
- 48.Somasundaram K, Jayaraman G, Williams T, Moran E, Frisch S, Thimmapaya B. Proc Natl Acad Sci USA. 1996;93:3088–3093. doi: 10.1073/pnas.93.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song C Z, Loewenstein P M, Toth K, Green M. Proc Natl Acad Sci USA. 1995;92:10330–10333. doi: 10.1073/pnas.92.22.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu W, Kerppola T K, Chen P L, Curran T, Chen-Kiang S. Mol Cell Biol. 1994;14:268–276. doi: 10.1128/mcb.14.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruder J M, Spaulding A J, Wierman M E. Mol Endocrinol. 1996;10:35–44. doi: 10.1210/mend.10.1.8838143. [DOI] [PubMed] [Google Scholar]
- 52.Peukert, K., Staller, P., Schneider, A., Carmichael, G., Haenel, F. & Eilers, M. (1997) EMBO J. in press. [DOI] [PMC free article] [PubMed]
- 53.Hopewell R, Li L, MacGregor D, Nerlov C, Ziff E B. J Cell Sci Suppl. 1995;19:85–89. doi: 10.1242/jcs.1995.supplement_19.12. [DOI] [PubMed] [Google Scholar]
- 54.Desbarats L, Gaubatz S, Eilers M. Genes Dev. 1996;10:447–460. doi: 10.1101/gad.10.4.447. [DOI] [PubMed] [Google Scholar]