Abstract
Application of stem cell biology to breast cancer research has been limited by the lack of simple methods for identification and isolation of normal and malignant stem cells. Utilizing in vitro and in vivo experimental systems, we show that normal and cancer human mammary epithelial cells with increased aldehyde dehydrogenase activity (ALDH) have stem/progenitor properties. These cells contain the subpopulation of normal breast epithelium with the broadest lineage differentiation potential and greatest growth capacity in a xenotransplant model. In breast carcinomas, high ALDH activity identifies the tumorigenic cell fraction, capable of self-renewal and of generating tumors which recapitulate the heterogeneity of the parental tumor. In a series of 577 breast carcinomas, expression of ALDH1 detected by immunostaining correlated with poor prognosis. These findings offer an important new tool for the study of normal and malignant breast stem cells and facilitate the clinical application of stem cell concepts.
Introduction
Although the concept that cancers arise from “stem” or “germ cells” was first proposed almost 150 years ago, it is only recently that advances in stem cell biology generated the experimental framework necessary to test this hypothesis (Reya et al., 2001; Sell et al., 2004). According to the cancer stem cell model, tumors originate in either tissue stem cells or progenitor cells, through deregulation of the normally tightly regulated process of self-renewal (Molofsky et al., 2004; Passegue et al., 2003). Self-renewal is the process by which stem cells generate progeny identical to themselves. Stem cells also differentiate to generate multipotent progenitors, which in turn give rise to committed progenitors and differentiated cells. Cancer stem cells share these properties with their normal counterparts: they have self-renewal capacity, driving tumorigenicity, recurrence and metastasis and they have the capacity to differentiate, albeit aberrantly, giving rise to a heterogeneous population of cancer cells. The differentiated cells constitute the bulk of the tumor, but they are not tumorigenic, due to their lack of self renewal capacity and limited proliferation potential. Experimental evidence supporting the cancer stem cell hypothesis was first generated in 1997 by Dicks’ group, who demonstrated that human leukemias are driven by a small population of leukemic stem cells capable of transferring the disease to NOD/scid mice (Bonnet and Dick, 1997). This concept was extended to solid tumors by Clarke and Wicha. They demonstrated that human breast cancers contain a cell population with stem cell properties, bearing the surface markers CD44+/CD24-/lin- (Al-Hajj et al., 2003). Subsequently, cancer stem cells have been identified and prospectively isolated from a variety of malignancies, including brain cancers, prostate cancer, melanoma, multiple myeloma, colon, pancreatic, and head and neck cancers (Collins et al., 2005; Fang et al., 2005; Li et al., 2007; Matsui et al., 2004; O’Brien et al., 2006; Prince et al., 2007; Ricci-Vitiani et al., 2006; Singh et al., 2004a; Singh et al., 2004b).
It is likely that cancer stem cells have a phenotype defined by the cell of origin (stem cells or early progenitor cells) and by the oncogenic events that contributed to transformation. Recent studies have provided evidence that supports this concept (Jamieson et al., 2004; Kelly et al., 2002). One approach for finding shared stem cell markers is to focus on conserved stem and progenitor cell functions. These functional markers may be inherited by the malignant stem cell compartment, across multiple histological subtypes of cancer from the same tissue of origin. A candidate marker which fits this description is aldehyde dehydrogenase 1 (ALDH1), a detoxifying enzyme responsible for the oxidation of intracellular aldehydes (Duester, 2000; Magni et al., 1996; Sophos and Vasiliou, 2003; Yoshida et al., 1998). ALDH may have a role in early differentiation of stem cells, through its role in oxidizing retinol to retinoic acid (Chute et al., 2006). It has been shown that murine and human hematopoietic and neural stem and progenitor cells have a high ALDH activity (Armstrong et al., 2004; Hess et al., 2004; Hess et al., 2006; Matsui et al., 2004). Increased ALDH activity has also been found in stem cell populations in multiple myeloma and acute myeloid leukemia (AML) (Matsui et al., 2004; Pearce et al., 2005). ALDH activity may thus provide a common marker for both normal and malignant stem and progenitor cells.
In the present study we demonstrate that cells with ALDH activity isolated from normal human breast have phenotypic and functional characteristics of mammary stem cells. Moreover, the ALDEFLUOR-positive cells isolated from human breast tumors contain the cancer stem cell population. We also demonstrate that both normal and malignant human mammary stem cells may be identified in situ, using immunostaining. Analyzing the expression of ALDH1 in 577 human breast carcinomas from two patient populations, we show that the expression of this stem/progenitor cell marker is a powerful predictor of poor clinical outcome. These findings provide support for the “cancer stem cell hypothesis” and open new possibilities for the study of mammary stem/progenitor cells and their role in mammary development and carcinogenesis. In addition, ALDH1 immunodetection is a simple method for identifying cancer stem/progenitor cells in situ, facilitating the clinical application of stem cell concepts.
Results
The ALDEFLUOR-positive population isolated from normal mammary epithelium has stem cell properties
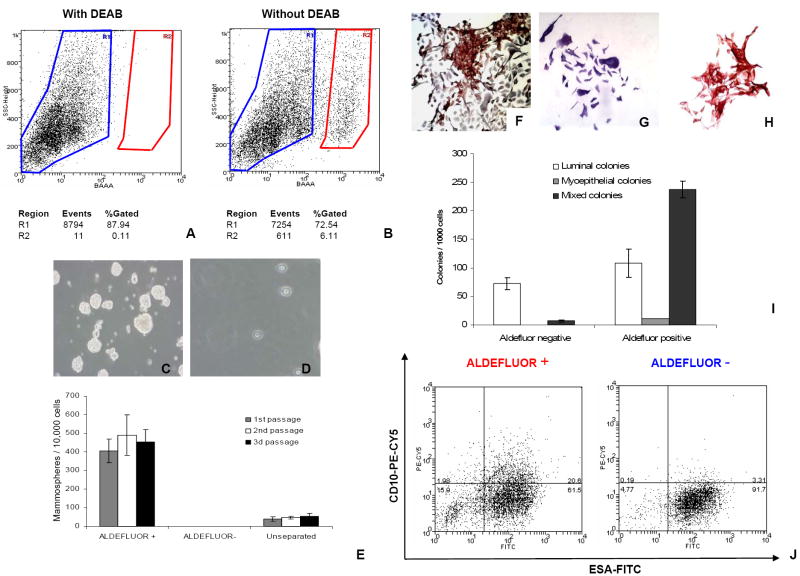
Single cell suspensions of normal mammary epithelial cells were obtained by mechanical and enzymatic digestion of breast reduction samples, as previously described (Stingl et al., 1998, Supplementary Methods 1). We utilized the ALDEFLUOR assay to assess the presence and size of the population with ALDH enzymatic activity in normal human breast epithelium. Analysis of breast reduction samples from 14 different patients showed an average of 8% (8.18 ± 4.31, n=14) ALDEFLUOR-positive population in normal mammary epithelial cells (Figure 1A-B). Using previously established in vitro and in vivo assays (Dontu et al., 2003; Kuperwasser et al., 2004; Stingl et al., 2006) we showed that functional characteristics associated with adult stem cells are displayed by the ALDEFLUOR-positive but not the ALDEFLUOR-negative population.
Figure 1. ALDEFLUOR positive cells from normal breast epithelium have stem cell properties.
A-B. Representative FACS analysis of normal breast epithelial cells using the ALDEFLUOR assay. Cells incubated with ALDEFLUOR substrate (BAAA) and the specific inhibitor of ALDH, DEAB, were used to establish the baseline fluorescence of these cells (R1) and to define the ALDEFLUOR-positive region (R2) (A). Incubation of cells with ALDEFLUOR substrate in the absence of DEAB induces a shift in BAAA fluorescence defining the ALDEFLUOR-positive population (B). In all experiments cells were first gated on PI negative cells (viable cells) which represented 93.4 ± 2.4% (Mean ± SDEV, n= 31) of the total population. C-E. ALDEFLUOR-positive cells sorted from fresh reduction mammoplasties were enriched in sphere initiating cells(C) with 451 ± 42 mammospheres (Mean ± SDEV, n= 6, derived from 3 different patients) generated by 10,000 cells plated, versus 50 ± 8 mammospheres (Mean ± SDEV, n= 6) generated by 10,000 unseparated cells (E). ALDEFLUOR-negative cells failed to grow in suspension (D-E). ALDEFLUOR-positive cells and unseparated cells were capable of self-renewal in vitro, as shown by similar mammosphere-initiating capacity in three passages (E). F-J. Evaluation of the differentiation potential of ALDEFLUOR-positive and ALDEFLUOR-negative cells. Sorted cells were grown in differentiating conditions for 12 days and stained by IHC with lineage-specific markers (ESA, CD10). The ALDEFLUOR-positive population generated 237±15 mixed colonies/1000 cells plated (67.2 ± 3.5% bi-lineage colonies) (ESA+ cells stained in brown and CD10+ stained in purple) (F), 11± 1 myoepithelial colonies/1000 cells plated (2.9 ± 0.5%) (CD10+) (G), and 108±25 luminal colonies/1000 cells plated (30.6 ± 5.4%) (ESA+) (H). The ALDEFLUOR-negative population produced 72±10 luminal colonies/1000 cells plated (90.8 ± 3.1%) (ESA+) (H), and only 7±2 mixed colonies/1000 cells plated (9.1±1.3%) (I). Data represent Means ± SDEV, n= 6, derived from 3 different patients. J. ALDEFLUOR-positive and ALDEFLUOR-negative cells grown in differentiating conditions were collected for flow cytometry analysis of lineage markers (ESA, CD10). ALDEFLUOR-positive cells generated uncommitted progeny (15.3±3.2%, CD10-/ESA-; 21.2±1.5%, CD10+/ESA+), luminal cells (63.2 ± 4.1%, CD10-/ESA+) and myoepithelial cells (2.1±0.3%, CD10+/ESA-), whereas ALDEFLUOR-negative cells generated predominantly luminal cells (93.5±3.4%, CD10-/ESA+). Data represent Means ± SDEV, n=3.
The ALDEFLUOR-positive population (Figure 1C-E), but not the ALDEFLUOR-negative population (Figure 1D-E) isolated from fresh mammoplasty samples was capable of generating mammospheres in suspension culture at a density of 5000 cells/ml, with a frequency of approximately 4% (Fig. 1 E), in three consecutive passages. Similar results were obtained when cells were plated in 96-well plates, 1 cell/well. The ALDEFLUOR-positive cells generated mammospheres with an efficiency of 10±3.5% whereas the ALDEFLUOR-negative cells generated no spheres and the unseparated cells generated spheres with an efficiency of 0.5±0.2%. The difference between the low density culture and single cell/well culture may be due to a degree of cell aggregation occurring in the former. These results are consistent with our previous findings showing that mammary epithelial cells that survive and proliferate in anchorage-independent conditions are likely to be breast stem cells with self-renewal capacity (Dontu et al., 2003). Similar results were reported by studies in different tissues (Li et al., 2003). ALDEFLUOR-positive cells sorted from dissociated mammospheres were capable of self-renewal in vitro as shown by similar mammosphere-initiating capacity in multiple passages (Figure 1E). The cell lineage composition of the mammospheres was conserved upon serial passages. ALDEFLUOR-positive cells were enriched 10-fold in clonogenicity in suspension, compared to unseparated cells.
In a clonogenic assay that assesses the lineage differentiation potential of single cells, the ALDEFLUOR-positive cells were enriched in bi-lineage progenitor cells that generated mixed ESA+/CD10+ colonies (Figure 1F-I). Figure 1I shows the numbers of myoepithelial, luminal epithelial and mixed lineage colonies generated by ALDEFLUOR-positive and ALDEFLUOR-negative cells. The mixed lineage colonies represented 67.2±3.5% of the total number of colonies generated by ALDEFLUOR-positive cells (237±15 mixed colonies/1000 cells plated), whereas they represented only 9.1±1.3% of the colonies generated by ALDEFLUOR-negative cells (7±2 colonies/ 1000 cells plated) (Figure 1I).
Differentiation potential of ALDEFLUOR-positive and -negative populations was also assessed by flow cytometry analysis of lineage-specific markers expressed in the progeny of these cells, generated in cultivation conditions that promote differentiation. Previous studies by Stingl et al. showed that in the human mammary epithelium uncommitted progenitors double negative for CD10 and ESA generate double negative progenitors bearing both markers. Subsequently, these give rise to the single positive, lineage-committed progenitors. (Stingl et al, 1998). Our results showed that, consistent with the findings of the clonogenic assay, the ALDEFLUOR-positive population was enriched in progenitors cells, which generateed un-commited progeny (15.3±3.2%, CD10-/ESA-; 21.2 ± 1.5%, CD10+/ESA+), myoepithelial (2.1±0.3%, CD10+/ESA-) and luminal epithelial cells (63.2±4.1%, CD10-/ESA+) (Figure 1J, left panel). The ALDEFLUOR-negative population was highly enriched in progenitors restricted to the luminal epithelial cell fate (93.5±3.4%, CD10-/ESA+) (Figure 1J, right panel).
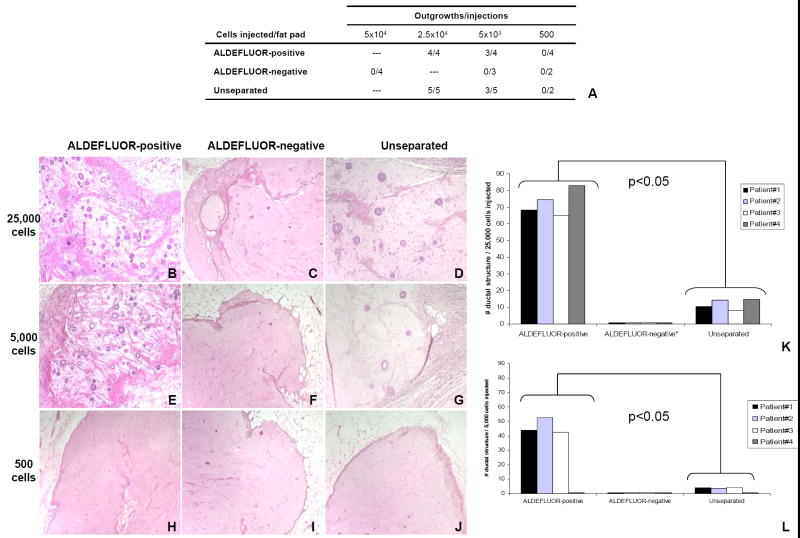
We utilized the mouse model described by Kuperwasser et al. to evaluate the ability of sorted cells from normal breast epithelium to grow and differentiate in vivo (Kuperwasser et al., 2004). ALDEFLUOR-positive, ALDEFLUOR-negative and unseparated cells were transplanted into humanized cleared mammary fat pads of NOD/scid mice (25,000, 5,000, and 500 ALDEFLUOR-positive cells, 50,000, 5,000 and 500 ALDEFLUOR-negative cells, and 25,000, 5,000, and 500 unseparated cells) (Figure 2A). Experiments were performed in triplicate. All the cell subpopulations, including the unseparated cells, were stained with propidium iodide (PI) for viability and sorted by side and forward scatter and viability, using flow cytometry (Supplementary Figure 5). Only ALDEFLUOR-positive and unseparated cells had outgrowth potential, as shown by duct formation upon implantation of 25,000 cells (Figure 2B-J). Moreover, the ALDEFLUOR-positive cell population was considerably enriched in in vivo outgrowth capability, because it consistently generated 10-fold more ducts in the humanized area of the mammary fat pad, compared to the unseparated population (Figure 2K-L). The ALDEFLUOR-negative population failed to repopulate the fat pads, even when 50,000 cells were injected (data not shown). We validated the human origin of the epithelial outgrowths by immunostaing with a human specific antibody anti-ESA (Figure 3A-D). As is the case in the human mammary tree, these small ducts generated in the animal host were composed of a luminal epithelial layer, expressing CK18 and an outer myoepithelial cell layer, expressing smooth muscle actin (SMA) (Figure 3E). This outer layer was not generated by myofibroblastic conversion, because it stained double positive for CK14 and SMA (Figure 3I). We also documented the cell lineage evolution by showing the presence of progenitor cells, identified by double positive staining with luminal and basal markers, such as CK14/CK18, CK18/CK17, and CK18/CK5/6 (Figure 3F-H) (Villadsen et al., 2007).
Figure 2. In vivo outgrowth potential of normal human breast epithelial cells sorted by the ALDEFLUOR assay.
A. Table showing the number of outgrowths generated in NOD/scid mouse fat pads by ALDEFLUOR-positive, ALDEFLUOR-negative, and Unseparated cells. B-J. Hematoxylin and eosin staining of ducts generated by ALDEEFLUOR-positive cells (B, E, H), ALDEFLUOR-negative cells (C, F, I), Unseparated cells (D, G, J). The number of cells injected is indicated on the left side (25,000 cells in B –D, 5,000 cells in E-G, 500 cells in H-J). We observed formation of ducts in the fat pads injected with 25,000 and 5,000 ALDEFLUOR-positive (B,E) or Unseparated cells (D,G). Only residual Matrigel and mouse tissue were observed in all the other fat pads (C, F, H-J). K-L. Evaluation of the number of ducts generated by each population (ALDEFLUOR-positive, ALDEFLUOR-negative, Unseparated). Only the ALDEFLUOR-positive and Unseparated cells produced outgrowths. The ALDEFLUOR-positive cells produced significantly more ducts than the Unseparated cells (p<0.05). (K, L)
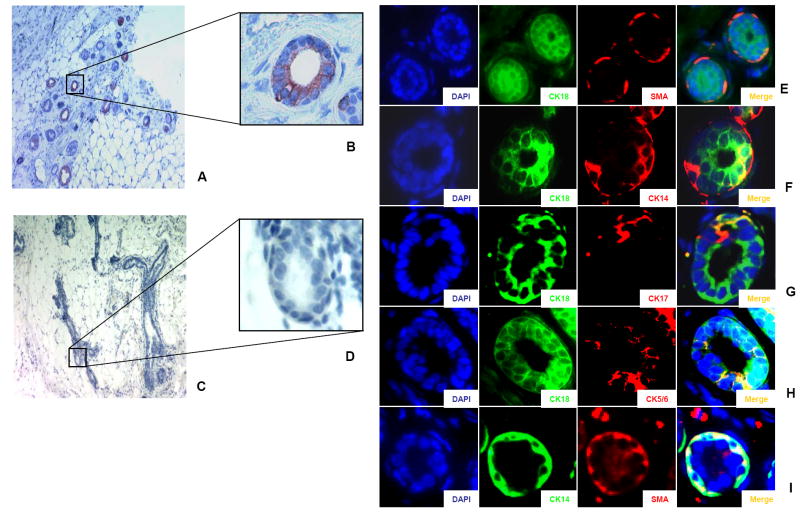
Figure 3. Characterization of the duct outgrowths generated in humanized NOD/scid cleared fat pads by ALDEFLUOR-positive cells from normal breast epithelium.
A-D. Evidence of the human origin of the epithelial ducts. Positive staining with a specific human antibody (anti-ESA) (A, B) which does not cross-react with mouse tissue (C, D) confirms the human origin of the ductal structure (red staining). E-I. Cell composition of ducts generated by ALDEFLUOR-positive cells. The ducts had a luminal layer (stained with anti-CK18, green signal) (and a myoepithelial layer (stained with SMA, red signal). Double staining with luminal-like cytokeratin (CK18; green signal) and basal-like cytokeratins (CK14, CK17, CK5/6, red signal) demonstrated a partial overlap between the luminal cells and the basal cells (yellow signal in the composite image, merge) suggesting a lineage evolution during duct formation. Double staining with CK14 (green signal) and SMA (red signal) showed cells positive for both markers (yellow signal) on the composite image (merge) (I). All nuclei were counterstained with DAPI.
Taken together, the results of the in vivo and in vitro assays indicate that the ALDEFLUOR-positive cells represent the cell population with the broadest differentiation potential in vitro and highest growth potential in vivo.
In situ characterization of ALDH1-positive cells in normal breast epithelium and mammosphere sections
We next investigated whether ALDH1 immunohistochemistry (IHC) could be used to detect mammary stem/progenitor cells in situ. We utilized flow cytometry analysis to determine the overlap between the cell population with a high ALDH enzymatic activity (ALDEFLUOR-positive) and the population immunostained by ALDH1. The ALDEFLUOR-positive and -negative populations from normal breast epithelium were isolated by FACS, fixed, and stained with an ALDH1 monoclonal antibody. The cells detected by immunostaining were all contained in the ALDEFLUOR-positive population, whereas the ALDEFLUOR-negative population contained no ALDH1-positive cells (Supplementary Figure 1A-C). We confirmed the results of this analysis by immunostaining separated ALDEFLUOR-positive and -negative cells on cytospins (Supplementary Figure 1 D-E).
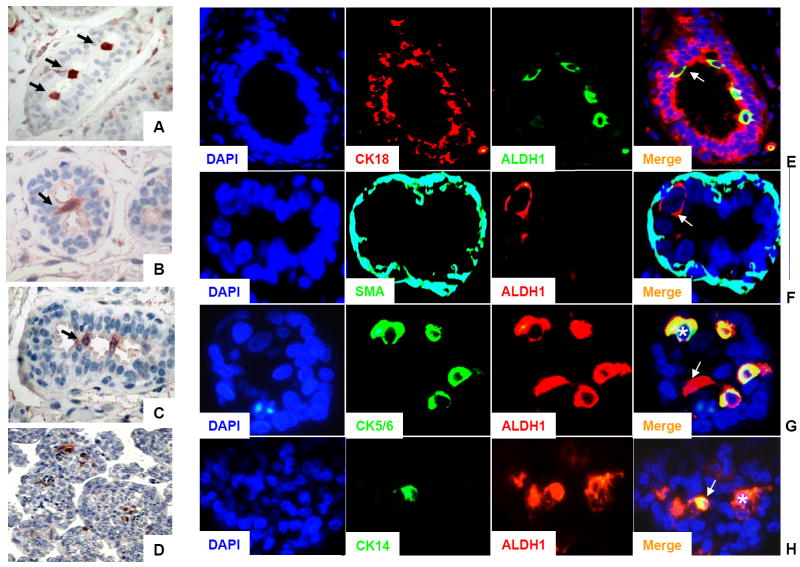
Immunostaining of paraffin-embedded sections of normal breast epithelium using the ALDH1 antibody identified a relatively rare population of ALDH1-positive cells located in the terminal ductal lobular units (TDLUs). ALDH1-positive cells appeared to form a bridge in the lumen that could be located at the bifurcation point of side branches in the TDLUs (Figure 4A-C and Supplementary Figure 2). This is consistent with recent published data that show that human stem/progenitor cells are localized in the ductal part of the TDLU structures (Villadsen et al., 2007). A stem cell marker would not be expected to colocalize with markers of mature differentiated mammary epithelial cells. We performed double staining with ALDH1 and CK18, a marker of luminal epithelial cells and ALDH1 and SMA, a marker of myoepithelial cells. The ALDH1-positive cells did not co-localize with CK18, or SMA in sections through normal human breast epithelium (Figure 4E-F). Although the phenotype of normal stem and/or progenitor cells from the human breast epithelium has not been identified, several markers including CK5/6 and CK14 have been found to be associated with in undifferentiated mammary epithelial cells (Boecker et al., 2002; Gudjonsson et al., 2002). We did not detect overlapping expression between CK5/6 or CK14 and ALDH1 in sections through normal human breast epithelium (data not shown). To determine if this was a result of the scarcity of these populations, we repeated the same analysis on mammosphere sections. We have shown previously that mammospheres generated from normal mammary epithelium are enriched in stem/progenitor cells (Dontu et al., 2003). The ALDH1-positive cells represented approx 5% of the mammosphere cell population (Figure 4D). A subset of these ALDH1-positive cells expressed CK5/6 or CK14 (Figure 4G-H). These results are consistent with the hypothesis that ALDH1-positive cells represent the stem/progenitor population of the normal human breast epithelium.
Figure 4. Characterization of ALDH1 positive-cells present in the normal breast epithelium and in mammosphere sections.
A-C. ALDH1 staining of normal breast epithelium. ALDH1-positive cells (red cytoplasmic staining) were in a luminal location, bridging across the lumen, probably at branching points of side-ducts (arrows). D. ALDH1 staining in mammospheres. Only 1-5 cells/mammosphere showed positive staining for ALDH1, (approximately 5% of the total population). E-F. Immunofluorescence of normal breast epithelium. E. Double staining with CK18 (red) and ALDH1 (green). Composite image (merge) showed absence of overlap between CK18 positive cells (mature luminal cells) and ALDH1-positive cells (arrow). F. Double staining with SMA (green) and ALDH1 (red). Composite image (merge) showed absence of overlap between SMA-positive cells (mature myoepithelial cells) and ALDH1-positive cells (arrow). G. Immunofluorescence of mammosphere sections. Double staining with CK5/6 (green) and ALDH1 (red). Composite image (merge) showed that only few ALDH1-positive cells displayed an exclusive red signal (arrow) whereas all the CK5/6-positive cells (asterisk) displayed a hybrid signal (yellow) corresponding to cells positive for ALDH1 and CK5/6. H. Double staining with CK14 (green) and ALDH1 (red). Composite image (merge) showed that most of the ALDH1-postive cells displayed an exclusive red signal (asterisk) whereas all the CK14 positive cells (arrow) displayed a hybrid signal (yellow) corresponding to cells positive for ALDH1 and CK14. All nuclei were counterstained with DAPI.
ALDEFLUOR-positive breast carcinoma cells display properties of cancer stem cells
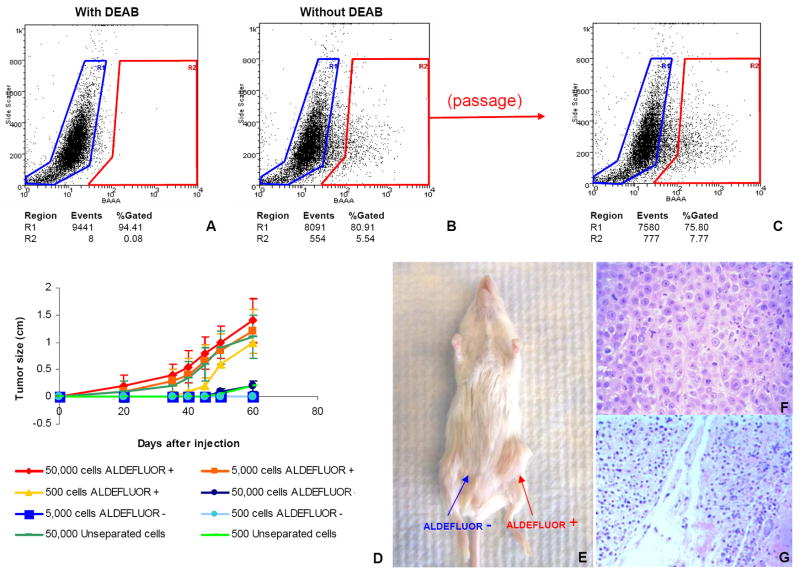
To investigate the tumorigenicity of the ALDEFLUOR-positive population in breast cancers we established xenotransplants from four independent human breast cancers (MC1, UM1, UM2, UM3). Cells from these tumors were transplanted orthotopically in the humanized cleared fat-pad of NOD/scid mice, without cultivation in vitro. The tumors were invasive ductal carcinomas, three ER-PR-ERBB2- (MC1, UM1, and UM3) and one ER+PR+ERBB2- (UM2). The tumorigenicity of the sorted ALDEFLUOR populations was assessed in early passages in animals. In contrast to assays that test tumorigenicity of sorted populations directly from patient tumors, this experimental design minimizes the bias introduced by the variable ability of breast cancers to xenotransplant. We found that the ALDEFLUOR-positive population in these three tumors represented 3% to 10% of the total cell population (Figure 5A-B and Supplementary Figure 3). We performed serial passages in vivo, using limiting dilutions of ALDEFLUOR-positive, -negative and unseparated cells (50,000 cells; 25,000 cells; 5,000 cells; 500 cells). Experiments were performed in triplicate. For each of the four tumors and for each of the three passages performed, only the ALDEFLUOR-positive population formed tumors, even when implanted in low numbers (Supplementary Table 1 and Supplementary Figure 3). As shown in Figure 5D, the size and latency of tumor formation correlated with the number of ALDEFLUOR-positive cells injected. Remarkably, 500 ALDEFLUOR-positive cells generated a tumor in as few as 40 days. ALDEFLUOR-negative cells failed to reproducibly generate tumors although limited growth was observed when 50,000 ALDEFLUOR-negative cells were injected. This is consistent with the presence of less than 0.01% contaminating ALDEFLUOR-positive cells, which is within the limits of FACS error. Alternatively, the growth may have been generated by progenitor cells with limited proliferation capacity. This would explain why these tumors could not be passaged more than once, when implanted as unseparated cells in the mouse fat-pad. H&E staining of the fat pad sections confirmed that tumors formed by ALDEFLUOR-positive cells contained malignant cells (Figure 5F) whereas only residual Matrigel, apoptotic cells and mouse tissue were seen at the sites of the ALDEFLUOR-negative cell injections (Figure 5G). No tumors were detected at these sites after 20–34 weeks. Consistent with the ALDEFLUOR-positive population having stem cell characteristics, tumors generated by this population recapitulated the phenotypic heterogeneity of the initial tumor, with a similar ratio of ALDEFLUOR-positive and negative cells (Figure 5C). This indicates that the ALDEFLUOR-positive cells were able to self-renew, generating ALDEFLUOR-positive cells and were able to differentiate, generating ALDEFLUOR-negative cells. We investigated the overlap between the ALDEFLUOR-positive population and the previously described breast cancer stem cell phenotype, CD44+/CD24-/lin- (Al-Hajj et al., 2003). Flow cytometry analysis of the xenografted tumors showed that these two phenotypes, ALDEFLUOR and CD44+/CD24-/lin-, identified a small overlapping cell fraction, representing 1.2%, 0.1%, and 0.9% in MC1, UM1 and UM2, respectively (Figure 6 A-B and Supplementary Figure 4). We tested tumorigenicity of the cells defined by both phenotypes in the MC1 tumor. The cell population bearing both cancer stem cell phenotypes had high tumorigenic capacity and generating a tumor from as few as 20 cells (Figure 6E). By contrast, the ALDEFLUOR-negative cells bearing the CD44+/CD24-/lin- phenotype, was not tumorigenic, even when implanted in numbers of 50,000 cells/fat pad (Figure 6D), suggesting that this population may lack tumorigenic cells. The fact that the unseparated cells from the same tumor sample were tumorigenic, when more than 500 cells were implanted (Figure 6C) supports this conclusion. The ALDEFLUOR-positive population that did not display the CD44+/CD24-/lin- phenotype was capable of generating tumors when implanted in numbers higher than 1500 cells (Figure 6F). We cannot exclude the possibility that these tumors are generated by contaminating highly tumorigenic cells. However ALDEFLUOR-negative cells did not generate tumors even from 50,000 cells. In addition, the tumors generated by 500 cells from the unseparated tumor population could not be passaged more than once. Taken together these findings suggest that the ALDEFLUOR-positive/lin-/Non CD44+/CD24- may contain progenitor cells with limited proliferation potential.
Figure 5. The ALDEFLUOR positive cell population from human breast tumors xeongrafted in NOD/scid mice has cancer stem cell properties.
A-B. Representative flow cytometry analysis of ALDH activity in cells derived from a human breast tumor, orthotopically xenotransplanted in NOD/scid mice. The ALDEFLUOR assay was performed as described above. In addition mouse of cell origin were eliminated from the analysis (see Supplementary methods 1 and Supplementary Figure 5). (A, B) All the ALDEFLUOR analyses on human breast tumor cells were first gated on PI negative cells (viable cells) which represented 73.6±1.8% (Mean ± SDEV, n=43) of the total population. C-G Only the ALDEFLUOR-positive population was tumorigenic. C. The ALDEFLUOR-positive population was capable of regenerating the phenotypic heterogeneity of the initial tumor after a passage in NOD/scid mice. D. Tumor growth curves were plotted for the numbers of cells injected (50,000 cells; 5,000 cells; 500 cells) and for each population (ALDEFLUOR-positive, ALDEFLUOR-negative, unseparated). Tumor growth kinetics correlated with the latency and size of tumor formation and the number of ALDEFLUOR-positive cells. E. Representative tumor grown in NOD/scid mouse at the ALDEFLUOR-positive cells’ injection site (5,000 cells injected). No tumor was detected at the ALDEFLUOR-negative cells’ injection site (5,000 cells injected). F-G. H & E staining of ALDEFLUOR-positive cells’ injection site, revealing presence of tumor cells (F). The ALDEFLUOR-negative cells’ injection site contained only residual Matrigel, apoptotic cells and mouse tissue (G). All the data presented in this figure were generated by analysis of the MC1 tumor. Similar results were obtained for three other tumors, generated from different patients (UM1, UM2, and UM3) tested (Supplementary Figure 4).
Figure 6. Tumorigenicity of the cells bearing the overlaping phenotype ALDEFLUOR-positive and CD44+/CD24-/lin-.
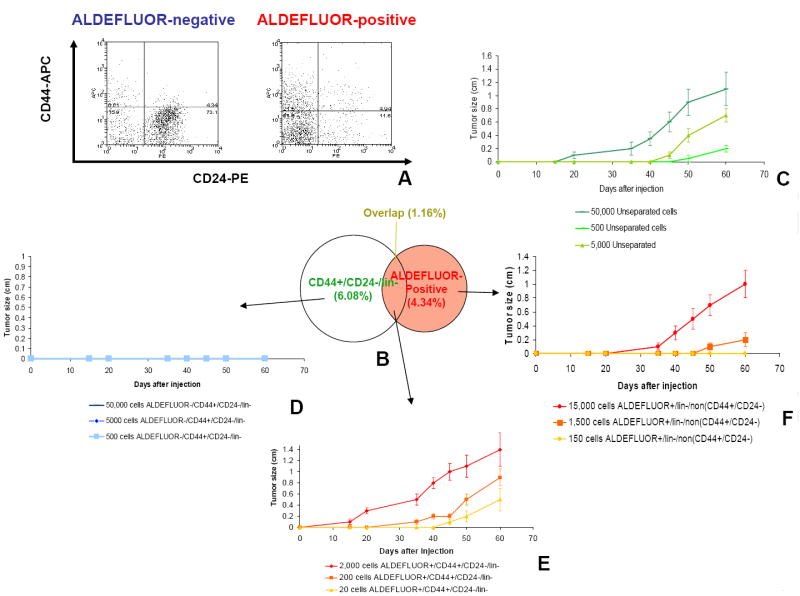
Cells were immunostained with a CD24–PE antibody, a CD44–APC antibody and antibodies for lineage markers labeled PE-Cy5, and subsequently stained with ALDEFLUOR. Cells were first gated based on viability and lin- markers, which represented 12.3±1.1% of the total population. Cells of mouse origin were also eliminated from the analysis. The four cell subpopulation defined by the ALDEFLUOR and CD44+/CD24-/lin- phenotypes were separated by FACS. A-B. The percentages shown in the diagram shows the representation of the cell sub-populations in the total tumor cell population and the overlap between the ALDEFLUOR phenotype and the CD24-/CD44+/lin- phenotype. C-F Tumorigenicity of the cell populations defined by the ALDEFLUOR and CD44-/CD24+/lin- phenotypes was tested using the xenotransplantation model described. Experiments were performed in triplicate. The unseparated cells generated tumors when implanted in numbers higher than 500 cells (C). The ALDEFLUOR-negative CD44+/CD24-/lin- cells were not tumorigenic, even at 50,000 cells/fat pad (D). The ALDEFLUOR-positive/CD44+/CD24-/lin- cells generated tumors from as few as 20 cells (E). The ALDEFLUOR-positive/lin-/NonCD44+/CD24- cells generated tumors when implanted in numbers higher than 1500 cells (F).
Analysis of ALDH1 protein on tissue microarrays (TMA) and correlation with histoclinical parameters
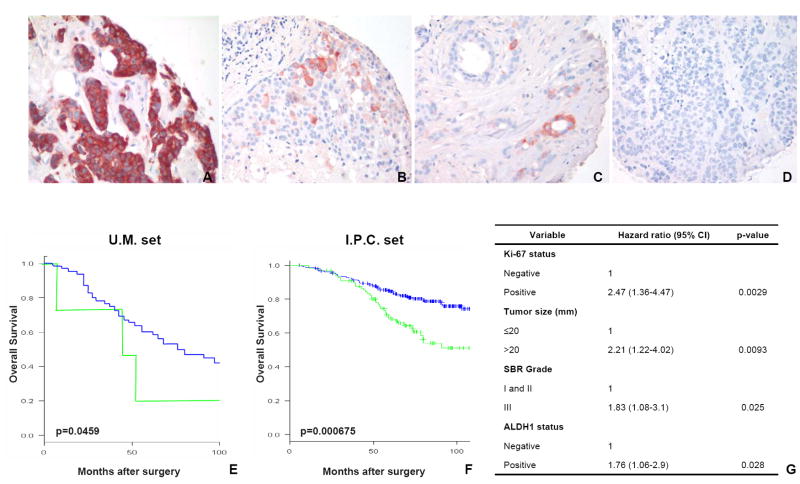
To assess the potential use of ALDH1 as a diagnostic and prognostic marker in breast cancer, we analyzed its expression by IHC in two independent sets of breast tumors (U.M. set, I.P.C. set), on tissue microarrays (TMAs). Among these two sets, 481 tumors were available for ALDH1 staining (136 cases from the U.M. set and 345 cases from the I.P.C. set). In the U.M. set, 24 tumors (19%) expressed ALDH1 and 122 tumors (81%) did not. Similar results were obtained in the I.P.C. set with 102 cases (30%) positive for ALDH1 staining and 243 cases negative (70%) (Figure 7A-D). Consistent with the idea that cancer stem cells constitute a minority of the tumor population, ALDH1-positive cells represented an average of 5% of cells in tumors expressing ALDH1. Only two of the 481 tumors had ALDH1 staining in the vast majority of the cell population (Figure 7A). We investigated whether ALDH1 expression correlates with the histoclinical characteristics of the breast cancers. We found similar results in both sets (Supplementary Table 2). ALDH1-positive tumors were associated with high histological grade (p<0.05 ; U.M. set, p<0.001 ; I.P.C. set, Fisher’s exact test), ERBB2 overexpression (p<0.05 ; U.M. set, p<0.001 ; I.P.C. set, Fisher’s exact test) and absence of estrogen and progesterone receptor expression (p<0.05 ; U.M. set, p<0.0001 ; I.P.C. set, Fisher’s exact test). No correlation was found with age, tumor size, and lymph node metastasis. We also found a correlation between expression of ALDH1 and that off the basal-like cytokeratins CK5/6 (p<0.05) and CK14 (p<0.01) (Supplementary Table 2).
Figure 7. Expression of ALDH1 in breast carcinomas, as shown by immunohistochemistry on tissue microarrays (TMA).
A-D. Example of ALDH1 staining in breast cancer. Only two of the 577 tumors analyzed were fully positive for ALDH1 (A). Representative examples of breast tumor cores positive for ALDH1 with 5-10% ALDH1-positive cells detected (B-C). Example of a tumor core with no detectable ALDH1 staining (D). E-F. Kaplan-Meier plot of patient overall survival: Survival differed significantly according to ALDH1 expression. Patients with tumors positive for ALDH1 staining (green curve) had a poor prognosis compared to patients with tumors negative for ALDH1 staining (blue curve). Similar results were observed in the U.M. set composed of 136 patients (p=0.0459) (E) and I.P.C. set composed of 341 patients (p=0.000675) (F). G. Cox multivariate analysis of overall survival for patients from I.P.C. set. When compared with known prognostic factors, ALDH1 status was an independent factor of prognosis, as was Ki-67 status, tumor size, SBR grade.
ALDH1 protein expression and clinical outcome
Analysis of overall survival (OS) showed a strong association of ALDH1-positive tumors with poor clinical outcome for both populations (p=0.0459 ; U.M. set, p=0.000675 ; I.P.C. set, log-Rank test) (Figure 7E-F). In the U.M. set, the 5-year OS was 19.8% [14.52-97.28] for patients with an ALDH1-positive tumor and 58.7% [33.22-100] for patients with an ALDH1-negative tumor. In the I.P.C. set, the 5-year OS was 69.59% [60.73-79.73] for patients with an ALDH1-positive tumor and 84.55% [80.02-89.33] for patients with an ALDH1-negative tumor.
We performed a Cox multivariate analysis of OS in which the values for ALDH1, tumor size, age, lymph node metastasis, histological grade, ER, PR, Ki-67 and ERBB2 were considered as categorical variables. ALDH1 expression was an independent prognostic factor, as was Ki-67 status, tumor size, and histological grade (Figure 7G). The relative risk of death due to cancer was 1.76 for patients with ALDH1-positive tumors compared to patients with ALDH1-negative tumors (p<0.028).
Discussion
The cancer stem cell hypothesis has fundamental implications for cancer biology in addition to clinical implications for cancer risk assessment, early detection, prognostication, and prevention. The development of cancer therapeutics based on tumor regression may have produced agents which kill differentiated tumor cells while sparing the small cancer stem cell population (Wicha et al., 2006). The development of more effective cancer therapies may thus require targeting this important cancer stem cell population. The success of these new approaches hinges on the identification, isolation and characterization of cancer stem cells. Recently, the phenotype of the mouse mammary stem cells was identified by several groups (Shackelton et al., 2006; Stingl et al., 2006). These studies showed that an entire, functional mammary gland can be regenerated in vivo in several serial passages, starting from a single cell (Shackelton et al., 2006). Also, considerable progress has been made recently towards identification of human mammary stem cells, although the phenotype of these cells has remained elusive (Clarke et al., 2006, Villadsen et al. 2007). Our study indicates that ALDH1 is a marker of stem/progenitor cells of the normal human breast and breast carcinomas. Utilizing in vitro assays we showed that ALDEFLUOR-positive cells contain the subpopulation of normal breast epithelium with the broadest lineage differentiation potential, capable of self-renewal. These cells also have the highest ability to grow in vivo, in a xenotransplantation animal model. In breast carcinomas, cells with high ALDH activity contain the tumorigenic cell fraction, able to self-renew and to recapitulate the heterogeneity of the parental tumor. With the caveat that xenotransplantation may change the properties of normal and cancer cells, this in vivo assay remains the gold standard for testing functional stem cell properties. The ALDEFLUOR-positive cell population has a small overlap with the previously described cancer stem cell, CD44+/CD24-/lin- phenotype. In the tumors we investigated, the overlap represented approximately 1% or less of the total cancer cell population. However, the cells bearing both phenotypes appeared to be highly enriched in tumorigenic capability, being able to generate tumors from as few as 20 cells.. It remains to be determined if these phenotypes are associated with stem cells in other breast cancers.
Identification of normal and malignant stem/progenitor cells by the same marker supports the concept that stem and progenitor cells are primary targets of transformation, and thus lends further support to the cancer stem cell hypothesis. In addition, the ability to identify stem/progenitor cells by this shared phenotypic trait, ALDH1 expression, permits analysis of cancer initiation and progression from the normal to the pre-malignant and then the malignant state. Unlike the previously described breast cancer stem cell phenotype, which requires the use of a combination of ten surface antigens (Al-Hajj et al., 2003), testing for ALDH1 expression is a simple method for identifying normal and cancer stem cells, amenable to clinical applications. We showed in the present study that ALDH1 expression is a powerful prognostic factor for breast cancer and it has direct or inverse correlation with known histoclinical parameters, such as tumor grade, ER/PR status, ERBB2 overexpression and basal-like cytokeratins (CK 5/6 and CK14).
In the vast majority of breast tumors analyzed in this study the ALDH1 positive cells represented a relatively small population, consistent with the notion that cancer stem cells constitute a minority of the tumor population. Remarkably, only two tumors out of 481 analyzed, had a predominant ALDH1 positive population. These tumors had a very aggressive clinical evolution and may have been driven by a stem cell population locked in self-renewal, undergoing little or no differentiation.
We propose that ALDH1 expression in a subset of tumors may reflect transformation of ALDH1-positive stem or early progenitor cells in these tumors. By contrast, ALDH1-negative tumors may be generated by the transformation of ALDH1-negative progenitor cells. In the ALDH1-positive tumors, the cancer stem cell population may inherit properties of normal stem cells that confer aggressiveness: ability to self-renew, high proliferation potential, resistance to damaging agents and chemoresistance. This hypothesis is consistent with the studies of AML (Bonnet et al., 1999). Alternatively, ALDH1-negative tumors may contain rare ALDH1-positive cells below the level of detection by immunostaining on TMAs. The detection of an ALDH1-positive population in TMA cores may be due to an increased self-renewal activity in these tumors. A recent study has shown that a gene expression signature associated with increased self-renewal of normal stem cells is a predictor of poor prognosis (Glinsky et al., 2005; Lahad et al., 2005). In agreement with our findings, previously described molecular signatures of breast cancer associated with a poor prognosis for breast cancer contain one or more ALDH isotypes (Alexe et al., 2006). Recently, a combinatorial analysis of gene expression data was used to re-analyze the van’t Veer breast cancer gene expression data set (van’t Veer et al., 2002). This analysis identified 17 genes associated with poor prognosis in breast cancer, two of which were ALDH isoforms. A recent study showed that granulocyte macrophage progenitor cells, transformed by the MLL-AF9 fusion protein, retained the global expression profile of their normal cells of origin and had only a subset of genes re-programmed. This set included 363 genes which were associated with self-renewal in normal hematopoietic stem cells including an ALDH isoform (Krivtsov et al., 2006).
In conclusion, our study lends support to the cancer stem cell hypothesis, by showing that both normal and malignant mammary stem cells share a common functional marker, ALDH1. Identification of ALDH1 as a potential marker of normal and malignant human breast stem cells opens important new avenues of research in normal breast development and breast carcinogenesis. Furthermore, our study suggests that ALDH1 expression may be used to detect both normal and malignant mammary stem cells in situ, in fixed paraffin-embedded sections. The clinical utility and relevance of this assay was demonstrated by a strong association of ALDH1 expression with clinical outcome in two independent tumor sets, totaling 577 patients. Since ALDH is also expressed in hematopoietic and neuronal stem cells, this marker may prove useful for the detection and isolation of cancer stem cells in other malignancies, thus facilitating the application of cancer stem cell biology to clinical practice.
Experimental Procedures
Dissociation of normal breast epithelium
Normal breast tissue from reduction mammoplasties was dissociated mechanically and enzymatically, as previously described (Stingl et al., 1998, Supplementary methods 1) To generate single cell suspension for the in vivo implantation, collagenase digestion time was limited to 6h. The mammoplasty samples were procured and utilized according to approved IRB protocols for research in human subjects.
Mammosphere culture
Mammosphere culture was performed as previously described (Dontu et al., 2003). Single cells were plated in ultra-low attachment plates (Corning, Acton, MA, USA) or plates coated with 1% agarose in PBS, at a density of 20,000 viable cells/ml in primary culture and 5000 cells/ml in subsequent passages. In the 1 cell/well experiments, conditioned medium from primary culture was used. Cells were plating using a cell sorter, during FACS. Ten plates were seeded from each of the ALDEFLUOR-positive, -negative and unseparated cell populations. For counting mammospheres, the content of all wells was collected, pooled and transferred on a collagen coated dish, in differentiating medium (see below). Mammospheres adhered in these conditions in approximately 48 h, after which they were stained with methyl blue and counted under low magnification.
Differentiating culture conditions
Single cell suspensions were plated on collagen-coated plates at a density of 2000 viable cells/10 cm diameter dish. Cells were grown in Ham’s F-12 medium (GIBCO™ INVITROGEN) with 5% fetal bovine serum (FBS), 5 μg/ml insulin, 1 μg/ml hydrocortisone, 10 μg/ml cholera toxin (Sigma, St Louis, MO, USA), 10 ng/ml epidermal growth factor (BD Biosciences) and 1X Pen/Strep/Fungizone Mix (GIBCO). Cells were fixed or collected for immunostaining after 12 days.
Flow cytometry
Cells were stained fresh or after fixation in methanol or RNA- later (Qiagen). Primary antibodies used were: ESA- FITC, CD10-PE (dilution 1:25, Novocastra, Newcastle, UK) and ALDH1 (dilution 1/100, BD Biosciences). Incubation was performed for 20 min. on ice in Hanks Balanced Salt Solution (HBSS, GIBCO) with 2% FBS, followed by washing in HBSS with 2% FBS. The secondary antibody used was anti-mouse IgG,- PE (dilution 1:250, Jackson Labs, MA, USA). After incubation, cells were washed once with HBSS and were re-suspended in HBSS supplemented with 5% FBS. For CD44/CD24/Lin staining the previously described protocol was followed (Al-Hajj., 2003, Supplementary methods 1). Fresh cells were stained with 1μg/ml PI (Sigma) for 5 min. for viability. Analysis was performed using a FACStarPLUS (Becton Dickinson, Palo Alto, CA, USA) flow cytometer.
Samples used for xenotransplantation
Human breast tumors were obtained as biopsy cores or pieces of tumors after surgery and implanted in humanized cleared fat pads of NOD/scid mice for establishing xenotransplants. The success of xenotransplantation was approximately 20%, similar to previous reports in the literature. Four xenotransplants were used: an ER-PR-ERBB2- tumor at the 15th passage in animals (MC1), an ER-PR-ERBB2- tumor at the 3rd passage (UM1), an ER+PR+ERBB2- tumor at the 4th passage (UM2), and an ER-PR-ERBB2- tumor at the 2nd passage (UM3). Two of the xenotransplants were generated from metastatic tumors (MC1, pleural effusion and UM2, ovarian metastasis) and two from primary tumors (UM1, UM3).
Aldefluor assay and separation of the ALDH positive population by FACS
The ALDEFLUOR kit (StemCell Technologies, Durham, NC, USA) was used to isolate the population with a high ALDH enzymatic activity. Cells obtained from freshly dissociated normal breast epithelium or breast cancer xenografts were suspended in ALDEFLUOR assay buffer containing ALDH substrate (BAAA, 1 μmol/l per 1×106 cells) and incubated during 40 minutes at 37°C. As negative control, for each sample of cells an aliquot was treated with 50mmol/L diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor. In order to eliminate cells of mouse origin from the xenotransplanted tumors, we used staining with an anti-H2Kd antibody (BD biosciences, 1/200, 20 min on ice) followed by staining with a secondary antibody labeled with phycoerythrin (PE) (Jackson labs, 1/250, 20 min on ice). The sorting gates were established using as negative controls the cells stained with PI only, for viability, the ALDEFLUOR-stained cells treated with DEAB and the staining with secondary antibody alone (see also Supplementary Figure 5 and 6).
Animal model
NOD/scid mice were used to assess the in vivo stem cell properties of the ALDEFLUOR-positive population, compared to the ALDEFLUOR-negative population and the unseparated population, from the normal breast epithelium and the four tumor xenografts. The animal model was described by Kuperwasser et al for xenotransplantation of normal mammary epithelial cells (Kuperwasser et al., 2004). The fat pads were cleared pre-puberty and humanized by injecting a mixture of irradiated and non-irradiated immortalized human fibroblasts (1:1 irradiated:non-irradiated, 50,000 cells/100μl Matrigel/fat pad). Irradiated fibroblasts (4Gy) support growth of normal and cancer epithelial cells by secreting a variety of growth factors, collagen and possibly directly interacting with the epithelial cells (Orimo et al., 2005; Tlsty et al., 2001). The immortalized fibroblasts were primary human mammary fibroblasts stably transfected with a retrovirus construct expressing telomerase. The fibroblast cell line is a generous gift from Dr. John Stingl and Dr. Connie Eaves (Terry Fox Laboratory, Vancouver, British Columbia, Canada). Estrogen pellets were implanted subcutaneously at the time of the clearing. The suspension of normal and malignant breast cells were obtained using the same dissociation method described above (see also Supplementary Methods 1), with the exception of collagenase incubation time, which was 1 h. Cells were mixed with Matrigel (BD biosciences) (1:1) and implanted in the cleared humanized fat pads 2-4 weeks later. The animals injected with normal breast cells were euthanized after 10 weeks. The animals injected with cancer were euthanized when the tumors were approximately 1.2 cm in the largest diameter, to avoid tumor necrosis and in compliance with regulations for use of vertebrate animal in research. A portion of each fat pad injected was fixed in formalin and embedded in paraffin for histological analysis. For the evaluation of the outgrowth potential of each cell population we analyzed thirty sections from each fat pad injected with sorted cells. H&E staining was performed every ten serial sections (4 slides/fat pad). For each H&E slide, the ductal structures present were counted under the microscope and averaged The animal studies were approved by the ULAM committee for research vertebrate animals.
Tissue Microarrays
The TMAs were provided by the Tissue Microarray Core laboratory at University of Michigan Medical School and by the Laboratoire d’Oncologie Moleculaire, Institut Paoli-Calmettes de Marseille. The first TMA contained 154 breast cancer cores from a consecutive population of patients treated at the University of Michigan Hospital, MI, USA (U.M. set) between 1984 and 1991 and the second TMA contained 552 breast cancer cores from a consecutive population of patients treated at the Institut Paoli-Calmettes, Marseille, France (I.P.C. set) between 1987 and 1999. Clinical and histopathological data are available for these patients (Jacquemier et al., 2005; Kleer et al., 2003).
Immunostaining
To assess the lineage composition of the colonies, cells were fixed on plates for 20 min in methanol, at -20°C, and were then stained using Peroxidase Histostain-Plus and Alkaline-phosphatase Histostain-Plus kits (Zymed, South San Francisco, CA, USA), according to the manufacturer’s protocol. The same fixation and staining protocol was used for cytospins. The primary antibodies, ALDH1, cytokeratin 18, ESA and CD10, were used at the dilutions indicated by the manufacturer. DAB (Zymed) and AEC were used as substrate for peroxidase, and NBT/BCIP (Gibco) for alkaline phosphatase.
For ALDH1 and ESA immunostaining, the paraffin-embedded sections through mammospheres, normal breast tissue and the TMAs were deparaffinized in xylene and rehydrated in graded alcohol. Antigen enhancement was done by incubating the sections in citrate buffer pH6 (Dakocytomation, Copenhagen, Denmark) as recommended. Staining was done using Peroxidase histostain-Plus Kit (Zymed) according to the manufacturer’s protocol. ALDH1 antibody (BD biosciences) was used at a 1/100 dilution and ESA antibody (Dakocytomation) was used at a 1/200 dilution. AEC (Zymed) was used as substrate for peroxidase. Slides were counter-stained with hematoxylin, and coverslipped using glycerin. TMA results were expressed in terms of percentage (P) and intensity (I) of positive cells as described previously (Ginestier et al., 2002). Results were scored by the quick score (Q) (Q = P × I). For the TMA, the mean of the score of minimum 2 core biopsies was calculated for each case.
For fluorescent double staining, the primary antibodies CK18, SMA, CK 5/6, CK14, and CK17 (Novocastra) were used at the dilutions indicated by the manufacturer. Texas-red and FITC labeled secondary antibodies (Jackson Labs) were used at the dilution 1/250 and incubated for 20 minutes. Nuclei were counterstained with DAPI/antifade (INVITROGEN) and coverslipped. Sections were examined with a fluorescent microscope (Leica, Bannockborn, IL, USA).
Statistical analysis
Distributions of molecular markers and other categorical variables were compared using standard chi2 tests or Fisher exact test. Statistical differences for the number of ductal structures were determined by using one-way ANOVA for independent samples. The overall survival interval was calculated from the date of diagnosis. For graphical presentation, follow-up was truncated at 100 months. Survival curves were derived from Kaplan-Meier estimates and the curves were compared by logrank tests. The influence of ALDH1 expression status was assessed in multivariate analysis by the Cox proportional hazard models with a stepwise selection. The model was adjusted for usual prognostic or predictive factors in breast cancer, including tumor size, age, lymph node metastasis, histological grade, ER, PR, Ki-67 and ERBB2 status. All statistical tests were 2-sided at the 5% level of significance, and were done using the R Version 2.3.0 software. Survival rates and relative risks (RR) are presented with their 95% confidence intervals (CI).
Supplementary Material
Acknowledgments
Thanks are due to Dr Thomas Giordano for tissue procurement, the University of Michigan Cancer Center Flow Cytometry core, Dr. W. Foulkes, Dr B. Boman, Dr. H. Korkaya, for advise and critical review of the paper. The fibroblast cell line is a generous gift from Dr. John Stingl and Dr. Connie Eaves, Terry Fox Laboratory, Vancouver, British Columbia, Canada. This work was supported by NIH grants CA66233 and CA101860, and in part by the University of Michigan Cancer Center NIH Support Grant 5 P 30 CA46592, and in part by Ligue Nationale Contre le Cancer (Label DB) and Institut National du Cancer.
Footnotes
Max Wicha has financial holdings and is a scientific advisor for OncoMed Pharmaceuticals.
References
- Alexe G, Alexe S, Axelrod DE, Bonates TO, Lozina II, Reiss M, Hammer PL. Breast cancer prognosis by combinatorial analysis of gene expression data. Breast Cancer Res. 2006;8:R41. doi: 10.1186/bcr1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, ito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, Lako M. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- Boecker W, Moll R, Dervan P, Buerger H, Poremba C, Diallo RI, Herbst H, Schmidt A, Lerch MM, Buchwalow IB. Usual ductal hyperplasia of the breast is a committed stem (progenitor) cell lesion distinct from atypical ductal hyperplasia and ductal carcinoma in situ. J Pathol. 2002;198(4):458–467. doi: 10.1002/path.1241. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Warren EH, Greenberg PD, Dick JE, Riddell SR. CD8(+) minor histocompatibility antigen-specific cytotoxic T lymphocyte clones eliminate human acute myeloid leukemia stem cells. Proc Natl Acad Sci USA. 1999;96:8639–8644. doi: 10.1073/pnas.96.15.8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, McDonnell DP. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;31:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer Stem Cells--Perspectives on Current Status and Future Directions: AACR Workshop on Cancer Stem Cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem. 2000;267:4315–4324. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- Ginestier C, Charafe-Jauffret E, Bertucci F, Eisinger F, Geneix J, Bechlian D, Conte N, Adelaide J, Toiron Y, Nguyen C, et al. Distinct and complementary information provided by use of tissue and DNA microarrays in the study of breast tumor markers. Am J Pathol. 2002;161:1223–1233. doi: 10.1016/S0002-9440(10)64399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson T, Villadsen R, Nielsen HL, Ronnov-Jessen L, Bissell MJ, Petersen OW. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 2002;16(6):693–706. doi: 10.1101/gad.952602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DA, Meyerrose TE, Wirthlin L, Craft TP, Herrbrich PE, Creer MH, Nolta JA. Functional characterization of highly purified human hematopoietic repopulating cells isolated according to aldehyde dehydrogenase activity. Blood. 2004;104:1648–1655. doi: 10.1182/blood-2004-02-0448. [DOI] [PubMed] [Google Scholar]
- Hess DA, Wirthlin L, Craft TP, Herrbrich PE, Hohm SA, Lahey R, Eades WC, Creer MH, Nolta JA. Selection based on CD133 and high aldehyde dehydrogenase activity isolates long-term reconstituting human hematopoietic stem cells. Blood. 2006;107:2162–2169. doi: 10.1182/blood-2005-06-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemier J, Ginestier C, Rougemont J, Bardou VJ, Charafe-Jauffret E, Geneix J, Adelaide J, Koki A, Houvenaeghel G, Hassoun J, et al. Protein expression profiling identifies subclasses of breast cancer and predicts prognosis. Cancer Res. 2005;65:767–779. [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, Sawyers CL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci USA. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahad JP, Mills GB, Coombes KR. Stem cell-ness: a “magic marker” for cancer. J Clin Invest. 2005;115:1463–1467. doi: 10.1172/JCI25455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu H, Heller S. Pluripotent stem cells from the adult mouse inner ea.r. Nature Medicine. 2003;9(10):1293–1299. doi: 10.1038/nm925. [DOI] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Magni M, Shammah S, Schiro R, Mellado W, Dalla-Favera R, Gianni AM. Induction of cyclophosphamide-resistance by aldehyde-dehydrogenase gene transfer. Blood. 1996;87:1097–1103. [PubMed] [Google Scholar]
- Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, Smith BD, Civin CI, Jones RJ. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103:2332–2336. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Morrison SJ. Diverse mechanisms regulate stem cell self-renewal. Curr Opin Cell Biol. 2004;16:700–707. doi: 10.1016/j.ceb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Moreb J, Zucali JR, Zhang Y, Colvin MO, Gross MA. Role of aldehyde dehydrogenase in the protection of hematopoietic progenitor cells from 4-hydroperoxycyclophosphamide by interleukin 1 beta and tumor necrosis factor. Cancer Res. 1992;52:1770–1774. [PubMed] [Google Scholar]
- O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2006;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Passegue E, Jamieson CH, Ailles LE, Weissman IL. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci USA. 2003;100(Suppl 1):11842–11849. doi: 10.1073/pnas.2034201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce DJ, Taussig D, Simpson C, Allen K, Rohatiner AZ, Lister TA, Bonnet D. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;23:752–760. doi: 10.1634/stemcells.2004-0292. [DOI] [PubMed] [Google Scholar]
- Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104(3):973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2006;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Sell S. Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol Hematol. 2004;51:1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004a;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004b;23:7267–7273. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- Sophos NA, Vasiliou V. Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact. 2003:143–144. 5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eaves CJ, Kuusk U, Emerman JT. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998;63(4):201–213. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Tlsty TD. Stromal cells can contribute oncogenic signals. Semin Cancer Biol. 2001;11:97–104. doi: 10.1006/scbi.2000.0361. [DOI] [PubMed] [Google Scholar]
- van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, Bissell MJ, Petersen OW. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem. 1998;251:549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.